Abstract
Previously, we have studied the minimal oligomer size of an aggregate amyloid seed and the mechanism of seed growth with a multilayer β-sheet model. Under high temperature simulation conditions, our approach can test the stability of possible amyloid forms. Here, we report our study of oligomers of Alzheimer's amyloid β-peptide (Aβ) fragments 16–22, 16–35, and 10–35 (abbreviated Aβ16–22, Aβ16–35, and Aβ10–35, respectively). Our simulations indicate that an antiparallel β-sheet orientation is the most stable for the Aβ16–22, in agreement with a solid state NMR-based model [Balbach, J. J., Ishii, Y., Antzutkin, O. N., Leapman, R. D., Rizzo, N. W., et al. (2000) Biochemistry 39, 13748–13759]. A model with twenty-four Aβ16–22 strands indicates a highly twisted fibril. Whereas the short Aβ16–22 and Aβ24–36 may exist in fully extended form, the linear parallel β-sheets for Aβ16–35 appear impossible, mainly because of the polar region in the middle of the 16–35 sequence. However, a bent double-layered hairpin-like structure (called hook) with the polar region at the turn forms parallel β-sheets with higher stability. An intra-strand salt-bridge (D23-K28) stabilizes the bent hairpin-like hook structure. The bent double-β-sheet model for the Aβ10–35 similarly offers oligomer stability.
Keywords: amyloid conformation, β-sheet, double-layered sheets, molecular dynamics simulation, protein folding
Changes in sequence or in shape of a protein may lead to a conformational disease. Conformational diseases are typically expressed in the appearance of amyloid fibrils. Understanding amyloid seed formation and elongation at the molecular level presents a major challenge, as it may lead to novel approaches in design and therapy (1). A relatively large number of protein conformational diseases have already been identified (2). Interestingly, at least in proteins related to Alzheimer's disease, prion protein-related encephalopathies, and type II diabetes, it has been discovered that certain short sequence fragments (5–40 residues) contained within the respective proteins can form amyloids, even in isolated peptide form. The human islet amyloid polypeptide (hIAPP), a 37-residue peptide hormone, is implicated in type II diabetes. Fibrils obtained from this hormone have been observed to be toxic to human and to rat islet β-cells in vitro (3). Experiments have demonstrated that a hexamer of hIAPP (residues 22–27, NFGAIL) and even a pentamer (residues 23–27, FGAIL) are already sufficient for amyloid formation and cytotoxicity (4). Prion proteins also contain such amyloidogenic peptide-fragments. Several fragments of the Syrian hamster prion protein (ShPrP) have been observed to form amyloids. These include residues 109–122, 178–191, and 202–218 (5). Within these, the most highly amyloidogenic peptide is AGAAAAGA, which corresponds to ShPrP residues 113–120. This fragment is conserved in all species whose PrP sequence has been determined (5). The β-peptide (Aβ, 42 residues) from Alzheimer's amyloid precursor protein, constitutes a particularly important example and is among the best studied amyloidogenic peptides. A key step in Alzheimer's disease involves proteolytic cleavage of Aβ (6). The fragment consisting of residues 25–35 in Aβ (GSNKGAIIGLM) has been shown to already form large β-sheet fibrils, essentially similar to those obtained by the full-length Aβ (refs. 7–11). Other fragments including 1–28, 16–22, and 10–35 have also been well studied (reviewed in ref. 12). Although experimental data on the β-sheet orientation have been elusive for Aβ25–35 (12, 13), solid state NMR experiments revealed that Aβ16–22 forms an antiparallel β-sheet (14), and that Aβ10–35 and the whole sequence of 1–40 form parallel β-sheet amyloids (15, 16). Although the IR spectra of the Aβ24–35 amyloid indicates the existence of an antiparallel β-sheet (17), x-ray diffraction failed to identify an ordered β-crystalline in the 24–35 fragment amyloid (12, 13).
That these disease-related short peptides are amyloidogenic and toxic makes them particularly useful for studies of amyloid formation and elongation. The kinetics of amyloid formation from such short peptides is similar to that of their larger parent proteins. Consequently, in principle, studies of such short amyloidogenic peptides may illuminate some of the fundamental processes taking place in amyloid formation in large protein systems. Although in principle, short peptide systems are considerably simpler than those of large proteins, obtaining atomic details on peptide amyloid formation from x-ray diffraction of amyloid fibrils has proven to be equally difficult. Amyloid fibrils from both short peptides and large proteins yield only limited information on the pattern of the β-sheet backbone within the fibril (12–16). Here, we study possible multilayer β-sheet oligomer organizations of the Aβ peptide fragments with high-temperature molecular dynamics (MD) simulations. Our simulations indicate that several parallel and antiparallel β-sheet orientations may be possible for the relatively small system of Aβ16–22, with one antiparallel β-sheet orientation being the most likely stable form for Aβ16–22, in agreement with solid state NMR data (14). Our 24-mer model of Aβ16–22 suggests a highly twisted fibril. Whereas the short Aβ16–22 and Aβ24–36 may exist in fully extended β-sheet form, the linear parallel β-sheets for Aβ16–35 appear impossible, mainly because of the polar region in the middle of the 16–35 sequence. However, a bent double-layered hairpin-like hook structure with the polar region at the turn forms a parallel β-sheet with higher stability. The bent, parallel double-sheet model for Aβ10–35 also displays conformational stability. Further, a model with the two bent sheets interlocking to produce an elongated form shows a good stability in the simulations. Conceptually, this model is similar to the one deduced from small-angle neutron scattering of the 1–40 micelle-like intermediates in the Aβ fibril assembly (18).
Here, we follow the Tycko and coworkers' short Aβ fragment notation (14).
Methods
Our model systems include the peptide oligomers solvated with thousands of water molecules in a rectangular box with periodic boundary conditions. All simulations were done at 330 K, which corresponds to the temperature often used to incubate the amyloid in experiments. The starting conformations of the peptide complex are generated to represent a β-sheet cluster. Next, the complex is allowed to relax in the MD simulations. MD simulations were performed in the canonical ensemble (NVT) for octamers using the program DISCOVER V.2.98. All atoms of the system were considered explicitly, and their interactions were computed by using the CFF91 force field with periodic boundary conditions. The potential energy functions include bond stretching, angle bending, torsion, and out-of-plane angle deformation terms and contain cross-terms to describe the couplings between bond–bond, angle–angle, bond–angle, bond–torsion, torsion–angle, and angle–angle–torsion (19). The nonbonded interactions include van der Waals interactions and electrostatic interactions. The hydrogen-bonding interactions are modeled by using the combined van der Waals interactions and electrostatic interactions between atomic pairs suitable to form hydrogen bonds. A distance cutoff of 11 Å was used for the nonbonded interactions. The time step in the MD simulations is 1 fs.
For the large 24-mer oligomers, simulations were done by using CHARMM polar hydrogen force field (20) and TIP3 water molecules, implemented in CHARMM (21) with NPT conditions. The time step used in the 24-mer simulations is 2 fs with SHAKE constraint on all bonds involving hydrogen atoms.
All of the starting conformations were built by using the INSIGHTII molecular modeling package (Accelrys, San Diego). The radial distribution functions were evaluated by using the Analysis module in the INSIGHTII package.
The Aβ16–22 octamers were simulated in a 45 × 45 × 45 Å3 box with 2618 water molecules with NVT condition at 330 K. The Aβ16–22 24-mer was simulated in NPT conditions of 1 atm (1 atm = 101.3 kPa) pressure, 330 K, and 4,654 water molecules, resulting in a box of 62.9 × 55.6 × 47.2 Å3. The box size for the Aβ16–35 octamers in linear conformation is 50 × 50 × 100 Å3 with 7,394 water molecules. For the bent hairpin Aβ16–35 octamer, a box of 40 × 50 × 60 Å3 with 3,187 water molecules is used. For the bent hairpin Aβ10–35 octamer, we use a box of 40 × 60 × 70 Å3 with 4,479 water molecules. The box size for the bent hairpin Aβ10–35 octamers in interlocked conformations is 50 × 50 × 100 Å3 with 7,216 water molecules.
Fragment 16–22: KLVFFAE.
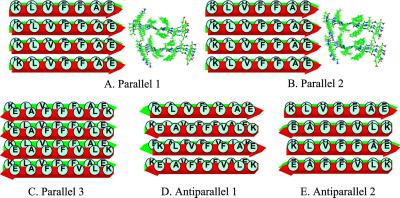
The Alzheimer's disease β-peptide fragment 16–22 is highly hydrophobic. Experimentally, the N- and C-termini are blocked by acetyl and NH2 groups, respectively (14). In our simulations, the exact same blocked peptide sequences are used. We simulated the KLVFFAE octamers, with three parallel and two antiparallel β-sheet arrangements, all with residues in-register (Fig. 1). For the three parallel octamers, we considered two arrangements with the relative position of the two layers also being parallel and one arrangement with the two layers being antiparallel. In one parallel model, the inter-layer hydrophobic core is formed by [-V-F-, -L-F-A-] (Parallel 2, Fig. 1B). In the second parallel model, the hydrophobic core residues are [-V-F-, -V-F-] (Parallel 1, Fig. 1A). In the third parallel model (Parallel 3, Fig. 1C), the hydrophobic core residues are also [-V-F-, -V-F-]. In the Parallel 3 model, there is a possibility of forming salt-bridges between Lys-16 and Glu-22 from the two β-sheet layers.
Fig 1.
Simulation of the peptide 16–22 fragment. Starting conformation and β-sheet orientations.
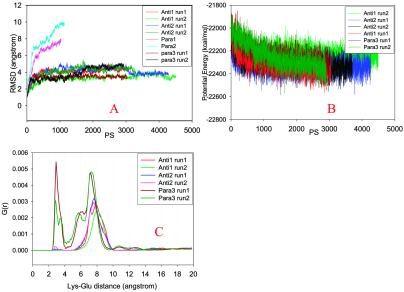
Both parallel octamers Parallel 1 and Parallel 2 dissociate quickly within 1 ns at 330 K, as indicated by the large rms deviation (rmsd) value observed immediately after starting the simulations (Fig. 2A). Dissociation of the two octamers is through separation of the layers, whereas each layer still has fairly good parallel β-strand interactions in Parallel 1. In Parallel 2, the β strands in one layer also separated (Fig. 3A). The poor interactions between the parallel β-sheet layers indicates that this two-parallel β-sheet organization is unlikely to be the constituent of amyloid fibrils.
Fig 2.
(A) rmsds of trajectories compared with the starting conformation. (B) Trajectories of the potential energies in simulations of the Antiparallel 1, Antiparallel 2, and Parallel 3 models. (C) Radial (distance) distribution functions of salt bridges in the octamers simulated. The distances between the nitrogen (Lys-16) and carboxylate carbon (Glu-22) are monitored.
Fig 3.
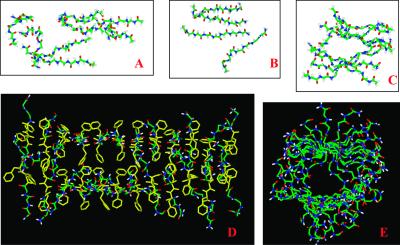
Snapshots from the simulations of Aβ16–22 fragment oligomers. (A) Parallel 2 model at 1.1 ns. (B) Parallel 3 model at 3 ns, run 2. Only one layer is shown for clarity. (C) Antiparallel 2 model at 4.2 ns, run 2. (D and E) The equilibrated structure of Antiparallel 3 24-mer at 2.5 ns. (D) A view perpendicular to the fibril axis. Backbone atoms are shown in color and Phe are shown in yellow. Other side chains are omitted for clarity. (E) A view along the fibril axis of the simulated structure. Only backbone atoms are shown for clarity.
Parallel 3 offers greater stability for the parallel octamers. Two 3 ns runs are simulated for the Parallel 3. In one simulation, the Parallel 3 octamer holds a stable β-sheets cluster throughout the simulation. In another run, the β-strands in one layer separated (Fig. 3B). However, the overall rmsd from the starting structure is still substantially lower than those for Parallel 1 and Parallel 2 (Fig. 2A).
Two antiparallel octamers, one with antiparallel sheets/antiparallel layers (Fig. 1D, Antiparallel 1), another with antiparallel sheets/parallel layers (Fig. 1E, Antiparallel 2), are much more stable than the parallel octamers. Two independent simulations are performed for each of the antiparallel octamers at 330 K, with all four simulations showing a small rmsd from the starting conformations (Fig. 2A). For the Antiparallel 1 model (antiparallel sheets/antiparallel layers), the two rmsd trajectories are separated with a small deviation. For the antiparallel sheet/parallel layer model (Antiparallel 2), both simulations show consistent trajectories for 4 ns at 330 K.
Simply by the β-strand stability of the octamers, the Antiparallel 1 and Antiparallel 2 models are more favored than the Parallel 3 model. We further examine the energetical stabilities of the octamers. If we assume that the octamers Antiparallel 1, Antiparallel 2, and Parallel 3 have the same entropical factors, the one with the lowest potential energy should have the lowest free energy and thus be the most stable oligomer. Table 1 compares the average potential energies from the simulations at 330 K. The Antiparallel 2 model is the most stable octamer. Energetically, one of the Antiparallel 1 model runs (run 1) has a similar average potential energy as the simulation runs for the Antiparallel 2 model (Table 1). However, the second Antiparallel 1 run (with the lowest rmsd in Fig. 1A) has a much higher (≈110 kcal/mol) average potential energy than the rest of the simulations. Two simulations of the Parallel 3 model consistently show higher potential energy. Note that the average potential energy we used is not free energy for the β-sheet oligomers. Therefore, the lower average potential energy for the Antiparallel 2 model is just an indication of its possible preference.
Table 1.
Statistics of potential energies (PE, kcal/mol) from MD simulations of selected octamers
| Run
|
Peptide 16–22 KLVFFAE | ||
|---|---|---|---|
| Average PE | Std. dev. | No. of frames | |
| Anti1 run1 | −22311 | 71.5 | 2543 |
| Anti1 run2 | −22220 | 66.4 | 3991 |
| Anti2 run1 | −22339 | 64.5 | 3767 |
| Anti2 run2 | −22324 | 67.5 | 3203 |
| Para3 run1 | −22235 | 98.2 | 3001 |
| Para3 run2 | −22242 | 112.7 | 2908 |
The potential energies from explicit water simulations with periodic boundary conditions, including water–water, water–peptides, and peptide–peptide interactions.
Salt bridges may contribute to the stabilities of the β-sheet oligomers. However, the salt bridges are less important than the hydrophobic interactions in the case of the Aβ-peptide fragment 16–22. Fig. 2C plots the pair distance distributions of Lys-16 and Glu-22 in the octamers. Specifically, we focus on the distances between the nitrogen (Lys-16) and carboxylate carbon (Glu-22). For the Parallel 3 model, the salt bridge distances peak around 3 Å. However, there is no close contact between Lys-16 and Glu-22 for the antiparallel models Antiparallel 1 and Antiparallel 2. For the parallel models, Parallel 3 may be stabilized by salt bridges as compared with the Parallel 1 and Parallel 2 models. For the antiparallel models, the hydrophobic interactions dominate.
Based on the β-strand stability and average potential energy, the antiparallel sheet/parallel layer model (Antiparallel 2) is more likely to be responsible for the experimentally observed amyloid structure. However, all three structures (Parallel 3, Antiparallel 1, and Antiparallel 2) may be correct for the relatively small system considered in these simulations, although one of the antiparallel structures would be expected to be the most stable for a much larger system.
To study the Antiparallel 2 arrangement further, we enlarged the simulation system to a 24-mer (a double-layered antiparallel β-sheet with 12 strands in each layer). At 330 K, a 2.5-ns equilibration of the 24-mer yields highly ordered and highly twisted (15°) oligomers (Fig. 3 D and E). This twist angle may reflect the interactions of Phe-19 and Phe-20 between the two parallel layers.
These results are consistent with a model based on solid-state NMR experiments, indicating an antiparallel β-sheet organization in the Aβ16–22 amyloid. X-ray diffraction patterns indicated that there are two strong periodicities in the Aβ16–22 fibrils: at 4.7 Å and at 9.9 Å (14). The 4.7-Å periodicity was assigned to the difference between the β-strands, and the 9.9 Å relates to the spacing between the layers (14). In our equilibrated 24-mer, we confirm the 4.7-Å inter-strand spacing. However, the distance between the layers is much wider, 12 Å in our model. This finding may reflect an inadequacy in our simulations of small oligomers as compared with the much larger fibrils.
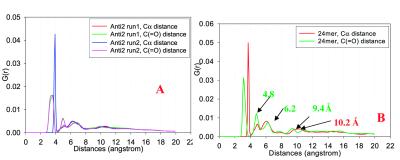
To compare further the simulated structure with the experimental results, we investigated the pair radial distribution functions for Cα-Cα distance and that between carbonyl carbons [C( O) – C(
O) – C( O)], as indicated in Fig. 4. Fig. 4A plots the distance distribution functions for octamer Antiparallel 2 models, averaged from trajectories of 4-ns simulations. Fig. 4B plots those for the 24-mers, averaged over 2.5-ns simulations. Both Fig. 4 A and B share the same feature. However, in Fig. 4B they are more resolved, the outcome of the 24-mer being more structured than the octamer. In general, the 3–4 Å and 6–7 Å peaks are the Cα-Cα and C… C distances within peptide strands. The peaks around 5 Å reflect the spacing between β-strands within the β-sheet. The peaks between 9–11 Å could be the separation between every other strand or the separation between layers. Generally, the Cα and C(
O)], as indicated in Fig. 4. Fig. 4A plots the distance distribution functions for octamer Antiparallel 2 models, averaged from trajectories of 4-ns simulations. Fig. 4B plots those for the 24-mers, averaged over 2.5-ns simulations. Both Fig. 4 A and B share the same feature. However, in Fig. 4B they are more resolved, the outcome of the 24-mer being more structured than the octamer. In general, the 3–4 Å and 6–7 Å peaks are the Cα-Cα and C… C distances within peptide strands. The peaks around 5 Å reflect the spacing between β-strands within the β-sheet. The peaks between 9–11 Å could be the separation between every other strand or the separation between layers. Generally, the Cα and C( O) have different (although very close) distributions, with the C(
O) have different (although very close) distributions, with the C( O) distance closer to the experimental observation of 4.7 Å inter-strand spacing.
O) distance closer to the experimental observation of 4.7 Å inter-strand spacing.
Fig 4.
Radial (distance) distribution functions for the Cα-Cα and C( O)… C(
O)… C( O) separations in the β-sheet oligomers of (A) octamer Antiparallel 2 model. (B) 24-mer of antiparallel sheets.
O) separations in the β-sheet oligomers of (A) octamer Antiparallel 2 model. (B) 24-mer of antiparallel sheets.
Aβ Peptides 16–35 and 10–35.
Aβ16–35 may be viewed as resulting from linking two previously simulated fragments, Aβ16–22 and Aβ24–35, with aspartic acid D23. In this work, we also simulated the β-sheet oligomers of Aβ24–36 and found that either parallel or antiparallel β-sheet may form stable oligomers. The details of the simulations of Aβ24–36 will be reported separately.
Experimentally, for the longer 10–35 sequence, the fibril adopts a parallel β-strand organization (16). Considering that the antiparallel β-sheets are stable for the two short fragments, the changing preference of the β-strand arrangements in the amyloid when the shorter segments are linked is important to understand. Clearly, their connection makes it impossible to keep an antiparallel interaction within each fragment. For the Aβ16–35 to adopt an antiparallel organization, Aβ16–22 has to interact with Aβ25–35, with interactions that are not necessarily optimized. In the current study, however, we did not explore the antiparallel β-sheet interactions in our simulations of the 16–35 and 10–35 peptides. Only parallel β-sheets have been considered.
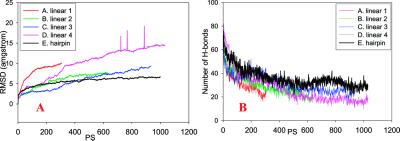
Extensive simulations are conducted to optimize the inter-layer matches (Fig. 5). In Aβ16–35 (and Aβ10–35), there are hydrophobic regions at both ends (residues 16–21 and 30–35), and the central region is rich in polar and charged residues. Because for neither Aβ16–22 nor Aβ10–35 are the parallel sheet/parallel layer organizations stable, the parallel sheet/parallel layer for the Aβ16–35 is also expected to be unstable, unless the central polar/charged region could stabilize the inter-layer interactions. However, in our simulations, the interactions at the central region are highly destabilizing. Hence, the parallel sheet/parallel layer cluster is unstable (Figs. 5A and 6). For an antiparallel layer organization, we consider nine possible inter-layer matches to see whether antiparallel layers (with resulting hydrophobic inter-layer interactions between residues -16–21- and -35–30-) can lead to a stable complex (three are reported in this work, Fig. 5 B–D). The inter-layer salt bridge between D23-K28 is also included in the complex (Fig. 5B). However, none of the complexes with the fully extended linear parallel β-strands survives the 330 K simulation, and all randomize rapidly (Fig. 6; Fig. 9, which is published as supporting information on the PNAS web site, www.pnas.org). Therefore, another type of inter-β-sheet interaction must be considered to stabilize the possible parallel β-sheets in the Aβ16–35 and Aβ10–35 fibrils.
Fig 5.
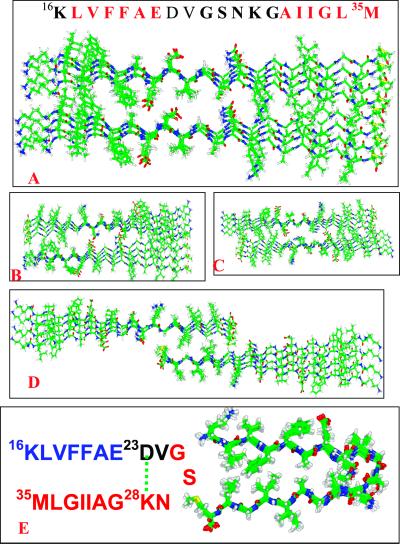
An illustration of the β-strand arrangements in the octameric models considered for the peptide 16–35 fragment. (A–D) Parallel β-sheet models with linear β-strands. (E) Parallel β-sheet with a bent hairpin-like hook conformation.
Fig 6.
Trajectories of the simulations of Aβ16–35 fragment octamers.
From the dissociation behavior of the linear complex, with the central polar/charged region always being destabilizing, this alternative structure must stabilize the central region. One possible solution to the problem is that the central polar/charged region forms a turn to facilitate the hydrophobic interactions between residues 16–22 and 30–35 within the same strands instead of between two strands (i.e., inter-layer interactions). This consideration implies bending the parallel stranded-sheet to produce a bent, double-layered β-sheet monomer. To create a bent structure, we have to choose a likely turn location. As a factor in the choice of the turn region in the monomeric structure, we consider a flexible region in the NMR studies (23), leading us to select the 24–27 region for the turn formation. Benzinger et al. (23) found that the inter-strand distance involving Gly-25 and Ser-26 was longer than at other locations. The additional advantage of a 24–27 turn is that it makes it possible to form an intra-strand salt bridge (D23–K28). Thus, we proceed to construct and test the stability of a one-layer parallel β-sheet folded into a double layer for Aβ16–35 (Fig. 5E). The self-folded β-sheet is greatly stabilized, compared with the rapid deformation of the linear β-strands (Fig. 6). The matched hydrophobic region is stabilized and so is the polar/charged turn in the 1-ns 330 K simulation (Fig. 9E).
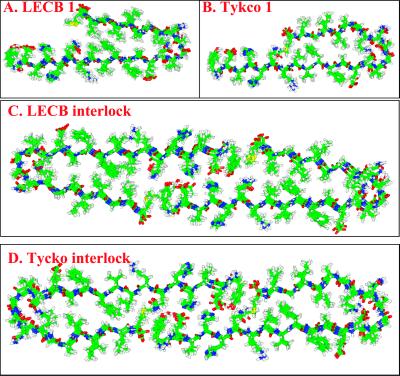
Next, we extend the stable model to test Aβ10–35. Two types of complexes are tested: a monomeric and an interlocked model (Fig. 7 A and C). Because in the Aβ16–35 we now create a hairpin-like hook structure, with an unmatched 7-residue tail (residues 10–16), the consideration in the construction of the interlocked species is to see whether the frayed 10–16 end needs a further inter-layer match to be stabilized.
Fig 7.
An illustration of the β-strand arrangements in the octameric models considered for the Aβ10–35 fragment with a parallel β-sheet with bent hairpin-like hook conformation.
This self-folded β-sheet layer feature is reinforced by the emerging solid-state NMR evidence reported in the 46th Biophysical Society meeting (R. Tykco, personal communication) simultaneously with the present work. Both our structure and the NMR structure point to the crucial role of the salt bridge and are consistent with a mutational study. Experimentally, an Asp-23→Lys mutation eliminates β-sheet formation (22). The obvious explanation is that the repulsive force breaks the salt bridge and destabilizes the turn, hence making it impossible to form a stable complex.
There is a slight difference between our model (called the LECB model, from our Laboratory of Experimental and Computational Biology) and the model from the Tycko group (called Tycko's model, Fig. 7). Our model forms a hydrophobic core with Leu-34, whereas in Tycko's model the Leu-34 faces outside. Subsequent simulations of Tycko's model show similar stabilities as our LECB model (Figs. 7 and 8; Fig. 10, which is published as supporting information on the PNAS web site).
Fig 8.
(A and B) Trajectories of the simulations of Aβ10–35 fragment octamers. (C) Radial (distance) distribution functions of salt bridges in the octamers simulated. The distances between the nitrogen (Lys-28) and carboxylate carbon (Asp-23) are monitored. (D and E) Radial (distance) distribution functions for the Cα-Cα and C( O)… C(
O)… C( O) separations in the bent β-sheet oligomers.
O) separations in the bent β-sheet oligomers.
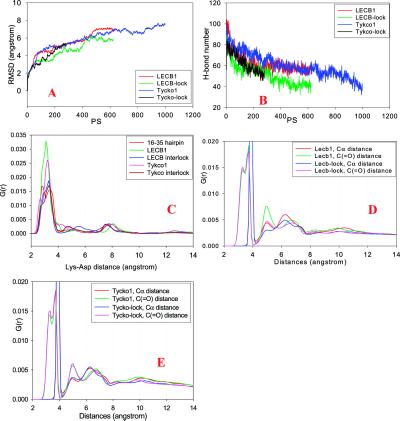
With the restriction of the turn, the salt bridges in all of the bent hairpin models are pretty stable. As may be seen in Fig. 8C, the salt bridge distances are mostly around 3 Å. In comparison, the salt bridges in the Parallel 3 model of Aβ16–22 (Fig. 2C) are more flexible (with large peaks around 3 Å, 6 Å, and 7 Å). The tight salt bridges indicate that the electrostatic interactions are important for the Aβ10–35 amyloid. Fig. 8 C and D plot the radial distribution functions for the LECB and Tykco models. All models show peaks around 5 Å and 10 Å, corresponding to the experimentally observed distances.
Both the monomer model and the interlocked model may form for the Aβ10–35. Experimentally, Aβ10–35 forms two types of amyloids at pH 5.6 and 7.4 (22). The one formed at pH 5.6 has a diameter of 90 Å, and that at pH 7.4 has an 80 Å diameter. Therefore, it appears that the interlocked conformation is the likely form at pH 5.6 and the monomer species is the likely form at pH 7.6. Conceptually, the interlocked model is similar to the one deduced from small-angle neutron scattering of the 1–40 micelle-like intermediates in the Aβ fibril assembly (18). However, it should be noted that this is not experimental evidence of a specific model, because the small-angle neutron-scattering data cannot give sufficient atomic detail.
Discussion
Because amyloid fibrils are insoluble aggregates, it is not possible to obtain detailed structures such as those obtained by x-ray crystallography or solution NMR. The structural information currently available is largely derived from data obtained from solid-state NMR experiments. These experiments have produced structural models for a number of peptide fragments, mostly derived from the Aβ. The structures for short fragments have been proposed to have an antiparallel organization. This is the case for Aβ16–22 and Aβ34–42, for example. Our explicit water simulations of short fragments A8, AGAAAAGA (24), Aβ16–22 (this work), and NFGAIL derived from the islet amyloid polypeptide (25) have shown a higher stabilization of antiparallel strands within the sheet, and a parallel arrangement between sheets. It appears that shorter peptides prefer an antiparallel organization, whereas, depending on their sequence, longer peptides may conceivably adopt either conformation (for example, Aβ24–36). For Aβ10–35, solid-state NMR data indicate a parallel organization within the sheet (16, 22). A recent solid-state NMR analysis of Aβ1–40 similarly indicates a parallel arrangement (15). Our simulations of the Aβ16–35 and Aβ10–35 reveal the underlying mechanism of the parallel organization.
A parallel organization for long peptides, such as those in the Aβ, has some important implications. For short peptides, a β-sheet organization has to involve strands derived from different peptides, hydrogen bonded to each other. In principle, longer peptides, such as the 40- or 42-residue long Aβ can conceivably organize in different ways. If the Aβ forms conventional, multi-stranded β-hairpin structures, each molecule can fold on itself to form a sheet, with the sheets running in parallel to the helix axis. However, such a molecular organization is inconsistent with a parallel arrangement of the strands observed by NMR, as a one-peptide sheet necessarily invokes at least a partial antiparallel organization. On the other hand, a parallel organization implies sheets similar in character to those observed for the shorter peptides, where each peptide corresponds to a strand. This can present problems, however, as a 40-residue strand produces very unstable extended sheets, as our explicit water simulations show. This work suggests that the sheet bends to create a double-layered sheet. Depending on the sequence, the bending may be asymmetrically positioned to create a hook-like double layer. Our simulations indicate that, consistent with models recently obtained from solid-state NMR, this type of structure is stable. The ends of two such hooks may interlock, to produce a stable, end-to-end interlocked double-layered β-sheet. Furthermore, amyloids are formed at high-molecular concentrations. At such concentrations, the chance of an inter-molecular encounter may not be very different from the peptide folding on itself, to create a single-peptide sheet.
Several additional observations also indicate the importance of a bent conformation in Aβ amyloid formation. Teplow and coworkers (26) have observed a helix intermediate involvement in the Aβ amyloid formation. Kallberg et al. (27) have identified a strong likelihood for a helix for residues 16–23. The C terminus may form a weak helix because of the Gly residues. Further, a helix-turn-helix to a β-structure conversion has been observed experimentally (28). The transition to a β-sheet is much slower than a coil-to-helix transition (29). We suspect that the involvement of a bent conformation in Aβ amyloid formation has both structural and kinetic significance.
Amyloid formation poses several intriguing questions. First, what is the conformation of the fibril? Second, what is the minimal seed size? Third, what is the driving force of amyloid formation? And fourth, what is the mechanism of amyloid growth? The final conformation is a cross-β, and the initial conformation is apparently a disordered state. At least in the cases of peptides such as the Alzheimer β peptide (Aβ), and the short peptides such as those derived from Aβ, for example, or from the islet amyloid polypeptide, there is evidence that in solution the peptides are disordered. However, the mechanism through which the disordered peptides assemble to yield the cross-β conformation is still unknown, even though considerable progress in our understanding has been made recently (26). Further, experiment suggests that seed formation is the rate-limiting step. What, then, is the conformation of the seed and its minimal size? And, what is the mechanism by which it grows? An answer to one of these problems would aid in illuminating the others.
The type of interlocked, double-layered bent sheet model suggested here implies a different mechanism than if the peptide self-folds to form a sheet, with sheet-packing. It suggests a possible seed (an interlocked monomer), with a minimal seed size (of about four strands), and a mechanism of fibril growth. Several lateral interlocked oligomers might provide an added stabilization. It would be interesting to see whether other peptides with appropriate size ranges form similar types of layer organizations. We note further that such a conformation and mechanism also appear consistent with the fact that all proteins can form amyloids under given conditions (30). It is not necessary for the entire protein to unfold and participate in the sheet. It is sufficient that a protein arm (or terminus) unfolds to contribute a strand to a growing fibril, with the rest of the protein potentially “hanging out” intact.
Supplementary Material
Acknowledgments
We thank the referee for suggesting the simulation of the Parallel 3 model of Aβ16–22. We thank Dr. Robert Tykco for providing the slides of the NMR-deduced amyloid structure before publication. We thank Drs. David Zanuy and K. Gunasekaran for making several thoughtful comments. We also thank Dr. Jacob V. Maizel for encouragement. The computation times are provided by the Advanced Biomedical Supercomputing Center, National Cancer Institute, Frederick, and by the National Institutes of Health Biowulf. The research of R.N. in Israel has been supported in part by a Magnet grant, by a Ministry of Science grant, and by the Center of Excellence in Geometric Computing and its Applications, which is funded by the Israel Science Foundation and administered by the Israel Academy of Sciences, and by the Adams Brain Center. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The publisher or recipient acknowledges the right of the U.S. Government to retain a nonexclusive, royalty-free license in and to any copyright covering the article.
Abbreviations
rmsd, rms deviation
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Carrell R. W. & Lomas, D. A. (1997) Lancet 350, 134-138. [DOI] [PubMed] [Google Scholar]
- 2.Chesebro B. (1998) Science 279, 42-43. [DOI] [PubMed] [Google Scholar]
- 3.Lorenzo A., Razzaboni, B., Weir, G. C. & Yankner, B. A. (1994) Nature 368, 756-760. [DOI] [PubMed] [Google Scholar]
- 4.Tenidis K., Waldner, M., Bernhagen, J., Fischle, W., Bergmann, M., Weber, M., Merkle, M., Voelter, W., Brunner, H. & Kapurniotu, A. (1999) J. Mol. Biol. 295, 1055-1071. [DOI] [PubMed] [Google Scholar]
- 5.Gasset M., Baldwin, M. A., Lloyd, D. H., Gabriel, J.-M., Holtzman, D. M., Cohen, F., Fletterick, R. & Prusiner, S. B. (1992) Proc. Natl. Acad. Sci. USA 89, 10940-10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haass C. & Strooper, B. D. (1999) Science 286, 916-919. [DOI] [PubMed] [Google Scholar]
- 7.Hughes E., Burke, R. M. & Doig, A. J. (2000) J. Biol. Chem. 275, 25109-25115. [DOI] [PubMed] [Google Scholar]
- 8.Iverson L. L., Mortishire-Smith, R. J., Pollack, S. J. & Shearman, M. S. (1995) Biochem. J. 311, 1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terzi E., Hölzmann, G. & Seelig, J. (1994) Biochemistry 33, 1345-1350. [DOI] [PubMed] [Google Scholar]
- 10.Pike C. J., Walencewicz-Wasserman, A. J., Kosmoski, J., Cribbs, D. H., Glabe, C. G. & Cotman, C. W. (1995) J. Neurochem. 64, 253-265. [DOI] [PubMed] [Google Scholar]
- 11.Shearman M. S., Ragan, C. I. & Iverson, L. L. (1994) Proc. Natl. Acad. Sci. USA 91, 1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serpell L. C. (2000) Biochim. Biophys. Acta 1502, 16-30. [DOI] [PubMed] [Google Scholar]
- 13.Inouye H., Fraser, P. & Kirschner, D. (1993) Biophys. J. 64, 502-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balbach J. J., Yoshita, I., Antzutkin, O. N., Leapman, R. D., Rizzo, N. W., Dyda, F., Reed, J. & Tycko, R. (2000) Biochemistry 39, 13748-13759. [DOI] [PubMed] [Google Scholar]
- 15.Antzutkin O. N., Balbach, J. J., Leapman, R. D., Yoshita, I., Rizzo, N. W., Reed, J. & Tycko, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13045-13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benzinger T. L. S., Gregory, D. M., Burkoth, T. S., Miller-Auer, H., Lynn, D. G., Botto, R. E. & Meredith, S. C. (1998) Proc. Natl. Acad. Sci. USA 95, 13407-13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konno T. (2001) Biochemistry 40, 2148-2154. [DOI] [PubMed] [Google Scholar]
- 18.Yong W., Lomakin, A., Kirkitadze, M. D., Teplow, D. B., Chen, S.-H. & Benedek, G. B. (2002) Proc. Natl. Acad. Sci. USA 99, 150-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maple J. R., Hwang, M. J., Jalkanen, K. J., Stockfisch, T. P. & Hagler, A. T. (1998) J. Comput. Chem. 19, 430-458. [Google Scholar]
- 20.Neria E., Fischer, S. & Karplus, M. (1996) J. Chem. Phys. 105, 1902-1921. [Google Scholar]
- 21.Brooks B. R., Bruccoleri, R. E., Olafson, B. D., Sate, D. J., Swaminathan, S. & Karplus, M. (1983) J. Comp. Chem. 4, 187-217. [Google Scholar]
- 22.Fraser P. E., McLachlan, D. R., Surewicz, W., Mizzen, C. A., Snow, A. D., Nguyen, J. T. & Kirschner, D. A. (1994) J. Mol. Biol. 244, 64-73. [DOI] [PubMed] [Google Scholar]
- 23.Benzinger T. L. S., Gregory, D. M., Burkoth, T. S., Miller-Auer, H., Lynn, D. G., Botto, R. E. & Meredith, S. C. (2000) Biochemistry 39, 3491-3499. [DOI] [PubMed] [Google Scholar]
- 24.Ma B. & Nussinov, R. (2002) Protein Sci. 11, 2335-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zanuy, D., Ma, B. & Nussinov, R. (2002) Biophys. J., in press.
- 26.Kirkitadze M. D., Condron, M. M. & Teplow, D. B. (2001) J. Mol. Biol. 312, 1103-1119. [DOI] [PubMed] [Google Scholar]
- 27.Kallberg Y., Gustafsson, M., Persson, B., Thyberg, J. & Johansson, J. (2001) J. Biol. Chem. 276, 12945-12950. [DOI] [PubMed] [Google Scholar]
- 28.Fezoui Y., Hartley, D. M., Walsh, D. M., Sekkoe, D. J., Osterhout, J. J. & Teplow, D. B. (2000) Nat. Struct. Biol. 7, 1095-1099. [DOI] [PubMed] [Google Scholar]
- 29.Muñoz V., Thompson, P. A., Hofrichter, J. A. & Eaton, W. A. (1997) Nature 390, 196-199. [DOI] [PubMed] [Google Scholar]
- 30.Chiti F., Taddei, N., Baroni, F., Capanni, C., Stefani, M., Ramponi, G. & Dobson, C. M. (2002) Nat. Struct. Biol. 9, 137-143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.