Abstract
Exposure to mating pheromone in haploid Saccharomyces cerevisiae cells results in the arrest of the cell cycle, expression of mating-specific genes, and polarized growth toward the mating partner. Proteins involved in signaling, polarization, cell adhesion, and fusion are localized to the tip of the mating cell (shmoo) where fusion will eventually occur. The mechanisms ensuring the correct targeting and retention of these proteins are poorly understood. Here we show that in pheromone-treated cells, a reorganization of the plasma membrane involving lipid rafts results in the retention of proteins at the tip of the mating projection, segregated from the rest of the membrane. Sphingolipid and ergosterol biosynthetic mutants fail to polarize proteins to the tip of the shmoo and are deficient in mating. Our results show that membrane microdomain clustering at the mating projection is involved in the generation and maintenance of polarity during mating.
Virtually all cell types generate asymmetries of one type or another at their plasma membrane. Epithelial cells establish junctional complexes between adjacent cells that function as barriers to prevent mixing of apical and basolateral membrane components (1). In other contexts, such as cell migration and immunological synapse formation, cells are able to generate and maintain highly polarized phenotypes without making use of permanent diffusion barriers (2, 3). In both cases membrane microdomains known as lipid rafts are a fundamental component of the polarization process. Lipid rafts are thought to be formed by the tight packing of the long and highly saturated acyl chains of sphingolipids together with sterols (4). They form platforms for polarized protein delivery and membrane compartmentalization (5). Different proteins [e.g., glycosylphosphatidylinositol (GPI)-anchored protein] specifically associate with lipid rafts and are thus sorted or retained in a polarized fashion.
Budding yeast Saccharomyces cerevisiae exhibits different types of cell polarity. During budding, growth is restricted to a zone adjacent to the previous budding site (haploids) or at either pole of the cell (diploids) (6). First, a hierarchy of positional signals defines the bud site (6). Then growth is focused there and restricted to the new bud by a diffusion barrier made of septins that is placed as a collar at the neck between mother and daughter cell (6–10). Yeast also exhibit polarized growth during mating. Binding of pheromone secreted by cells of the opposite mating type to specific membrane receptors results in the stimulation of a mitogen-activated protein kinase signaling cascade (11, 12) and polarized growth toward the mating partner (13). Pheromone signaling leads to the arrest of the cell cycle, induction of mating-specific genes, and the recruitment of signaling, polarity establishment, and cell adhesion proteins to the site of growth (14–17). Polarized growth leads to the formation of a mating projection toward the mating partner, thus bringing the cells in direct contact (18). Then, after the cell wall in the contact zone between both cells is removed, fusion factors promote fusion of the cells. Several fusion mutants have been isolated (19–22). Interestingly, most of the proteins required for cell fusion have been shown to be localized to the mating projection. However, little is known about the mechanisms responsible for the polarization of these proteins. Previously, we have demonstrated that ergosterol-sphingolipid rafts are required for the correct sorting of the major plasma membrane +H-ATPase Pma1p (23, 24). In this work, we show that lipid rafts are clustered at the tip of the mating projection. Proteins destined to the mating projection partition into lipid rafts and are thus retained and segregated from the rest of the membrane.
Materials and Methods
Strains and Plasmids.
The following strains are derived from RH690-15D (a his4 ura3 leu2 lys2 bar1) (23) and were generated by fusion of the indicated genes with the coding sequence of the IgG-binding domain of Staphylococcus aureus protein A (PA) as described (25): MBY205 (YPL176c), MBY206 (FUS1), MBY211 (FIG 1), MBY212 (FUS2), MBY213 (STE6), MBY214 (SHO1), MBY222 (PRM1), MBY242 (HXT2). RH3804 (23), RH3622 (26), CKY698 (27), AAY1017 (28), and 2239 (28) have been described. RH690–13B (a lcb1–100 lcb3:KanMx his4 ura3 leu2 lys2 bar1) was obtained from H. Riezman (University of Basel, Basel), and 2339 (a ura3 leu2 kar1-1) was obtained from M. Rose (Princeton University, Princeton). FUS1-GFP chromosomal fusions in RH690-15D (MBY229) and CKY698 (MBY1202) were done with a PCR-based method as described (25) by using pMBQ41 as template. FUS1-PA fusions in RH3622 (MBY240) and RH690-13B (MBY223) were done as indicated above. MBY245–2c (a lcb1–100 ura3 leu2 lys2 bar1) was generated by using standard techniques (28). To construct pMBQ30 and pMBQ32, FUS1 and YPL176c, respectively, were amplified by PCR with XbaI/BamHI (FUS1) or XbaI/EcoRI (YPL176c) sites at the ends and ligated with GFP (flanked by BamHI and HindIII sites) into p416 (29). pMBQ31 was constructed by replacing the central portion (300–1,260) of GAS1 with GFP and ligated into p416. pMBQ41 was generated by replacing PA in pBS1365 (PA-tagging cassette) with GFP. All yeast media were prepared as described (28). Gap1-GFP was expressed from pCK230 (27).
Lipid Analysis.
Cells were grown in yeast extract/peptone/dextrose (YPD) to OD600 = 1 and then treated with α-factor (5 μM) for 3 h. Then cells were lysed as described (24), and half of the sample was extracted with 1% Triton X-100 (TX100) or 1% 3-[(3-cholamidopropyl)dimethyl-ammonio]1-propane sulfonate (CHAPS) at 4°C for 30 min and the rest was incubated only with buffer. After Optiprep density gradient centrifugation, lipids were extracted from the upper fraction and ergosterol was quantified as described (23). The percentage of detergent-resistant membrane (DRM)-associated ergosterol corresponds to the ratio of detergent resistant vs. total ergosterol in membranes. To determine the pattern of DRM-associated phospholipids, cells were labeled with [32P]orthophosphate with or without the addition of α-factor for 3 h. Cells were lysed, and the fraction of the indicated lipid species in DRM (in CHAPS) was determined as described (23).
Microscopy.
Except for the rhodamine–phalloidin staining, cells were imaged live in synthetic medium. For visualization of sterol-rich domains, cells were incubated in complete synthetic medium with filipin (9 μg/ml prepared in DMSO) for 15 min at room temperature. Then cells were washed three times in medium and mounted for microscopy in the same medium. Images were captured in an Olympus BX61 microscope equipped with a charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI) with a ×100 lens.
DRM Association, Immunoprecipitation, and Immunobloting.
Western blotting and immunoprecipitation of PA-tagged proteins were done as described (25). Fus1-GFP and Gap1-GFP were immunoprecipitated as described (24) by using anti-GFP-specific Abs raised in our lab. DRM association was analyzed by using TX100 or CHAPS as described (24).
Results
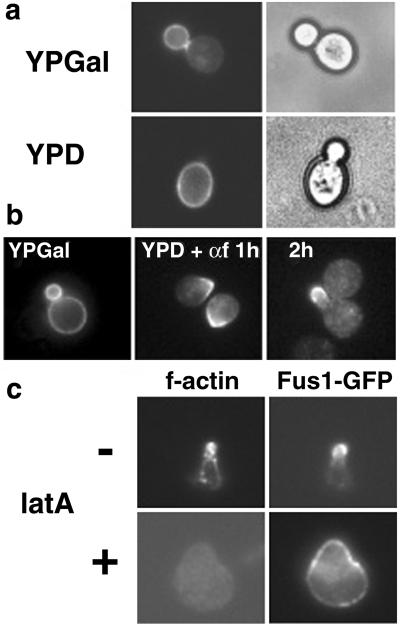
Fus1p is a type I membrane protein localized to the tip of mating projections and is required for cell fusion (19, 30). To investigate how the polarized localization is achieved, we expressed Fus1 tagged with GFP (Fus1-GFP) under the control of a truncated (weak) version of the inducible GAL1 promoter (GALS) (29). On induction of expression in vegetative growing cells, Fus1-GFP fluorescence was concentrated in the bud with little signal in the mother cell (Fig. 1a). After turning off protein production, the pool of Fus1-GFP already present in the membrane did not move into the plasma membrane of the new daughter cell (Fig. 1b). This result indicates that diffusion of Fus1-GFP along the membrane plane between mother and bud was prevented as also shown before for GFP-Ist2p (9). When α-factor was added after Fus1-GFP synthesis had been turned off, cell surface Fus1-GFP polarized toward the presumptive mating-projection site after 1 h and localized to the tip of the process after 2 h (Fig. 1b). This result shows that on addition of pheromone, polarization of Fus1-GFP happened in the absence of polarized secretion. Consistent with previous results (31), the maintenance of this polarization was actin-dependent. After complete disruption of the actin cytoskeleton, Fus1-GFP was localized in dots that were distributed along the entire surface of the cell (Fig. 1c).
Fig 1.
Reorganization of the plasma membrane in α-factor-treated cells. (a) WT cells (RH690-15D) were grown in YPD at 30°C and shifted to YPGal to induce expression Fus1-GFP under the control of the GALS (29) promoter (pMBQ30). Then cells were shifted to YPD for 2 h to turn off Fus1-GFP expression. (b) Fus1-GFP was expressed as in a, and cells were shifted to YPD for 1 h and then treated with α-factor (3 μM) for the indicated times. (c) Actin-dependent localization of Fus1-GFP. Cells were processed as in b and after 2 h of α-factor (3 μM) treatment, and latrunculin A (latA) was added for 1 h more. Cells were fixed and f-actin stained with rhodamine-phalloidin.
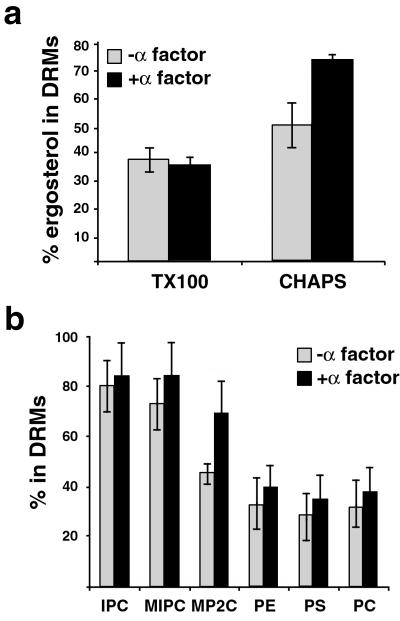
The highly polarized structure of the mating projection is reminiscent of the mammalian immunological synapse (3). On activation the T cell receptor becomes detergent resistant. A cluster of proteins and lipid rafts are recruited to the site of contact with the antigen-presenting cell whereas detergent-soluble proteins are excluded (5). Similar to yeast mating cells, the stability of the ensemble is kept for several hours by an actin scaffold. To analyze whether lipid rafts also are involved in yeast mating, we first investigated the biosynthetic levels of ergosterol, phospholipids, and sphingolipids by labeling control and α-factor-treated cells with [14C]acetate or [32P]orthophosphate. Although synthesis of phospholipids remained unchanged, a 30% increase for ergosterol biosynthesis was observed (data not shown). Then we analyzed the DRM association of these lipids in TX100 and CHAPS. Interestingly, although the amount of TX100-insoluble ergosterol did not change upon α-factor treatment, a 50% increase in CHAPS-insoluble ergosterol was found in α-factor-treated cells (Fig. 2a). Next, we checked the behavior of [32P]orthophosphate-labeled glycerophospholipids and sphingolipids in α-factor-treated cells by using CHAPS. Similar to the results with TX100, all sphingolipid species were mostly CHAPS-resistant (≈80%) in α-factor-treated and control cells (Fig. 2b). Interestingly, mannose-(inositol phosphorus)2-ceramide (MP2C) DRM association was increased by ≈50% after α-factor treatment. In contrast, glycerophospholipids were mostly soluble, although to a lesser extent than in TX100. Therefore we decided to use CHAPS for studying the role of lipid rafts in mating.
Fig 2.
(a) DRM association of ergosterol in vegetative (−α-factor) and pheromone-treated (+α-factor) cells. The percentage of DRM-associated ergosterol corresponds to ratio of detergent resistant vs. total ergosterol in membranes. The values correspond to the means of three experiments with SD. (b) To determine the pattern of DRM-associated phopholipids, cells were labeled with [32P]orthophosphate with or without the addition of α-factor for 3 h. Cells were lysed, and the fraction of the indicated lipid species in DRM (in CHAPS) was determined as described (23). IPC, inositol-phosphorylceramide; MIPC, mannose-inositol-phosphorylceramide; MP2C, mannose-(inositol phosphorus)2-ceramide; PE, phosphatidylethanolamine; PS, phosphatidylserine; PC, phosphatidylcholine (n = 3). In ref. 23 PS and # (symbol shown in ref. 23 figures) were swapped.
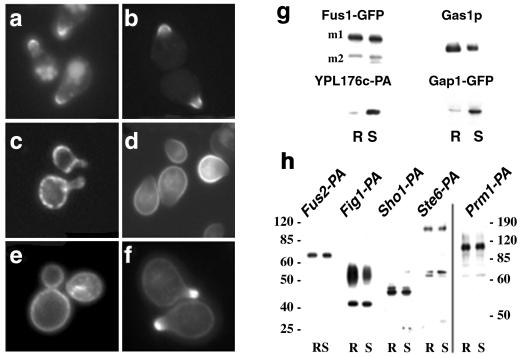
Next, we investigated whether Fus1-GFP associated with DRMs. Approximately 50% of Fus1-GFP was immunoprecipitated from the detergent-resistant fraction (R) after density gradient centrifugation (Fig. 3g). Similar to Fus1-GFP, GFP-GPI, a Gas1-derived marker for DRMs (23), also concentrated at the tip of the mating projection (Fig. 3b), even though the protein was expressed before the addition of pheromone. Next, we investigated the fate of detergent-soluble proteins. We have previously demonstrated that human transferrin receptor does not associate with DRMs when expressed in yeast (23). A GFP fusion of yeast transferrin receptor-like YPL176c (YPL176-GFP) localized in a punctate pattern along the surface of the body of the cell, mostly excluded from the shmoo tip (Fig. 3c), and also was detergent-soluble (Fig. 3g). We also examined the amino acid permease Gap1. To prevent vacuolar targeting of Gap1-GFP, we used a bul1Δ bul2Δ mutant strain in which the protein is efficiently targeted to the surface (27). In contrast to GFP-GPI, Gap1-GFP was localized to the surface membrane around the body of the cell and was excluded from the tip of the shmoo (Fig. 3d); Fus1-GFP, and GFP-GPI localization in this background remained polarized as in WT cells (data not shown). Importantly, most (88%) of Gap1-GFP was found in the detergent-soluble (S) fraction (Fig. 3g). These data suggest that a lipid raft-dependant mechanism regulates the segregation of different proteins at the surface of α-factor-polarized cells. Next, we examined the distribution of sterols at the plasma membrane by using filipin. In unpermeabilized control cells, filipin stained more or less evenly the plasma membrane (Fig. 3e). In contrast, in α-factor-treated cells sterols were concentrated at the tip of the shmoo (Fig. 3f), indicating that sterol-rich domains are clustered there. Next we investigated whether other membrane proteins known to be localized to the mating projection also would be DRM-associated. Mating projection-localized Fus2 (32), Fig1 (22), Sho1 (33), Ste6 (34), and Prm1 (21) all were found to be significantly detergent-resistant (Fig. 3h).
Fig 3.
Shmoo tip-targeted proteins become DRM-associated. (a) Fus1-GFP localized to the tip of shmoo in WT cells (MBY229) after α-factor (3 μM) induction. A pool of Fus1-GFP localized to the vacuole, probably because of turnover (see Fig. 5). (b) Gas1-derived GFP-GPI (RH690-15D[pMBQ31]). (c) Transferrin receptor-like YPL176c-GFP (RH690-15D). (d) Gap1-GFP (pCK230) in bul1Δ bul2Δ mutant cells (CKY698). Fus1-GFP and GFP-GPI were localized as in WT cells in bul1Δ bul2Δ mutant cells not (not shown). (e) Sterol distribution in control and α-factor polarized cells (f) was visualized after incubation with filipin (9 μg/ml) for 15 min. (g) Cells (MBY229 and CKY698[pCK230]) were preincubated with 5 μM α-factor for 15 min and pulse-labeled with [35S]methionine for 15 min, and after a 45-min chase, the cells were lysed. After extraction with 1% CHAPS and Optiprep density gradient centrifugation, Fus1-GFP and Ga1p-GFP were immunoprecipitated from detergent-resistant (R) and soluble (S) fractions and analyzed by SDS/PAGE and phosphorimaging (m1 and m2 are two mature products of Fus1-GFP). YPL176c-PA (MBY205) and Gas1p (MBY229) DRM-association was analyzed by immunoblotting. Gas1p and Fus1-GFP remained DRM associated as in WT cells in the bul1Δ bul2Δ background (not shown). (h) Fus2, Fig1, Sho1, St6, and Prm1 were tagged with the IgG-binding domain of PA and processed as before and detected by immunoblotting. At least 30% of the protein floated after density gradient centrifugation whereas the bulk of YPL176c-PA remained soluble under the same conditions. The molecular weight is indicated.
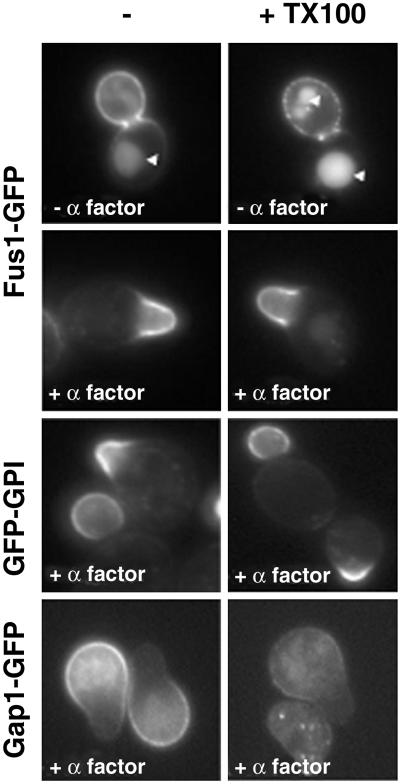
We next addressed the issue of whether the shmoo tip-localized markers are detergent resistant in situ. To do so we treated cells expressing Fus1-GFP with cold TX100 (35). After treatment with detergent, Fus1-GFP remained on the cell surface in vegetative growing cells. The staining became patchy, indicating that the detergent-resistant pool of Fus1-GFP aggregated when the detergent-sensitive membrane was extracted. When the experiment was performed in α-factor-polarized cells, Fus1-GFP remained as a continuous detergent-resistant domain at the tip of the shmoo (Fig. 4). Consistent with this finding, GFP-GPI also remained resistant to extraction with cold detergent. In contrast, Gap1-GFP was solubilized. These data, together with the concentration of sterols at the tip of the shmoo, demonstrate that lipid rafts are clustered at the tip of mating projections.
Fig 4.
DRMs are clustered at the tip the mating projections. The distribution of DRM-associated Fus1-GFP was investigated by treating the cells with cold TX100 (1%) for 30 min. In budding cells, Fus1-GFP was partially extractable with cold detergent; the DRM-associated pool was found in patches along the entire surface of the bud. The arrowheads mark the vacuole. In contrast, in α-factor-treated cells Fus1-GFP remained as a continuous detergent-resistant domain at the tip of shmoos. Gap1-GFP, but not GFP-GPI, was sensitive to extraction in α-factor-treated cells.
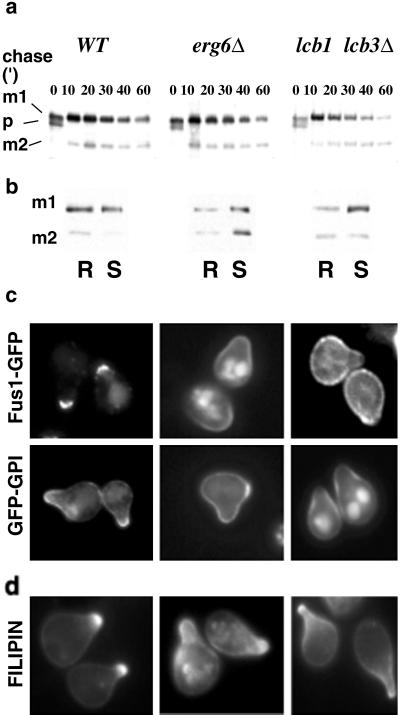
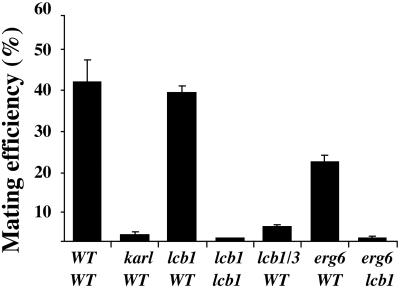
To further analyze the possible role of lipid rafts in shmoo polarization, we used the erg6Δ and lcb1–100 lcb3Δ lipid mutants. The erg6Δ mutant, which cannot make ergosterol, is defective in endocytosis (26) and, interestingly, mates with reduced efficiency (36). The lcb1–100 lcb3Δ double mutant has reduced sphingolipid levels at 24°C (shows a total biosynthetic block at 37°C) and also is impaired in endocytosis at 37°C (37). To examine the maturation of PA-tagged Fus1 (Fus1-PA) in WT and mutant cells, we performed a pulse–chase experiment. In WT cells, at the beginning of the chase Fus1-PA is present as a precursor (p), endoplasmic reticulum (ER) form (30), and a more glycosylated (m1), Golgi form (30) that is further processed to yield an additional mature form (m2) (Fig. 5a). The protein was expressed and processed similarly in mutant cells (Fig. 5a), indicating that pheromone signaling and induction of mating-specific genes were normal and, that no defect in ER to Golgi transport of Fus1-PA occurred at 24°C. Under these conditions, Fus1-PA DRM association was significantly reduced in both mutants compared to WT cells (Fig. 5b). We next analyzed the localization of Fus1-GFP and GFP-GPI. In WT cells, both Fus1-GFP and GFP-GPI were found in the tip of the projection in most (94% and 78%, respectively) of the cells (Fig. 5c). In contrast, in erg6Δ, only a small fraction of the cells showed a polarized localization of the markers (30% for Fus1-GFP and 23% for GFP-GPI). Fus1-GFP was localized in a very broad region of the shmoo or randomly distributed in the entire surface (40% and 30% of the cells, respectively); the Fus1-GFP signal in the vacuole was also more prominent (Fig. 5c). This defect was even larger for the lcb1–100 lcb3Δ double mutant; only 21% for Fus1-GFP and 5% for GFP-GPI of the cells showed polarized distribution of the markers (Fig. 5c). This polarization defect was also present at the membrane lipid level because sterols also were distributed in a less polarized fashion than in WT cells (Fig. 5d). These results indicate that in permissive conditions (24°C, YPD, 3 μM α-factor) the mutants still formed shmoos with normal frequency (data not shown), expressed Fus1, and exhibited a polarized actin cytoskeleton (data not shown); however, they could not retain the marker proteins Fus1-GFP and GFP-GPI in the tip of the projection (Fig. 5). The clustering of mating-specific proteins to the fusion zone suggests that their function in cell fusion may depend on their effective concentration in the region of cell-to-cell contact. Therefore, we decided to test the effect of different lipid biosynthetic mutants on mating efficiency by using quantitative mating assays (19). A requirement for sterols in mating has been shown (38). The mating efficiency of erg6Δ cells with a WT partner was reduced 2-fold compared to WT cells (Fig. 6). Interestingly, when erg6Δ cells were mated to an lcb1–100 partner that did not show any defect when mated to a WT strain the mating efficiency dropped >80-fold. A similar bilateral mating defect was observed when both partners had the lcb1–100 allele. Finally, the mating efficiency of lcb1–100 lcb3Δ cells with a WT partner was reduced 12-fold. Thus, mutants with reduced levels of DRM-associated lipids (23) showed impaired mating although conditions were permissive for other aspects of the pheromone response.
Fig 5.
Processing and sorting of shmoo-tip markers in lipid biosynthetic mutants. (a) WT (MBY206), erg6Δ (MBY240), and lcb1–100 lcb3Δ (MBY223) cells were preincubated with 5 μM α-factor for 15 min, pulse-labeled with [35S]methionine for 5 min, and chased for various times at 24°C. Fus1-PA maturation was analyzed by immunoprecipitation, SDS/PAGE, and autoradiography. The position of precursor (p) and to mature products (m1 and m2) is indicated. (b) DRM association of Fus1-PA in WT (MBY206), erg6Δ (MBY240), and lcb1–100 lcb3Δ (MBY223) cells was analyzed as in Fig. 2. (c) Localization of Fus1-GFP (pMBQ30) and GFP-GPI (pMBQ31) in WT (RH690-15D), erg6Δ (RH3622), and lcb1–100 lcb3Δ (RH690-13B) cells. Cells were grown on YPGal and treated with 3 μM α-factor for 3 h. (d) Sterol distribution.
Fig 6.
Lipid biosynthetic mutants have reduced mating efficiency. Quantitative matings were performed at 24°C for 3 h as described (19). The mating efficiency (diploids/total cells) is given as the means of three experiments with SD. AAY1017 was used as a WT tester. Mating mutant kar1-1 strain (2339) was used as a control.
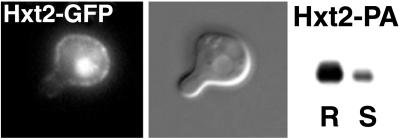
Altogether our results suggest that polarized localization of proteins to the mating projection uses lipid rafts as a platform for membrane segregation. Yet, an additional sorting step between different types of raft-associated proteins is necessary. Although all of the mating projection-localized membrane proteins that we tested were DRM-associated (Fig. 3), not all DRM-associated proteins are retained in the mating projection. The glucose transporter Hxt2 was resistant to detergent extraction but is nevertheless excluded from the tip of the shmoo (Fig. 7).
Fig 7.
DRM-associated Hxt2 is excluded from the tip of the shmoo. Hxt2-GFP (MBY238) and Hxt2-PA (MBY242) were induced by shifting the cells to low (0.1%) dextrose. Cells were treated with 3 μM α-factor for 3 h and processed as in Fig. 3. Most of Hxt2-PA was found in the DRM fraction (R).
Discussion
In this study, we have investigated the process of polarization during yeast mating. Altogether our results show that on exposure to pheromone the plasma membrane of yeast undergoes a process of reorganization that results in the clustering of lipid rafts at the mating projection. Proteins are destined to the tip of the mating projection partition into lipid rafts and become thus polarized at the cell surface.
In the mating response, cells polarize toward a mating partner with which they will fuse. Polarization and fusion require the correct targeting of specific proteins to the tip of the mating projections. Our results show that lipid rafts are required for the segregation of different domains at the surface of mating cells. The compartmentalization of raft and non-raft membrane domains could start as early as the endoplasmic reticulum (ER) because raft association starts there (23) and Gap1p and GPI-anchored Gas1p are sorted into different populations of ER to Golgi vesicles (39). The process of polarization of the plasma membrane during mating is illustrated by the dramatic change in the distribution of sterol-rich domains on exposure to pheromone (Fig. 3 e and f). Importantly, the tip of the mating projection appears to be a cluster of DRMs. Treatment of cells with cold TX100 reveals that Fus1-GFP-containing DRMs are interspersed at the surface of budding cells with detergent-soluble membrane domains (Fig. 4). In contrast, in mating cells DRMs form a highly polarized cluster at the tip of the mating projections that seems to be continuous (Fig. 4). Although polarized secretion is a major driving force for the localization of proteins to the mating projection, several lines of evidence indicate that additional mechanisms are involved. Previously expressed and surface localized Fus1-GFP becomes polarized on addition of pheromone in conditions that prevent new synthesis (Fig. 1b). Moreover, in lipid biosynthetic mutants that affect lipid raft function, Fus1-GFP and GFP-GPI are secreted in a polarized fashion but cannot be retained at the tip of the mating projection (Fig. 5). This is not the result of a defect in endocytosis or actin polarity because the experiments were done in permissive conditions in which both endocytosis and actin polarity were not significantly affected. In addition, several different endocytosis mutants do not show mating defects (20). Nevertheless, we cannot rule out some role for endocytosis and recycling in the polarization of the membrane during mating.
Polarized distribution of membrane proteins during mating differs from budding in the sense that the polarization of the cell is not relying mainly on polarized secretion and the establishment of a diffusion barrier. In mating cells, proteins destined to the tip of the shmoo are selectively retained by a lipid raft-dependent mechanism.
Ergosterol-sphingolipid rafts seem to be the major phase in the plasma membrane (24). This finding has to be reconciled with the fact that the mating projection where they cluster (Fig. 3f) accounts for only a small part of the cell surface. This issue is explained by the existence of different types of rafts; one type is clustered in the mating projection and contains shmoo tip-localized proteins; another type is not clustered and is intercalated with detergent-soluble membranes (Fig. 7). A similar phenomenon has been observed in migrating T cells where leading edge and uropod components segregate into specific rafts (40). The question of how different lipid rafts are segregated in α-factor-polarized cells remains open. We propose that specific clustering of proteins destined to the tip of the shmoo might mediate their separation from the rest of the plasma membrane. Further work is needed to identify the proteins mediating this clustering in the tip of the shmoo.
Acknowledgments
We thank H. Riezman, C. Kaiser, and B. Seraphin for strains and plasmids and C. Eroglu, S. Eaton, C. Walch-Solimena, and members of the Simons laboratory for comments and suggestions.
Abbreviations
GPI, glycosylphosphatidylinositol
CHAPS, 3-[(3-cholamidopropyl)dimethyl-ammonio]1-propane sulfonate
DRM, detergent-resistant membrane
PC, phosphatidylcholine
TX100, Triton X-100
PA, protein A
YPD, yeast extract/peptone/dextrose
References
- 1.van Meer G., Gumbiner, B. & Simons, K. (1986) Nature 322, 639-641. [DOI] [PubMed] [Google Scholar]
- 2.Manes S., Mira, E., Gomez-Mouton, C., Lacalle, R. A., Keller, P., Labrador, J. P. & Martinez, A. C. (1999) EMBO J. 18, 6211-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bromley S. K., Burack, W. R., Johnson, K. G., Somersalo, K., Sims, T. N., Sumen, C., Davis, M. M., Shaw, A. S., Allen, P. M. & Dustin, M. L. (2001) Annu. Rev. Immunol. 19, 375-396. [DOI] [PubMed] [Google Scholar]
- 4.Brown D. A. & London, E. (1998) Annu. Rev. Cell Dev. Biol. 14, 111-136. [DOI] [PubMed] [Google Scholar]
- 5.Simons K. & Toomre, D. (2000) Nat. Rev. Mol. Cell. Biol. 1, 31-39. [DOI] [PubMed] [Google Scholar]
- 6.Chant J. (1999) Annu. Rev. Cell Dev. Biol. 15, 365-391. [DOI] [PubMed] [Google Scholar]
- 7.Faty M., Fink, M. & Barral, Y. (2002) Curr. Genet. 41, 123-131. [DOI] [PubMed] [Google Scholar]
- 8.Barral Y., Mermall, V., Mooseker, M. S. & Snyder, M. (2000) Mol. Cell 5, 841-851. [DOI] [PubMed] [Google Scholar]
- 9.Takizawa P. A., DeRisi, J. L., Wilhelm, J. E. & Vale, R. D. (2000) Science 290, 341-344. [DOI] [PubMed] [Google Scholar]
- 10.Nern A. & Arkowitz, R. A. (2000) Mol. Cell 5, 853-864. [DOI] [PubMed] [Google Scholar]
- 11.Jackson C. L., Konopka, J. B. & Hartwell, L. H. (1991) Cell 67, 389-402. [DOI] [PubMed] [Google Scholar]
- 12.Elion E. A. (2001) J. Cell Sci. 114, 3967-3978. [DOI] [PubMed] [Google Scholar]
- 13.Jackson C. L. & Hartwell, L. H. (1990) Cell 63, 1039-1051. [DOI] [PubMed] [Google Scholar]
- 14.Madden K. & Snyder, M. (1998) Annu. Rev. Microbiol. 52, 687-744. [DOI] [PubMed] [Google Scholar]
- 15.Leberer E., Thomas, D. Y. & Whiteway, M. (1997) Curr. Opin. Genet. Dev. 7, 59-66. [DOI] [PubMed] [Google Scholar]
- 16.Chenevert J., Corrado, K., Bender, A., Pringle, J. & Herskowitz, I. (1992) Nature 356, 77-79. [DOI] [PubMed] [Google Scholar]
- 17.Guo B., Styles, C. A., Feng, Q. & Fink, G. R. (2000) Proc. Natl. Acad. Sci. USA 97, 12158-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson C. L. & Hartwell, L. H. (1990) Mol. Cell. Biol. 10, 2202-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trueheart J., Boeke, J. D. & Fink, G. R. (1987) Mol. Cell. Biol. 7, 2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brizzio V., Gammie, A. E. & Rose, M. D. (1998) J. Cell Biol. 141, 567-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heiman M. G. & Walter, P. (2000) J. Cell Biol. 151, 719-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erdman S., Lin, L., Malczynski, M. & Snyder, M. (1998) J. Cell Biol. 140, 461-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagnat M., Keranen, S., Shevchenko, A. & Simons, K. (2000) Proc. Natl. Acad. Sci. USA 97, 3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagnat M., Chang, A. & Simons, K. (2001) Mol. Biol. Cell 12, 4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puig O., Rutz, B., Luukkonen, B. G., Kandels-Lewis, S., Bragado-Nilsson, E. & Seraphin, B. (1998) Yeast 14, 1139-1146. [DOI] [PubMed] [Google Scholar]
- 26.Munn A. L., Heese-Peck, A., Stevenson, B. J., Pichler, H. & Riezman, H. (1999) Mol. Biol. Cell 10, 3943-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helliwell S. B., Losko, S. & Kaiser, C. A. (2001) J. Cell Biol. 153, 649-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burke D., Dawson, D. & Stearns, T., (2000) Methods in Yeast Genetics: A Laboratory Course Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 29.Mumberg D., Muller, R. & Funk, M. (1994) Nucleic Acids Res. 22, 5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trueheart J. & Fink, G. R. (1989) Proc. Natl. Acad. Sci. USA 86, 9916-9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayscough K. R. & Drubin, D. G. (1998) Curr. Biol. 8, 927-930. [DOI] [PubMed] [Google Scholar]
- 32.Elion E. A., Trueheart, J. & Fink, G. R. (1995) J. Cell Biol. 130, 1283-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raitt D. C., Posas, F. & Saito, H. (2000) EMBO J. 19, 4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuchler K., Dohlman, H. G. & Thorner, J. (1993) J. Cell Biol. 120, 1203-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seveau S., Eddy, R. J., Maxfield, F. R. & Pierini, L. M. (2001) Mol. Biol. Cell 12, 3550-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaber R. F., Copple, D. M., Kennedy, B. K., Vidal, M. & Bard, M. (1989) Mol. Cell. Biol. 9, 3447-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zanolari B., Friant, S., Funato, K., Sutterlin, C., Stevenson, B. J. & Riezman, H. (2000) EMBO J. 19, 2824-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomeo M. E., Fenner, G., Tove, S. R. & Parks, L. W. (1992) Yeast 8, 1015-1024. [DOI] [PubMed] [Google Scholar]
- 39.Muniz M., Morsomme, P. & Riezman, H. (2001) Cell 104, 313-320. [DOI] [PubMed] [Google Scholar]
- 40.Gomez-Mouton C., Abad, J. L., Mira, E., Lacalle, R. A., Gallardo, E., Jimenez-Baranda, S., Illa, I., Bernad, A., Manes, S. & Martinez, A. C. (2001) Proc. Natl. Acad. Sci. USA 98, 9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]