Abstract
On phosphorylation of Y221 by Abelson (Abl) kinase, the Crk-II adapter protein undergoes an intramolecular reorganization initiated by the binding of its own Src homology 2 (SH2) domain to the pY221 site. Conformational changes induced by phosphotyrosine recognition promote the binding of the Src homology 3 (SH3) domain of the Abl tyrosine kinase to a proline-rich loop located between the βD and βE strands of the SH2 domain (DE loop). We have determined the NMR solution structure of the ternary complex of the Abl SH3 domain with the Crk SH2 domain bound to a Crk pY221 phosphopeptide. The SH2 domain bridges two ligands that bind at distinct sites. The interaction between the Abl SH3 domain and the Crk SH2 domain is localized to a canonical eight-residue site within the DE loop. From 15N relaxation experiments, the DE loop of the SH2 domain in the complex displays a significant degree of conformational freedom. The structural and dynamic data therefore indicate that these SH2 and SH3 domains do not assume a unique orientation with respect to one another; rather, they appear to be only tethered via the DE loop. Thus, SH2 domain–SH3 domain interactions do not require additional tertiary contacts or restriction of domain orientation when a recognition motif is presented in a mobile loop. This complex between the Abl SH3 domain, Crk SH2 domain, and Crk phosphopeptide is an example of the extremely modular nature of regulatory proteins that provides a rich repertoire of mechanisms for control of biological function.
Keywords: NMR, signal transduction, domain orientation, modular binding domain
Our understanding of the regulation of biological processes by protein interactions has been significantly enhanced by the description of modular binding domains and their preferred target sequences (1, 2). In particular, the Src homology 2 (SH2) and Src homology 3 (SH3) domains of cytoplasmic tyrosine kinases and associated adapter proteins demonstrate a wide variety of regulatory mechanisms involving both intramolecular and intermolecular interactions with target peptide regions (3, 4). Whether additional contacts exist beyond those presented by canonical peptide models is an important issue, particularly as they can potentially modulate binding affinity and specificity (5, 6). An interesting example in this context is provided by the protein interactions observed between the Abelson (Abl) tyrosine kinase and the adapter protein Crk-II.
Abl has a wide range of functions including signal transduction, cytoskeletal and cell cycle regulation, neural development, and reaction to oxidative stress (7). In addition to its catalytic activity, it possesses an SH2 domain, an SH3 domain, nuclear translocation sequences, a DNA binding domain, and an actin binding domain. Chimeric Abl variants activated as a result of chromosomal translocations (Bcr-Abl, Tel-Abl) or by fusion to viral sequences (v-Abl) are transforming (8, 9) through activation of Ras, phosphoinositol 3-kinase, PKC, and Janus kinase (JAK)/signal transducers and activators of transcription (STAT) pathways (10). Abl can also be activated by deletion of its SH3 domain, supportive of its negative regulatory role (11).
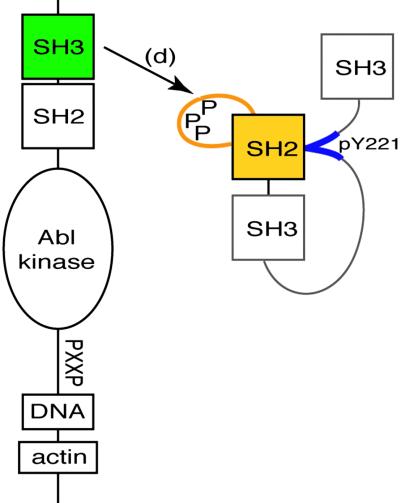
A binding partner and substrate of c-Abl is the Crk-II adapter. This isoform of the Crk protooncoprotein is composed of one SH2 domain and two SH3 domains (12, 13). It links phosphotyrosine-containing motifs (recognized by the SH2 domain) to SH3-binding proteins such as DOCK180 and C3G, which in turn regulate small GTPases involved in cell adhesion and movement (14). Abl kinase interacts with the Crk adapter protein in distinct ways depending on the isoform and the phosphorylation state of Y221 in Crk. Initially, unphosphorylated Crk binds Abl via the N terminal of its two SH3 domains at one of several proline-rich sequences that lie adjacent to the Abl catalytic domain (15). This association promotes the tyrosine phosphorylation of Crk (16). Autorecognition of pY221 by the Crk SH2 domain induces an intramolecular reorganization (17) with small conformational changes in the SH2 domain that promote the exposure of a large 20-residue loop (DE loop, residues 65–85) located between the βD and βE strands (18). This extended DE loop, which is rich in proline residues, is unique to the mammalian Crk SH2 domain and can serve as a binding site for the regulatory SH3 domain of Abl (18) (Fig. 1). As a consequence, another interaction mode for Crk and Abl that may modify Abl signaling is presented.
Fig 1.
The complex described in this study includes the Abl SH3 domain (green), the Crk SH2 domain (yellow), and a 13-aa phosphopeptide corresponding to the Crk SH2 internal binding site (blue).
In this article, we examine this second regulatory interaction by NMR structural studies of a ternary complex consisting of the Abl SH3 domain and a phosphopeptide-activated Crk-II SH2 domain. This structure of the Crk SH2 domain is particularly significant as Crk is one of the prototypic SH2 domain-containing adapter proteins. Intermolecular nuclear Overhauser effect (NOE) observations localized the Abl SH3 domain exclusively to the DE loop of the Crk SH2 domain. Because of the size of the loop and lack of additional observed surface interactions, the two molecules appear to be only tethered and therefore relatively unrestricted in their individual motions as supported by additional NMR dynamics studies. Furthermore, we describe specific structural and binding characteristics of the Crk SH2 and Abl SH3 domains and compare them to other well-characterized SH2 and SH3 domains.
Materials and Methods
Cloning and Expression of the Crk SH2 and Abl SH3 Domains.
Construction of an overexpression vector fusing GST to the human Crk-II SH2 domain (residues 6–125) has been described (18). Soluble GST-SH2 was expressed in BL21:DE3 cells (Novagen) and purified by affinity chromatography. After cleavage with human thrombin (Sigma), the liberated SH2 domain (with Gly-Ser N-terminally appended) was purified to homogeneity by gel filtration on a Sephacryl S-200 column. A PCR fragment corresponding to the SH3 domain (residues 62–121) of human Abl kinase, was inserted into the NcoI and BamHI sites of pET11d (Novagen). After expression, the domain was purified by anion-exchange and gel filtration chromatography. Isotopically labeled Abl and Crk for NMR studies were obtained by fermentations in minimal media containing 99% [15N]-ammonium chloride and/or 99% 13C-glucose as sole carbon and nitrogen sources.
Cloning and Expression of the Crk Phosphopeptide.
An oligonucleotide cassette corresponding to Crk residues 217–229 (EPGPYAQPSNTDK) was ligated into pAED4-MMHb (19). Expression of this plasmid in minimal media supplemented with 13C/15N substrates produced a fusion protein consisting of an insoluble TrpLE carrier protein followed by 9×-His, a unique Met and the Crk peptide sequence. On purification by denaturing Ni2+-chelating chromatography and C4 reverse-phase HPLC, the fusion protein was dissolved in 0.1 M HCl and cleaved with 50 mg of CNBr at room temperature for 3 h. After lyophilization to remove excess CNBr, the cleaved products were resuspended in 4 ml water. On raising the pH to 7.0, the carrier protein precipitated, leaving the cleaved peptide in the supernatant that was subsequently purified to homogeneity by C18 reverse-phase chromatography. Quantitative phosphorylation was achieved by incubating 1 mg of peptide with 5,000 units of purified Abl kinase (New England Biolabs) in the manufacturer's buffer supplemented with 10 mM dATP at 30°C for 1 day.
Determination of Binding Affinities.
The change in intrinsic fluorescence of a 1 μM solution of Abl SH3 domain (20 mM sodium phosphate, pH 6.8, 100 mM NaCl) on addition of 0–350 μM Crk[65–82] synthetic peptide was measured by using a Varian Eclipse fluorescence spectrophotometer (excitation 280 nm, emission 350 nm; 5-nm slit widths). The value of the dissociation constant was determined directly from the binding curve by using the profit software package for Macintosh (Quantum Software, Zurich). The dissociation constant describing the binding of the Abl SH3 domain with a Crk SH2 domain/Crk phosphopeptide complex was determined by measuring the amide chemical shift changes observed in six samples of 0.47 mM 15N-labeled Abl SH3 domain supplemented, respectively, with 0, 0.25, 0.50, 0.75, 1.0, and 1.5 molar equivalents of unlabeled Crk SH2 domain/Crk phosphopeptide.
NMR Experiments and Structure Generation.
Ternary complexes consisting of eight different combinations of 15N/13C-labeled and unlabeled Crk SH2 domain, Abl SH3 domain, and Crk phosphopeptide were mixed at stoichiometric ratios and concentrated to 0.6–1.5 mM in 50 mM sodium phosphate, pH 6.8 and 0.02% sodium azide. NMR spectra were collected at 30°C on 500-, 600-, and 800-MHz Varian Unity Inova spectrometers equipped with pulsed-field gradient accessories. Resonance assignments and intramolecular distance restraints were based on the following NMR experiments (20, 21) performed (i) on 15N-labeled samples: 1H-15N heteronuclear single quantum correlation (HSQC), NOESY-HSQC (125-ms mixing time), and total correlation spectroscopy (TOCSY)-HSQC (50-ms mixing time) and (ii) on 15N/13C-labeled samples: 1H-13C HSQC, HNCACB, CBCA(CO)NH, H(CCO)NH-TOCSY, C(CO)NH-TOCSY, HCCH-TOCSY, and simultaneous 15N-13C-NOESY-HSQC (50-, 125-ms mixing time). Methyl prochiral assignments were made according to the method of Neri et al. (22). Stereospecific Hβ resonance assignments were obtained from analysis of short mixing time (50 ms) simultaneous 15N-13C-NOESY-HSQC spectra. Assignments and intramolecular NOEs were obtained by 13C/15N- or 15N-labeling one component in a background of two unlabeled components. Intermolecular distance restraints were obtained from reverse half-filtered 2D and 3D NOESY spectra (300-ms mixing time) on samples in 99% D2O.
Amide 15N relaxation parameters for the Crk SH2 and Abl SH3 domains were measured and analyzed as described in Farrow et al. (23). Amide 15N T1 values were obtained from inversion recovery spectra acquired with relaxation delays of 5, 71, 141, 232, 323, 444, and 606 ms. Amide 15N T2 values were derived from offset corrected amide 15N T1ρ spectra acquired with relaxation delays of 8, 16, 24, 40, 56, 72, and 88 ms. Steady-state heteronuclear 1H{15N}-NOE spectra (24) were acquired with and without 3 s of 1H saturation and a total recycle delay of 7 s.
NMR data were processed with nmrpipe (25) and interpreted with pipp software (26). Structure calculations were performed according to the ARIA protocol (27) by using CNS 1.0 software (28) and structural information obtained from NOEs and φ/ψ backbone dihedral angle restraints from the TALOS database (29). The Crk SH2 domain/Crk phosphopeptide and Abl SH3 domain structures were initially solved independently followed by assembly of the ternary complex in two stages. An initial Cartesian dynamics stage allowed the Abl SH3 domain to dock itself onto a Crk SH2 domain/Crk phosphopeptide complex whose coordinates were fixed. Afterward, a second Cartesian dynamics stage was then allowed to proceed through eight successive ARIA iterations to produce the final ensemble. Structures were assessed by using procheck (30). Figures were made with molmol (31), molscript (32), and RASTER3D (33). The lowest energy structure was deposited at the Protein Data Bank under ID code . NMR assignments for the complex were deposited in the BioMagResBank (34) under accession no. .
Results
General Features of the Ternary Complex.
The ternary structure of the Crk SH2 domain bound to a Crk-derived phosphopeptide and the Abl SH3 domain was determined by NMR spectroscopic approaches. Based on titrations of the Crk phosphopeptide into labeled Crk SH2 domain (data not shown), binding at this interface is in the slow exchange regime, indicative of tight binding in accord with the 1 μM or better affinity of SH2 domain-phosphopeptide complexes. In contrast, chemical shift changes observed during titrations of unlabeled Crk SH2 domain/Crk phosphopeptide into 15N-labeled Abl SH3 domain (data not shown) demonstrate fast exchange relative to the chemical shift time scale. A dissociation constant of 30 ± 11 μM has been calculated based on amide chemical shift changes in residues G76, D77, N78, and D79 that occupy key positions in the RT-Src loop. For this Kd value, we estimate that 87 ± 2% of the protein will be in the complexed state at 1.5 mM concentration. This finding points to no significant effect from free protein in our structural study, consistent with no resonances of unbound protein and no inconsistent structural data being observed. Of interest, the Kd value for the binding of the Abl SH3 domain to an 18-residue proline-rich peptide derived from the Crk SH2 domain DE loop determined by intrinsic fluorescence methods is 370 ± 30 μM (data not shown). The apparent 12-fold difference in affinities suggests that the Crk SH2 domain DE loop imposes conformational constraints that significantly diminish the entropic cost of binding to the Abl SH3 domain.
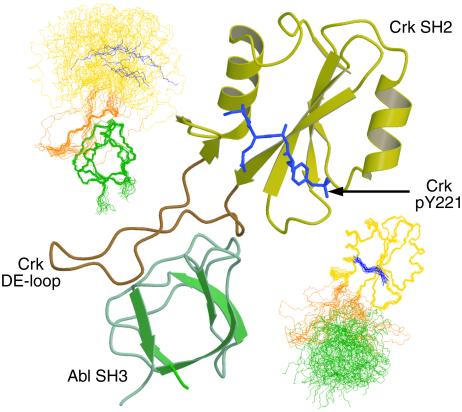
SH3 domains represent a diverse class of modular protein–protein interaction motifs (35) that generally bind a consensus target sequence of PxxP (36). Previous scanning mutagenesis studies established that the Abl SH3 domain binding site was located in the proline-rich amino terminal portion of the DE loop of the Crk SH2 domain (18). In agreement with this study, intermolecular NOEs were observed only to protons within the stretch of residues from 67–75 in the Crk SH2 domain. One representative structure of the ternary complex is shown in Fig. 2. When either the Crk SH2 domain or Abl SH3 domain is exclusively superimposed in the ternary complex, the conformational mobility of the two domains caused by the DE loop is apparent. Spread to its maximum circumference, the DE loop spans a diameter of ≈30 Å, comparable to the diameter of the Abl SH3 domain. Titrations of unlabeled Abl SH3 domain into the 15N-Crk SH2 domain/unlabeled Crk-phosphopeptide complex revealed only minor amide chemical shift perturbations for resonances S55 and E102 outside of the DE loop region. With the lack of intermolecular NOE observations beyond the canonical binding site and the lack of major chemical shift changes on ternary complex formation, Abl SH3 domain binding appears to be limited to the DE loop. (Note that chemical shift and NOE data described throughout are not shown.) Because of the size of the DE loop, the Abl SH3 and Crk SH2 domains are able to experience a significant degree of conformational freedom in the ternary complex.
Fig 2.
Ternary complex of the Crk SH2 domain (yellow/orange, ribbon representation), Crk phosphopeptide (blue, heteroatom representation), and the Abl SH3 domain (green, ribbon representation). The loop in the Crk SH2 domain important for binding the Abl SH3 domain (DE loop) is labeled. Main-chain representation of the 20 lowest energy structures superposed according to the Abl SH3 domain (Upper Left) and the Crk SH2 domain/Crk phosphopeptide (Lower Right).
Dynamics of the Ternary Complex.
A correlation time of 9.0 ns for the Crk SH2 domain was obtained from a relaxation analysis of 15N T1 and T2 relaxation times and heteronuclear 1H{15N}-NOE enhancements (dynamic data are available in Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Measurements for the Crk SH2 domain/phosphopeptide complex in the absence of the Abl SH3 domain were not performed. Significant resonance overlap combined with prolines situated at nine of 20 aa positions made it possible to measure dynamic data for only two amides in the DE loop. However, from the longer than average T1 and T2 relaxation times and negative heteronuclear NOE enhancements for these two residues, it is apparent that the DE loop experiences a range of additional motions at rates faster than global tumbling. Sharp resonances observed for proline residues of the DE loop in HACAN and CBCA(CO)N(CA)HA experiments (37) corroborate the amide relaxation study in a qualitative sense.
Analysis of the relaxation data for 15N-labeled Abl SH3 domain (data not shown) demonstrates that on binding the Crk SH2 domain/phosphopeptide complex, the correlation time of the Abl SH3 domain increased from 4.6 ns to 6.3 ns. The small increase in the correlation time for the Abl SH3 domain in the complex, relative to what might be expected in a rigid structure (≈10 ns), reflects to a large extent the relatively unrestricted nature of the SH3 and SH2 domains in the complex.
Structural Details of the Abl SH3 Domain and Ligand Binding.
SH3 domains are compact, globular modules composed primarily of two β-sheets arranged in a “handshake” configuration (35). On binding, most ligands of SH3 domains adopt a polyproline type II (PP-II) helical conformation of ≈10 aa in length. The symmetry of the PP-II helix positions the side chains such that forward (type 1) or reverse (type 2) binding orientations are possible (38). Based on two high-resolution crystal structures of Abl complexes (39, 40), the ligand may be functionally divided into (i) a specificity region of four residues, (ii) a hinge residue, and (iii) a binding region of five residues that adopt a PP-II helix conformation.
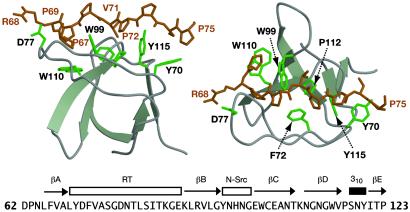
Fig. 3 highlights the Abl SH3 domain and its interaction with the Crk DE loop in our structure. A series of aromatic residues (Y70, F72, W99, W110, and Y115) generate three shallow grooves that make complementary contacts with side chains presented with appropriate periodicity by the polyproline type II helix. These complementary contacts, evidenced by 37 intermolecular NOEs, involve residues P67, P69, V71, P72, and P75 of the Crk SH2 domain. These combined with torsion angle data led to a backbone precision of 0.44 Å from the ensemble of Abl SH3 domain structures (Table 1).
Fig 3.
Detail of the Abl SH3 domain bound to the Crk SH2 domain (residues 68–75) in the ternary complex (two orientations rotated 90° toward the viewer). Side chains of residues identified to make contacts to Crk SH2 domain are highlighted. D71 of Abl may participate in an ionic interaction with R68 of Crk. Secondary structure elements are labeled above the sequence, following the standard nomenclature for SH3 domains.
Table 1.
Statistics of the 20 lowest energy structures in the final ensemble
| rms deviation from experimental restraints | ||
| Distance restraints, Å | ||
| Unambiguous (4,247) | 0.018 ± 0.010 | |
| SH2-SH2 (2,411) | 0.015 ± 0.008 | |
| SH3-SH3 (1,628) | 0.018 ± 0.016 | |
| SH2-SH3 (37) | 0.031 ± 0.006 | |
| SH2-pY (64) | 0.005 ± 0.012 | |
| Ambiguous (96) | 0.032 ± 0.016 | |
| Hydrogen bonds (50) | 0.035 ± 0.002 | |
| Direct 3JHNHα couplings, ° (54) | 0.267 ± 0.015 | |
| φ-ψ dihedral angle, ° (166) | 0.221 ± 0.017 | |
| Deviations from ideal geometry | ||
| Bonds, Å | 0.002 ± 0.00004 | |
| Angles, ° | 0.272 ± 0.003 | |
| Impropers, ° | 0.136 ± 0.004 | |
| Ramachandran map analysis | ||
| Most favored regions | 79.3% | |
| Additional allowed regions | 19.3% | |
| Generously allowed regions | 1.3% | |
| Disallowed regions | 0.1% | |
| rms deviation from mean structure | ||
| Backbone | All atoms | |
| SH2 only (secondary structure) | 0.49 ± 0.11 Å | 1.11 ± 0.13 Å |
| SH2 only (13–64, 86–118) | 0.68 ± 0.13 Å | 1.29 ± 0.13 Å |
| SH3 only (secondary structure) | 0.31 ± 0.15 Å | 1.00 ± 0.15 Å |
| SH3 only (64–119) | 0.42 ± 0.07 Å | 1.21 ± 0.12 Å |
| Phosphopeptide only (221–224) | 0.66 ± 0.17 Å | 1.32 ± 0.16 Å |
| SH2, SH3, and phosphopeptide | 6.78 ± 3.13 Å | 7.10 ± 3.09 Å |
Numbers of experimental restraints are given in parentheses.
For intramolecular and intermolecular NOEs, numbers were not corrected for redundant observations.
Ramachandran analysis follows the parameters defined by procheck (44).
Ligand binding is enhanced by a variety of contributions from nearby side chains residing in two flexible loops designated RT and n-Src. Major amide chemical shift perturbations in the Abl SH3 domain on binding to the Crk SH2 domain were localized to residues G76–T79 in the RT loop. Amide resonances belonging to the n-Src loop (N94, H95, and N96) on the opposite side of the same binding pocket experienced line broadening in addition to minor chemical shift changes. As was observed in other binding studies of the Abl SH3 domain (41), line broadening of H95 was severe enough to prevent detection.
A comparison of this structure with other Abl SH3 domain complexes reveals rms deviation values of 1.19 Å to the Abl SH3 domain/p41-ligand complex (39) and 1.22 Å to the Abl SH3 domain/3BP1-ligand complex (40), based on superposition of the Cα atoms of residues 65–121. The high affinity (Kd = 1.5 μM) for the p41 ligand (APSYSPPPPP) is attributed to the tyrosine at position 4, which makes numerous hydrophobic contacts with the RT-Src loop and forms two stabilizing hydrogen bonds to the side chains of D77 and S81 (according to the residue numbering of the Abl SH3 domain in this article) via the ring hydroxyl group (39). In the phosphoinositol 3-kinase SH3 domain, the mutation D21N (analogous to D77N in the Abl SH3 domain) induces structural changes throughout the amino terminal binding cleft (42). Whereas the corresponding specificity region in Crk differs significantly at position 4 (GPRPPVPPSP versus APSYSPPPPP in p41 and APTMPPPLPP in 3BP1), a hydrogen bond to D77 is still possible through the guanidinium group of R68 at position 3. Although we have no explicit evidence for an SH3-D77 to SH2-R68 hydrogen bond from intermolecular side-chain NOEs, the structure suggests that it could be present to stabilize the interaction.
Structural Details of the Crk SH2 Domain and Phosphopeptide Binding.
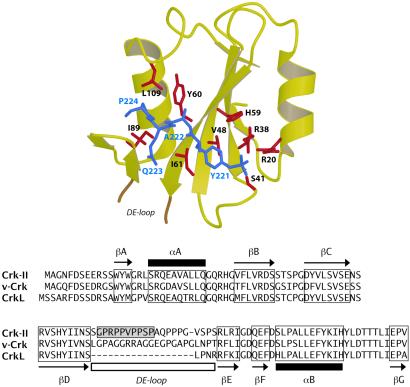
Fig. 4 illustrates the Crk SH2 domain and its interaction with the Crk pY221 peptide in our structure. SH2 domains typically bind 4–7 residue sequences containing phosphotyrosine (43) although interactions that do not require phosphotyrosine, such as to the SH2D1A/SAP SH2 domain, have been reported (44, 45). Chemical shift changes observed from titrations of unlabeled SH2 domain into a sample of 13C,15N-labeled Crk phosphopeptide were localized to only four residues, pY221-P224. Consistent with binding, resonances of these residues were broadened, in particular pY221, whose HN and Hα resonances were not observed.
Fig 4.
(Upper) Detail of the Crk SH2 domain bound to the Crk phosphopeptide (residues 221–224) in the ternary complex. Residues observed to make contacts are highlighted. (Lower) Sequence comparison of Crk-II with homologues v-Crk and CrkL. The sequence of murine Crk-II corresponds to the protein fragment described in this study. Only Crk-II contains a binding site for the Abl SH3 domain (shaded box).
A total of 64 intermolecular NOEs were observed between the Crk SH2 domain and residues pY221–P224 of the Crk phosphopeptide. Reminiscent of Src (43), the Crk SH2 domain binds the phosphopeptide in a shallow cleft. P224 at position pY+3 is buried in a pocket lined by residues from the jaw-like EF and BG loops. Important nonpolar contacts are contributed by Y60, I61, I89, and L109. As the majority of NOEs were observed either between the phosphotyrosine pY221 or P224, the interior two positions A222 and Q223 may not be as critical for binding.
Numerous hydrogen bonds bind the phosphate group in a deep ionic pocket containing R38. This arginine, at position βB5, is invariant across SH2 domains owing to its significant function. Two of the Hη protons of R38 (position βB5) and one of the Hη protons for R20 (position αA2) were significantly downfield-shifted (Hη shifts of 5.79/10.24 and 5.93/9.62 ppm for R38 and 5.88/10.05 ppm for R20), suggesting participation in hydrogen bonds with the phosphate group (46). A very high (1.0 Å) resolution crystal structure of the SH2 domain of Lck kinase in complex with a phosphopeptide target revealed two distinct conformations for the arginine in the αA2 position. Conformational exchange of the arginine at position βB5 was noted from the NMR studies of the C-terminal phospholipase Cγ1 and Src SH2 domain complexes (47, 48). As several resonances in the Crk phosphopeptide were severely broadened in the complex, conformational exchange at the phosphate binding site may be a hallmark of SH2 domain/phosphopeptide interactions.
A comparison of the βB-BC amide shifts of Crk with those of the N-terminal SH2 domain of Shp2 (previously called Syp) phosphatase (49) revealed similar downfield-shifted amide resonances at position BC1 (S41 in Crk, K35 in Shp2). From the crystal structure of Shp2, a hydrogen bond occurs between the BC1 position and the pTyr phosphate (49). It is likely that a similar hydrogen bond exists between the BC1 position of Crk SH2 domain and pY221. Hydrogen bonds between position BC1 and the pTyr also occur in Src SH2 domain complexes (47).
Discussion
Although modular binding domains interact with their peptide targets in regions defined primarily by the consensus motifs, binding to a target within the context of a folded protein raises the possibility of additional “tertiary” interactions. Contacts at noncontiguous sites of an SH3 domain target have been observed in Hck and Fyn SH3 domain complexes with the HIV Nef protein (5, 50). Two additional contacts beyond the canonical site are made by the Csk SH3 domain on association with the PEST sequence of proline-enriched tyrosine phosphatase (51). Finally, tertiary contacts stabilize the intramolecular SH3 domain-linker interface in the structures of the Src and Hck tyrosine kinases (52–55). The additional interactions outside of those to the proline-rich sequence provide specificity and higher affinity. For domain–domain interactions, such tertiary interactions might be expected. However, in the case of this ternary complex involving the Abl SH3 domain, the Crk-II SH2 domain, and a Crk phosphopeptide, interactions of the Abl SH3 domain to Crk outside of those to the consensus proline-rich target motif are not observed.
The lack of tertiary interactions and the flexibility in the DE loop binding region lead to a poor definition of the relative orientation of the SH2 and SH3 domains with respect to each other. This observed mobility is reminiscent of the dynamic behavior that has been reported between tandem SH2–SH3 domains of Abl kinase (56, 57), Hck kinase (52, 58), Lck kinase (59), and Src kinase (60). The recent solution structure of the SH3–SH2–SH3 adapter protein Grb2 also demonstrates a lack of rigid domain orientation (61, 62). Functionally, this flexibility may be necessary to facilitate binding to many different independent or consolidated phosphotyrosine-containing and proline-rich ligands from disparate components of the signal transduction machinery.
Abl kinase exploits both intramolecular and intermolecular SH2 and SH3 domain interactions to achieve a high degree of regulation (11). On phosphorylation of Crk by Abl, the regulatory SH3 domain of Abl could be decoupled from another regulatory site through intermolecular competition with the exposed DE loop of Crk. Our results demonstrate that there is no unique domain–domain orientation for the Crk–Abl interaction in the ternary complex. By exploiting a loop for binding instead of a surface, Crk may be able to interact with other signal transduction proteins in addition to Abl. The large size of the DE loop could facilitate the approach and binding of multidomain proteins such as Abl kinase. Importantly, this structural analysis shows how an SH2 domain can simultaneously interact with two distinct protein ligands, one of which engages the conventional phosphopeptide-binding site, whereas the other recognizes an entirely different binding motif. Although the proline-rich DE loop is unique to the mammalian Crk SH2, these observations may have a more general significance. For example, the SH2D1A/SAP protein, composed almost exclusively of an SH2 domain, can bind both to a (phospho)tyrosine-containing peptide from the CD150/SLAM receptor and simultaneously to the Fyn tyrosine kinase (63).
SH3 domain-ligand interactions have affinities in the μM range, providing for relatively specific interactions while still allowing for competition. In multiprotein signal transduction complexes that use SH3 domain–ligand interactions, the overall affinity is likely to be much greater in combination with additional contacts from SH2 and other modular binding domains. In this respect, there may be no benefit to Abl kinase in using specific surface interactions with Crk beyond the canonical SH3 domain binding site in the DE loop. In an alternative functional context, if the role of the DE loop is to simply sequester the SH3 domain of Abl, it alone should be sufficient.
Immunoprecipitation assays have previously shown that the DE loop is more accessible for binding when the Crk SH2 domain is occupied by its own endogenous pY-containing ligand (18). Precise structural details of how the DE loop is sequestered by the unliganded SH2 domain are unknown, because no other structure of the Crk SH2 domain is available. However, a model in which the DE loop has significant dynamic behavior in both states is supported on three grounds (17, 18): (i) backbone chemical shift changes in the DE loop on pY221 binding are limited to only a few residues, (ii) resonances are particularly sharp in this region, indicating mobility, and (iii) multiple conformations of the DE loop were observed in the unliganded SH2 domain. These observations suggest that phosphopeptide binding may promote a shift in a conformational equilibrium toward an SH3 domain-accessible state. Because the Abl SH3 domain has an intrinsic ability to sample binding partners rapidly, a shift in the conformational equilibrium of the DE loop may be sufficient to sustain a protein partnership between Abl and Crk.
Mutations that disrupt the ability of the Crk SH2 domain to bind its own endogenous phosphorylated ligand also disrupt its ability to bind the Abl SH3 domain via the DE loop (18). Furthermore, in the intact protein, phosphotyrosine binding induces a modular reorganization that prevents the first of its two SH3 domains from interacting with targets (64, 65). Potentially, phosphotyrosine binding initiates a new regulatory program by specifying a new set of biological partners for Crk. In this regard, it is of interest that a variant of Crk-II with a Y221F substitution binds constitutively to conventional SH2- and SH3-binding partners, but is compromised in its ability to induce attachment-induced activation of the Rac GTPase because of a failure in membrane localization (66). Crk-II therefore undergoes a complex series of interactions on phosphorylation of Y221, potentially involving the proline-rich insert in the SH2 domain. The observation that the Crk homologue CrkL differs from Crk-II by the absence of the SH3 binding site in the DE loop suggests that Crk-II has evolved a specialized role in signal transduction by virtue of its extended DE loop. The absence of the regulatory tyrosine in v-Crk provides another means to avoid regulation, leading to an oncogenic form. This study highlights but a single example of the variety of regulatory mechanisms possible when using multiple modular binding domains and target motifs in the context of intramolecular and intermolecular interactions.
Supplementary Material
Acknowledgments
We thank Dr. Ranjith Muhandiram for assistance with the NMR experiments. This work was supported by grants from the National Cancer Institute of Canada with funds from the Canadian Cancer Society (to J.D.F.-K., L.E.K., and T.P.).
Abbreviations
Abl, Abelson
HSQC, heteronuclear single quantum correlation
NOE, nuclear Overhauser effect
SH2, Src homology 2
SH3, Src homology 3
Data deposition: The structure has been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ). The NMR resonance assignments have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. ).
References
- 1.Pawson T. & Nash, P. (2000) Genes Dev. 14, 1027-1047. [PubMed] [Google Scholar]
- 2.Pawson T. & Scott, J. D. (1997) Science 278, 2075-2080. [DOI] [PubMed] [Google Scholar]
- 3.Gmeiner W. H. & Horita, D. A. (2001) Cell Biochem. Biophys. 35, 127-140. [DOI] [PubMed] [Google Scholar]
- 4.Yaffe M. B. (2002) Nat. Rev. Mol. Cell. Biol. 3, 177-186. [DOI] [PubMed] [Google Scholar]
- 5.Grzesiek S., Bax, A., Clore, G. M., Gronenborn, A. M., Hu, J. S., Kaufman, J., Palmer, I., Stahl, S. J. & Wingfield, P. T. (1996) Nat. Struct. Biol. 3, 340-345. [DOI] [PubMed] [Google Scholar]
- 6.Lee C. H., Saksela, C., Mirza, U. A., Chait, B. T. & Kuriyan, J. (1996) Cell 85, 931-942. [DOI] [PubMed] [Google Scholar]
- 7.Van Etten R. A. (1999) Trends Cell. Biol. 9, 179-186. [DOI] [PubMed] [Google Scholar]
- 8.Franz W. M., Berger, P. & Wang, J. Y. (1989) EMBO J. 8, 137-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer B. J. & Baltimore, D. (1994) Mol. Cell. Biol. 14, 2883-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou X. & Calame, K. (1999) J. Biol. Chem. 274, 18141-18144. [DOI] [PubMed] [Google Scholar]
- 11.Barila D. & Superti-Furga, G. (1998) Nat. Genet. 18, 280-282. [DOI] [PubMed] [Google Scholar]
- 12.Reichman C. T., Mayer, B. J., Keshav, S. & Hanafusa, H. (1992) Cell Growth Differ. 3, 451-460. [PubMed] [Google Scholar]
- 13.Feller S. M., Posern, G., Voss, J., Kardinal, C., Sakkab, D., Zheng, J. & Knudsen, B. S. (1998) J. Cell. Physiol. 177, 535-552. [DOI] [PubMed] [Google Scholar]
- 14.Feller S. M. (2001) Oncogene 20, 6348-6371. [DOI] [PubMed] [Google Scholar]
- 15.Ren R., Ye, Z. S. & Baltimore, D. (1994) Genes Dev. 8, 783-795. [DOI] [PubMed] [Google Scholar]
- 16.Amoui M. & Miller, W. T. (2000) Cell. Signalling 12, 637-643. [DOI] [PubMed] [Google Scholar]
- 17.Rosen M. K., Yamazaki, T., Gish, G. D., Kay, C. M., Pawson, T. & Kay, L. E. (1995) Nature 374, 477-479. [DOI] [PubMed] [Google Scholar]
- 18.Anafi M., Rosen, M. K., Gish, G. D., Kay, L. E. & Pawson, T. (1996) J. Biol. Chem. 271, 21365-21374. [DOI] [PubMed] [Google Scholar]
- 19.Schumacher T. N., Mayr, L. M., Minor, D. L. J., Millhollen, M. A., Burgess, M. W. & Kim, P. (1996) Science 271, 1854-1857. [DOI] [PubMed] [Google Scholar]
- 20.Kay L. E. (1995) Prog. Biophys. Mol. Biol. 63, 277-293. [DOI] [PubMed] [Google Scholar]
- 21.Sattler M., Schleucher, J. & Griesinger, C. (1999) Prog. NMR Spectrosc. 34, 93-158. [Google Scholar]
- 22.Neri D., Szyperski, T., Otting, G., Senn, H. & Wüthrich, K. (1989) Biochemistry 28, 7510-7516. [DOI] [PubMed] [Google Scholar]
- 23.Farrow N. A., Muhandiram, R., Singer, A. U., Pascal, S. M., Kay, C. M., Gish, G., Shoelson, S. E., Pawson, T., Forman-Kay, J. D. & Kay, L. E. (1994) Biochemistry 33, 5984-6003. [DOI] [PubMed] [Google Scholar]
- 24.Barbato G., Ikura, M., Kay, L. E., Pastor, R. W. & Bax, A. (1992) Biochemistry 31, 5269-5278. [DOI] [PubMed] [Google Scholar]
- 25.Delaglio F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277-293. [DOI] [PubMed] [Google Scholar]
- 26.Garrett D. S., Powers, R., Gronenborn, A. M. & Clore, G. M. (1997) J. Magn. Reson. 95, 214-220. [DOI] [PubMed] [Google Scholar]
- 27.Nilges M., Macias, M. J., O'Donoghue, S. I. & Oschkinat, H. (1997) J. Mol. Biol. 269, 408-422. [DOI] [PubMed] [Google Scholar]
- 28.Brünger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 29.Cornilescu G., Delaglio, F. & Bax, A. (1999) J. Biomol. NMR 13, 289-302. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski R. A., MacArthur, M. W., Mossk, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 31.Koradi R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51-55. [DOI] [PubMed] [Google Scholar]
- 32.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 33.Merritt E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505-524. [DOI] [PubMed] [Google Scholar]
- 34.Seavey B. R., Farr, E. A., Westler, W. M. & Markley, J. L. (1991) J. Biomol. NMR 1, 217-236. [DOI] [PubMed] [Google Scholar]
- 35.Mayer B. J. (2001) J. Cell. Sci. 114, 1253-1263. [DOI] [PubMed] [Google Scholar]
- 36.Ren R., Mayer, B. J., Cicchetti, P. & Baltimore, D. (1993) Science 259, 1157-1161. [DOI] [PubMed] [Google Scholar]
- 37.Kanelis V., Donaldson, L. W., Muhandiram, D. R., Rotin, D., Forman-Kay, J. D. & Kay, L. E. (2000) J. Biomol. NMR 16, 253-259. [DOI] [PubMed] [Google Scholar]
- 38.Lim W. A., Richards, F. M. & Fox, R. O. (1994) Nature 372, 375-379. [DOI] [PubMed] [Google Scholar]
- 39.Pisabarro M. T., Serrano, L. & Wilmanns, M. (1998) J. Mol. Biol. 281, 513-521. [DOI] [PubMed] [Google Scholar]
- 40.Musacchio A., Saraste, M. & Wilmanns, M. (1994) Nat. Struct. Biol. 1, 546-551. [DOI] [PubMed] [Google Scholar]
- 41.Xu R., Cahill, S. & Cowburn, D. (1999) J. Biomol. NMR 14, 187-188. [DOI] [PubMed] [Google Scholar]
- 42.Okishio N., Tanaka, T., Fukuda, R. & Nagai, M. (2001) Biochemistry 40, 119-129. [DOI] [PubMed] [Google Scholar]
- 43.Songyang Z., Shoelson, S. E., Chauduri, M., Gish, G., Pawson, T., Haser, W. G., King, F., Roberts, T., Ratnofsky, S. & Lechleider, R. J. (1993) Cell 72, 767-778. [DOI] [PubMed] [Google Scholar]
- 44.Li S. C., Gish, G., Yang, D., Coffey, A. J., Forman-Kay, J. D., Ernberg, I., Kay, L. E. & Pawson, T. (1999) Curr. Biol. 9, 1355-1362. [DOI] [PubMed] [Google Scholar]
- 45.Poy F., Yaffe, M. B., Sayos, J., Saxena, K., Morra, M., Sumegi, J., Cantley, L. C., Terhorst, C. & Eck, M. J. (1999) Mol. Cell 4, 555-561. [DOI] [PubMed] [Google Scholar]
- 46.Pascal S. M., Yamazaki, T., Singer, A. U., Kay, L. E. & Forman-Kay, J. D. (1995) Biochemistry 34, 11353-11362. [DOI] [PubMed] [Google Scholar]
- 47.Xu R. X., Word, J. M., Davis, D. G., Rink, M. J., Willard, D. H., Jr. & Gampe, R. T., Jr. (1995) Biochemistry 34, 2107-2121. [DOI] [PubMed] [Google Scholar]
- 48.Pascal S. M., Singer, A. U., Gish, G., Yamazaki, T., Shoelson, S. E., Pawson, T., Kay, L. E. & Forman-Kay, J. D. (1994) Cell 77, 461-472. [DOI] [PubMed] [Google Scholar]
- 49.Lee C. H., Kominos, D., Jacques, S., Margolis, B., Schlessinger, J., Shoelson, S. E. & Kuriyan, J. (1994) Structure (London) 2, 423-438. [DOI] [PubMed] [Google Scholar]
- 50.Arold S., Franken, P., Strub, M. P., Hoh, F., Benichou, S., Benarous, R. & Dumas, C. (1997) Structure (London) 5, 1361-1372. [DOI] [PubMed] [Google Scholar]
- 51.Ghose R., Shekhtman, A., Goger, M. J., Ji, H. & Cowburn, D. (2001) Nat. Struct. Biol. 8, 998-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sicheri F., Moarefi, I. & Kuriyan, J. (1997) Nature 385, 602-609. [DOI] [PubMed] [Google Scholar]
- 53.Williams J. C., Weijland, A., Gonfloni, S., Thompson, A., Courtneidge, S. A., Superti-Furga, G. & Wierenga, R. K. (1997) J. Mol. Biol. 274, 757-775. [DOI] [PubMed] [Google Scholar]
- 54.Xu W., Harrison, S. C. & Eck, M. J. (1997) Nature 385, 595-602. [DOI] [PubMed] [Google Scholar]
- 55.Moarefi I., LeFevre-Bernt, M., Sicheri, F., Huse, M., Lee, C.-H., Kuriyan, J. & Miller, W. T. (1997) Nature 385, 650-653. [DOI] [PubMed] [Google Scholar]
- 56.Gosser Y. Q., Zheng, J., Overduin, M., Mayer, B. J. & Cowburn, D. (1995) Structure (London) 3, 1075-1086. [DOI] [PubMed] [Google Scholar]
- 57.Fushman D., Xu, R. & Cowburn, D. (1999) Biochemistry 38, 10225-10230. [DOI] [PubMed] [Google Scholar]
- 58.Engen J. R., Smithgall, T. E., Gmeiner, W. H. & Smith, D. L. (1999) J. Mol. Biol. 287, 645-656. [DOI] [PubMed] [Google Scholar]
- 59.Eck M. J., Atwell, S. K., Shoelson, S. E. & Harrison, S. C. (1994) Nature 368, 764-769. [DOI] [PubMed] [Google Scholar]
- 60.Tessari M., Gentile, L. N., Taylor, S. J., Shalloway, D. I., Nicholson, L. K. & Vuister, G. W. (1997) Biochemistry 36, 14561-14571. [DOI] [PubMed] [Google Scholar]
- 61.Maignan S., Gulloteau, J.-P., Fromage, N., Arnoux, B., Becquart, J. & Ducruix, A. (1995) Science 268, 291-293. [DOI] [PubMed] [Google Scholar]
- 62.Yuzawa S., Yokochi, M., Hatanaka, H., Ogura, K., Kataoka, M., Miura, K., Mandiyan, V., Schlessinger, J. & Inagaki, F. (2001) J. Mol. Biol. 306, 527-537. [DOI] [PubMed] [Google Scholar]
- 63.Latour S., Gish, G., Helgason, C. D., Humphries, R. K., Pawson, T. & Veillette, A. (2001) Nat. Immunol. 2, 681-690. [DOI] [PubMed] [Google Scholar]
- 64.Feller S. M., Knudsen, B. & Hanafusa, H. (1994) EMBO J. 13, 2341-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okada S., Matsuda, M., Anafi, M., Pawson, T. & Pessin, J. E. (1998) EMBO J. 17, 2554-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abassi Y. A. & Vuori, K. (2002) EMBO J. 21, 4571-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.