Abstract
The goal of this work is to define the structural and sequence features common to sandwich-like proteins (SPs), a group of very different proteins now comprising 69 superfamilies in 38 protein folds. Analysis of the arrangements of strands within main sandwich sheets revealed a rigorously defined constraint on the supersecondary substructure that holds true for 94% of known SP structures. The invariant substructure consists of two interlocked pairs of neighboring β-strands. It is even more typical for centers of SP than the well-known “Greek key” strands arrangement for their edges. As homology among these proteins is not usually detectable even with the most powerful sequence-comparing algorithms, we employed a structure-based approach to sequence alignment. Within the interlocked strands we found 12 positions with fixed structural roles in SP. A residue at any of these positions possesses similar structural properties with residues in the same position of other SPs. The 12 positions lie at the center of the interface between the β-sheets and form the common geometrical core of SPs. Of the 12 positions, 8 are occupied by only four hydrophobic residues in 80% of all SPs.
Proteins of 69 superfamilies in 38 protein folds have been described as sandwich-like proteins (SPs) [see folds 1.2.1–1.2.38 in Structural Classification of Proteins (SCOP) (1), release 1.59] . Spatial structures of SPs are composed of β-strands, which form two main β-sheets that pack face-to-face. Although the general architecture of SPs is relatively uniform, the number of strands and the arrangement of the strands varies widely (2–7). Some SPs, in addition to two “main” sandwich sheets, contain “auxiliary” β-sheets. Comparison of proteins in different superfamilies does not show either functional homology or significant sequence homology. In fact, some SPs share so little homology (less than 10–15%) that it cannot be detected even with the most advanced homology search algorithms such as hhms (8) or psi blast (9).
The goal of this research is to define the structural and sequence features that these very different proteins have in common. Early attempts to find common structural characteristics of β proteins came from Richardson (10), and Sternberg and Thornton (11). Richardson originally described the rule applicable to supersecondary structures, the so-called Greek key supersecondary structural unit, in 1977. In other investigations the analysis of a vast number of widely divergent β protein structures revealed a number of structural regularities that govern the folding motif of the β proteins (2, 5, 7, 12–14).
In previous communications we presented the results of an analysis of sequence conservation and structural features of two SP families: immunoglobulin variable domains and cadherins (15–17). These investigations showed that functionally dissimilar proteins with no significant homology nonetheless do share a number of common sequence and structural features.
In the present paper we extend comparative analysis to all SP superfamilies. Our investigation of structural and sequence features common to SPs consists of two parts: the search for “structural determinants” of SPs, residues that have the same structural properties across SPs, and the search for “sequence determinants,” a subset of residues chosen among the structural determinants that share both structural and chemical properties in all SPs.
Our first task is to analyze the supersecondary substructure of SPs to determine whether they have features that are invariant. The analysis discovered that despite a seemingly unlimited number of arrangements of strands resulting in sandwich-like structure, there exists a rigorously defined constraint on supersecondary structure that applies to almost all SPs. This constraint can be termed “the rule of two interlocked pairs of strands.” These four strands form a small sandwich-like substructure within a protein.
Another aspect of our research involves finding positions in sequences that are occupied by similar residues in all SPs. The usual methods of searching for conserved positions in protein families by using the alignment algorithms (18–21) cannot be easily applied to a set of structurally and sequence dissimilar proteins that are not derived from a common ancestor. However, in the case of SP, a structure-based alignment of these sequences from different superfamilies and protein folds is made possible by the discovery of their common substructure. Analysis of four interlocked strands revealed eight hydrophobic positions conserved across all SPs.
Methods and Results
Strand Definition and Arrangement of Strands in SPs.
Definitions of strands adopted in our work coincide for the most part with the definitions of the PDBsum database (22). One difference between the two systems of definition is that we consider two PDBsum-defined strands to be a single strand with a small bulge if the following conditions hold true: (i) the two PDBsum strands are neighbors in a sequence; (ii) both share H bonds with the same strand; (iii) they are parallel to each other; and (iv) the number of interposed residues between the two PDBsum strands is no more six.
Analysis of H bonds between main-chain atoms allowed us to determine the arrangements of the strands in space and to identify those strands that make up the two main sandwich sheets. We then sequentially numbered those strands that constitute main sheets, omitting the strands of the auxiliary sheets. In essence, we abstracted sandwich structures from the actual molecules and analyzed strand arrangements only within the main sheets.
The “Rule of Two Interlocked Pairs of Strands” Strand Packing of SPs.
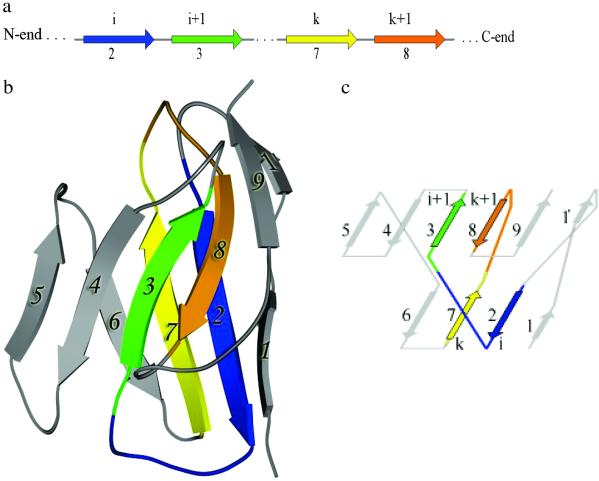
Analysis of the arrangements of strands in two main β-sheets of structures of 298 protein entry domains comprising 38 protein folds revealed a definite rule that is valid for almost all SPs. This rule can be stated as follows: in any given sandwich-like protein structure there exist two pairs of strands: i, i + 1 and k, k + 1 meaning that: (i) strands of each pair are adjacent to each other in a sequence (Fig. 1a); (ii) strand i is located in one main sheet and i + 1 in the other main sheet; (iii) similarly, strand k is found in one main sheet and k + 1 in the other; (iv) strands i and k are located within the same sheet; they are antiparallel to each other and linked by hydrogen bonds; and (v) likewise, strands k + 1 and i + 1 are located within another main sheet and are antiparallel to each other and H bonded (Fig. 1 b and c).
Fig 1.
The schematic representation of a typical variable domain of the immunoglobulins. (a) β-Sheet strands are numbered as they are presented in a sequence. Strands 2, 3, 7, and 8 are shown. (b) A chain fold of the immunoglobulin variable domain of the heavy chain (Protein Data Bank code ). The drawing is done with the MOLSCRIPT program (23). β-Sheet strands are shown as ribbons. (c) Arrangement of the strands in two main β-sheets. Two interlocked pairs of strands, (i, i + 1) and (k, k + 1), correspond to strands 2 and 3, and 7 and 8, respectively.
These two interlocked pairs form a sandwich-like substructure within SPs, and are usually found in the middle of the sheets. The number of strands interposed between strands i and k varied from 1 to 10, but in 80% of the cases the number of interposed strands was 2–4 (see Figs. 1c and 2b, where the number of strands between the strands i and k are equal to 4 and 2, respectively). Two interlocked pairs were detected in 94% of all analyzed structures.¶ Thus, this investigation led to the discovery of a central feature of the supersecondary structure that is invariant in almost all SPs.
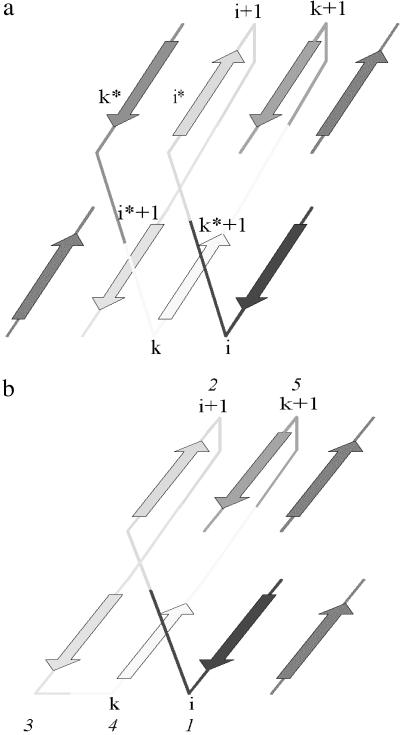
Fig 2.
Arrangement of interlocked pairs of strands. (a) Schematic representation of two sets of interlocked pairs of strands. Strands of the first set of interlocked pairs are named i, i + 1 and k, k + 1, and strands of the second set of interlocked pairs, i*, i* + 1 and k*, k* + 1. (b) Strands of the interlocked pairs of strands participate in the complete Greek key (two overlapped Greek keys). Strands 1, 4, 3, and 2 form one Greek key, and strands 5, 2, 3, and 4 form another. The interlocked pairs of strands are 1, 2, 4, and 5 (i, i + 1 and k, k + 1, respectively). Strands are numbered beginning with the i strand.
The structures of 75 SP domains contain an extra set of interlocked pairs; i.e., in addition to the i, i + 1 and k, k + 1 pairs of strands, we observed another set of two interlocked pairs of strands (i*, i* + 1 and k*, k* + 1). It is important to note that in all structures the interlocked pairs of strands were always arranged in the same way (the strands k = k* + 1 and i + 1 = i*) are shown in Fig. 2a.
Comparison of the “Rule of Interlocked Pairs” and the Greek Key Topology.
The analysis of the strands' arrangement showed that ≈70% of known SP domains contain the Greek key, a type of the supersecondary substructure originally described 25 years ago (10). Thus, the interlocked pairs, which were found in almost all structures, are much more typical for SPs than the Greek key. Generally, interlocked pairs are found at centers of SPs, whereas Greek keys are found at their edges. The two types of invariant substructures are independent of each other. For example, the immunoglobulin shown in Fig. 1 contains the interlocked pairs (strands 2, 3, 7, and 8) but no Greek key, whereas the protein shown in Fig. 2b contains both interlocked pairs (strands 1, 2, 4, and 5, and two Greek keys (strands 1, 2, 3, and 4, and 2, 3, 4, and 5).
Structurally Based Sequence Alignment of Nonhomologous Proteins.
The essential element of our method of structurally based sequence alignment is that it involves an alignment not of whole sequences, but of corresponding strands in their respective proteins. The selection of corresponding strands in a group of homologous proteins (protein family) with the same number of strands and the similar arrangement of the strands in space is a straightforward problem. Structurally similar strands are collected from all proteins of the group under consideration. The multiple alignment is conducted separately for each set of the corresponding strands.
In contrast to the analysis of homologous proteins, the determination of the corresponding strands in a group of nonhomologous proteins can be a complicated problem to attempt to solve. However, in the case of SPs, the delineation of an invariant supersecondary substructure made it possible to identify and align strands with structurally analogous roles from very diverse proteins with a dissimilar arrangement of strands and variable numbers of the strands. Thus i strands of all SP structures were aligned with each other, then all i + 1 strands, and so forth.
To find conserved positions in i, i + 1, k, and k + 1 strands, each residue in these strands was characterized with respect to (i) its residue–residue contacts (residues are considered to be in contact if the distances between any heavy atoms are closer than 5 Å), (ii) hydrogen bonds with the main-chain atoms of residues in other strands, and (iii) residue surface exposure. This analysis enabled us to align residues with similar structural properties in different proteins.
Carried out in this way, the alignment of strands maximizes the number of positions occupied by structurally similar residues. It is important to note that no gaps within strands are allowed, because strands are viewed as indivisible structural units. Adjacent residues within a strand are always assigned sequential position numbers. However, gaps between strands are a common occurrence. The advantage of this structurally based approach is that it makes possible a common system of numbering for sequences from different superfamilies. It allowed us to compare nonhomologous SPs.
For the alignment of residues from i, i + 1, k, and k + 1 strands we used all of the structures of 279 SP domains. Examples of the multiple alignment are presented in Table 1. Because the strand alignments are based on structural properties of residues, the first residue in the ith strand of one sequence can possess similar structural properties as (and be aligned with), for example, the third residue of the ith strand of another sequence. See, for example, the first residue S in the i strand (Protein Data Bank code in Table 1) and the third residue T in the i strand (Protein Data Bank code ). Thus, in the common system of numbering, the i strand starts at position 3 in the protein. It is clear that the length of the i strand in one protein may not be identical to length of the i strand in another protein. A consequence of introducing a common numbering system based on the structural alignment of residues is that strands can start at positions other than position 1 and their lengths can vary for different sequences.
Table 1.
Structurally based sequence alignment of the i, i + 1, k, and k + 1 strands
| Fold
|
Str
|
i | i + 1 | k | k + 1 | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
| 1_1_1 | 1INE | S | L | K | L | S | C | A | A | M | S | W | V | R | Q | N | L | Y | L | Q | L | N | A | L | Y | F | C | A | S | |||||||||||||||
| 2_2_2 | 1TF4 | G | T | F | R | Y | W | F | T | L | I | T | V | S | S | A | Y | D | L | Y | Y | V | E | I | D | C | R | R | E | V | Q | F | R | I | A | |||||||||
| 3_1_1 | 1CGT | Q | V | T | V | R | F | V | V | N | N | G | T | L | Y | L | T | G | N | V | W | Y | Y | D | V | S | V | K | Q | L | E | F | K | F | F | K | K | |||||||
| 6_1_1 | 1HOE | Y | S | Q | A | D | N | T | V | T | V | K | V | V | Y | Q | I | T | T | V | G | H | A | R | Y | L | A | R | C | |||||||||||||||
| 7_1_1 | 1AAC | D | T | V | T | W | I | N | V | H | F | V | A | Q | A | Y | S | L | T | F | G | T | Y | D | Y | H | C | |||||||||||||||||
| 8_1_1 | 1RLW | H | K | F | T | V | V | V | L | R | P | Y | V | E | L | F | I | P | V | W | N | E | T | F | E | F | I | V | L | E | I | T | L | M | ||||||||||
| 9_1_1 | 1CZY | T | H | L | S | L | F | F | K | V | T | L | M | L | L | L | F | C | P | V | A | I | F | I | K | A | I | |||||||||||||||||
| 11_1_4 | 1SVA | S | V | A | R | I | P | L | I | L | M | W | E | A | V | T | V | K | D | S | L | Y | V | S | A | R | Y | F | K | I | T | L | R | K | R | |||||||||
| 12_1_5 | 1G6E | V | N | W | V | E | S | V | T | L | Q | F | G | S | W | N | P | E | I | L | S | I | ||||||||||||||||||||||
| 13_1_1 | 1LOX | V | Y | R | V | C | V | S | T | K | V | E | L | W | L | V | G | E | E | E | F | K | V | N | L | L | F | V | R | L | R | K | K | |||||||||||
| 14_2_1 | 1PGS | V | K | T | I | K | M | F | I | K | N | Y | A | N | V | Y | V | K | L | E | I | D | V | G | N | T | E | L | K | I | Y | T | ||||||||||||
| 15_1_1 | 1DKV | T | F | L | V | G | L | I | G | F | G | I | Y | E | R | F | K | L | G | E | Y | I | L | V | P | S | ||||||||||||||||||
| 16_1_1 | 1JOB | A | E | S | V | Y | R | L | F | D | H | W | N | V | V | L | D | F | L | I | T | F | P | A | N | L | K | A | S | V | V | |||||||||||||
| 18_1_1 | 1SLU | M | K | R | Q | V | I | Q | L | L | K | V | E | L | L | I | P | I | V | V | Y | T | D | V | K | Y | R | V | W | K | ||||||||||||||
| 19_1_1 | 1FUX | L | T | W | S | K | S | F | A | V | T | V | Y | T | Y | L | P | V | Y | Q | F | K | V | W | A | L | ||||||||||||||||||
| 20_1_1 | 1G13 | G | N | V | T | L | S | V | — | G | K | V | D | L | V | L | E | K | E | G | T | Y | S | L | P | K | S | E | F | V | V | S | L | V | S | E | I | R | Y | N | G | |||
| 21-1-1 | 1I9B | N | E | V | D | V | V | F | D | L | A | A | Y | P | S | I | R | Q | R | F | S | C | A | T | C | R | I | K | I | G | S | W | ||||||||||||
| 22_1_1 | 1BHG | T | V | V | L | R | I | Y | A | I | V | W | V | F | E | A | D | I | R | L | R | I | T | I | A | I | ||||||||||||||||||
| 23-1-1 | 1BVP | A | R | G | D | V | M | M | I | Y | L | L | H | V | H | N | Q | V | V | F | Y | |||||||||||||||||||||||
| 24_1_1 | 1AOL | V | L | T | P | D | L | F | Y | V | C | N | P | L | A | I | H | Y | W | G | L | R | ||||||||||||||||||||||
| 26_1_1 | 1ALY | Y | I | Y | A | Q | V | T | F | C | S | F | I | A | S | L | C | L | G | Q | S | I | H | L | G | G | S | V | F | V | N | V | T | D | ||||||||||
| 27_1_1 | 1SPP | T | E | C | V | W | T | L | Q | K | L | L | V | S | I | P | T | M | T | V | K | Y | K | R | E | I | I | F | L | R | D | |||||||||||||
| 28_1_1 | 1CB8 | M | V | Q | A | I | F | Y | T | G | I | E | I | E | T | D | C | A | V | L | I | K | H | K | T | A | V | L | S | I | R | D | L | |||||||||||
| 29_1_1 | 1IAZ | R | K | I | A | V | G | V | D | N | T | W | T | A | L | N | T | Y | A | L | L | Y | N | G | Q | K | V | G | V | L | A | Y | L | M | S | |||||||||
| 30_1_1 | 1DU5 | N | L | D | F | F | D | I | S | P | M | S | F | L | Y | S | T | F | T | C | P | A | N | Y | K | V | V | F | C | |||||||||||||||
| 31_1_1 | 1YGS | V | W | V | R | C | L | S | V | F | Q | S | A | Y | I | K | V | F | L | R | M | S | F | |||||||||||||||||||||
| 33_1_1 | 1P35 | V | L | M | M | F | N | I | G | P | I | R | S | V | Y | Y | V | A | V | C | V | L | L | S | F | E | Y | N | ||||||||||||||||
| 34_1_1 | 1NLS | E | W | V | R | V | G | L | I | L | S | W | S | F | T | S | K | L | G | R | A | L | F | Y | A | V | V | A | S | F | E | A | T | F | T | |||||||||
| 36_1_1 | 1HS6 | T | Y | T | A | E | V | S | V | S | L | V | A | L | M | S | K | I | Y | K | F | I | Q | I | A | L | V | V | G | |||||||||||||||
| 38_1_1 | 1QEX | S | V | D | I | P | L | F | H | M | A | K | L | L | V | T | C | Q | S | Y | V | T | A | T | I | S | M | R | A | A | V | K | V | A | ||||||||||
Fold, the proteins are classified as in SCOP database (release 1.59), i.e., with three numbers that identify the protein in SCOP: protein fold, superfamily, and family, respectively. Str, Protein Data Bank codes of the structure (24). The residues at the conserved hydrophobic positions are boldface. Each vertical column in the table, starting with the third one, corresponds to a particular position in one of the four (i, i + 1, k, and k + 1) strands.
Sequence Characteristics of the i, i + 1, k, and k + 1 Strands.
Analysis of the structurally aligned sequences in this common system of numbering revealed that in the i strands, only positions 6–8 are always occupied in all known SPs. This finding means that residues found at position 6 of SP sequences all share similar structural characteristics. The same applies to residues at positions 7 and 8. In the other three strands of the invariant substructure, the following positions are structurally similar to residues in all SP structures: in i + 1 strands, positions 4–6; in k strands, positions 8–10; and in k + 1 strands, positions 6–8. These 12 positions are occupied by residues with structurally similar properties in their respective SP structures. According to the definition above, these residues are the structural determinants of the interlocked pairs. The structural determinants lie at the center of the interface between the β-sheets and form the common geometrical core of SP structures.
Inspection of amino acid frequencies in these 12 positions showed that two of three positions in each strand are the conserved hydrophobic positions of SP: positions 6 and 8 in i strands, 4 and 6 in i + 1 strands, 8 and 10 in k strands, and 6 and 8 in k + 1 strands. They are occupied by either aliphatic (A, V, L, and I), aromatic (W, Y, and F) or nonpolar residues (M and C). Residues at these eight positions are termed the SP sequence determinants. Eighty percent of all SP sequence determinants are V, L, I, and F residues.
Structural Features of the Sequence Determinants.
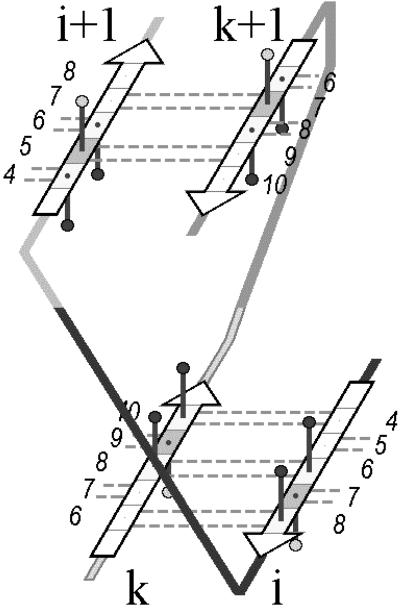
Relative positions of the eight sequence determinants in i, i + 1, k, and k + 1 strands are common in all SP structures that contain interlocked pairs (Fig. 3). The strands i and k are oriented toward each other in such a way that residues at position 6 in strand i always form main-chain hydrogen bonds with residues at position 8 in strand k, and side chains of both these residues seem to be located within the hydrophobic interior of SPs. The orientation of strands i + 1 and k + 1 is also fixed: residues at position 6 in i + 1 and position 8 in k + 1 strands are located opposite of each other in the sheet, but do not form H bonds. Side chains of both residues look inside the hydrophobic interior of SPs. This observation can be accounted for by a well known stereochemical relationship between directions of strands, interstrand H bonds, and the direction of residues' side chains. Relative positions of the SP sequence determinants in these four strands are shown schematically in Fig. 3.
Fig 3.
Relative positions and orientations of the key triplets of residues in two interlocked pairs of strands. Each of four triplets contains two conserved residues contributing to the hydrophobic interior and one nonconserved residue between them that seem to be located outside the core. The well seen shift of core-forming sites (shaded triplets) within each of the sheets results from a usual right-handed twist of β-sheets (14).
Conclusion
Commonly used approaches to uncovering common sequence features on the basis of sequence homology cannot be used when dealing with a group of such different proteins as SPs. Meanwhile, it is reasonable to suppose that proteins grouped together on the basis of common architecture would, on close scrutiny, reveal some commonality on the level of supersecondary and primary structure as well. The knowledge of the structural and sequence determinants of such seemingly dissimilar sequences arising from different ancestors would shed light on the most important question as to how structure is defined from sequence and, in perspective, would allow one to classify proteins with no known homologs and predict their known tertiary structure.
Acknowledgments
We are grateful to Dr. C. Chothia for very important and stimulating discussions, and Drs. I. Kister, P. Ehrlich, and Yu. Vasiliev for critical review of the manuscript. We thank L. Pogost for performing computer calculation, and N. S. Bogatyreva for drawing some of the figures. I.M.G. and A.E.K. thank M. Goldman for continuous encouragement. A.V.F. is supported by an International Research Scholar's Award from the Howard Hughes Medical Institute and the Russian Foundation for Basic Research.
Abbreviations
SP, sandwich-like protein
The interlocked pairs were not found in the structures of the superfamily neurophysin II (fold 10), the families killer toxin-like protein SKLP, plant antimicrobial protein MIAMP1 (fold 12), the superfamily HSP20-like chaperones (fold 17), the superfamily ecotin, trypsin inhibitor (fold 18), the family coagulation factor C2 domain (fold 22), the family adenovirus fiber protein head domain (knob domain; fold 25), and structures of several other domains.
References
- 1.Murzin A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995) J. Mol. Biol. 247, 536-540. [DOI] [PubMed] [Google Scholar]
- 2.Chothia C. & Finkelstein, A. V. (1990) Annu. Rev. Biochem. 57, 1007-1039. [DOI] [PubMed] [Google Scholar]
- 3.Woolfson D. N., Evans, P. A., Hutchinson, E. G. & Thornton, J. M. (1993) Protein Eng. 6, 461-470. [DOI] [PubMed] [Google Scholar]
- 4.Yee D. P. & Dill, K. A. (1993) Protein Sci. 2, 884-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chothia C., Hubbard, T., Brenner, S., Barns, H. & Murzin, A. (1997) Annu. Rev. Biophys. Biomol. Struct. 26, 597-627. [DOI] [PubMed] [Google Scholar]
- 6.Salem G. M., Hutchinson, E. G., Orengo, C. A. & Thornton, J. M. (1999) J. Mol. Biol. 287, 969-981. [DOI] [PubMed] [Google Scholar]
- 7.Efimov A. V. (1998) FEBS Lett. 437, 246-250. [DOI] [PubMed] [Google Scholar]
- 8.Durbin R., Eddy, S., Krogh, A. & Mitchison, G., (1998) Biological Sequence Analysis: Probabilisitic Models of Proteins and Nucleic Acids (Cambridge Univ. Press, Cambridge, U.K.).
- 9.Schaffer A. A., Aravind, L., Madden, T. L., Shavirin, S., Spouge, J. L., Wolf, Y. I., Koonin, E. V. & Altschul, S. F. (2001) Nucleic Acids Res. 29, 2994-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson J. (1977) Nature 268, 495-500. [DOI] [PubMed] [Google Scholar]
- 11.Sternberg M. J. E. & Thornton, J. M. (1976) J. Mol. Biol. 105, 367-382. [DOI] [PubMed] [Google Scholar]
- 12.Efimov A. V. (1982) Mol. Biol. (Moscow) 16, 799-806. [PubMed] [Google Scholar]
- 13.Chirgadze Yu. N. (1987) Acta Crystallogr. A 43, 405-417. [Google Scholar]
- 14.Chothia C. (1984) Annu. Rev. Biochem. 53, 537-572. [DOI] [PubMed] [Google Scholar]
- 15.Gelfand I. M. & Kister, A. E. (1995) Proc. Natl. Acad. Sci. USA 92, 10884-10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chothia C., Gelfand, I. M. & Kister, A. E. (1998) J. Mol. Biol. 278, 457-479. [DOI] [PubMed] [Google Scholar]
- 17.Kister A. E., Roytberg, M. A., Chothia, C., Vasiliev, Yu. M. & Gelfand, I. M. (2001) Protein Sci. 10, 1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ptitsyn O. B. & Ting, K. L. (1999) J. Mol. Biol. 291, 671-682. [DOI] [PubMed] [Google Scholar]
- 19.Mirny L. A. & Shakhnovich, E. I. (1999) J. Mol. Biol. 291, 177-196. [DOI] [PubMed] [Google Scholar]
- 20.Friedberg I. & Margalit, H. (2002) Protein Sci. 11, 350-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W. W., Reddy, B. V., Tate, J. G., Shindyalov, I. N. & Bourne, P. E. (2002) Nucleic Acids Res. 30, 409-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskowski R. A. (2001) Nucleic Acids Res. 29, 221-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 24.Berman H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. N. (2000) Nucleic Acids Res. 28, 235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]