Abstract
Transport of macromolecules between the cell nucleus and cytoplasm occurs through the nuclear pores and is mediated by soluble carriers known as karyopherins (Kaps), transportins, importins, or exportins. We report that Kap β2B (transportin-2) forms complexes with the mRNA export factor TAP in the presence of RanGTP, as shown by coimmunoprecipitation from HeLa cells. The interaction strictly depends on the presence of RanGTP. In digitonin-permeabilized cells, Kap β2B mediates TAP-GFP export from the nuclei in the presence of RanGTP. A TAP mutant that does not coimmunoprecipitate with Kap β2B is also not exported by Kap β2B. In the permeabilized cells assay, TAP is also exported independently of Kap β2B by direct interaction with nucleoporins, in agreement with previous reports. The export rate is, however, significantly lower than the Kap β2B-mediated pathway. Both Kap β2B and TAP are present and enriched in the poly(A)+ RNA complexes isolated from HeLa cell nuclear lysates. Poly(A)+ RNA strongly accumulates in the nuclei of HeLa cells treated with Kap β2B short interfering RNA, indicating that Kap β2B is involved in the export of at least a large proportion of the mRNA species. The export of β-actin and GAPDH mRNA is also inhibited, whereas 28S RNA is not affected. The data support the conclusion that Kap β2B participates directly in the export of a large proportion of cellular mRNAs, and TAP connects Kap β2B to the mRNAs to be exported.
Trafficking of proteins and nucleic acids in or out of the cell nucleus occurs through the nuclear pores (1–5). Macromolecules below a certain size limit (40–60 kDa for proteins) can diffuse through the pores, whereas species above the size limit (and some smaller species) are transported by a complex system of soluble carriers generally known as karyopherins (Kaps), transportins, importins, or exportins (6–10). The carriers bind their cargoes either in the cytoplasm or in the nucleoplasm, dock them to components of the nuclear pore complexes, and assist their passage across the nuclear envelope. Most Kaps belong to the Kap β family and are characterized by the ability to bind the small GTPase Ran that regulates the interaction of Kaps with their cargoes (11, 12). Ran is present in the nucleus in the GTP-bound form and has opposite action on the import versus the export complexes. Ran-GTP induces release of the cargoes from the Kaps that mediated their import and facilitates binding of the export Kaps to the cargoes that are subsequently exported.
In mRNA export several conserved factors are required in both metazoans and yeast: Mex67 (the orthologue of the mammalian TAP/NXF1), Mtr2, Yra1, Sub2, Dbp5, Gle1, Gle2, members of the IP6 pathway, pre-mRNA-processing factors, and the metazoan counterparts (12–23). Among these proteins TAP plays a prominent role by mediating the translocation step across the nuclear pore of cellular and viral mRNAs by direct interaction with nucleoporins (24–30).
Contrary to most nucleocytoplasmic export pathways characterized to date, no Kap β family member was found to be directly involved in mRNA export. We previously identified and cloned Kap β2B (GenBank accession no. AF007748) on the basis of its very high sequence similarity with Kap β2A (transportin-1), which functions in nuclear import of heterogeneous ribonucleoproteins (31–33). However, it was established that Kap β2B does not function in this pathway (31). Kap β2B is ubiquitously expressed in mammalian tissues, as indicated by our Northern blot experiments (data not shown) and by expression information present in the Unigene database. Here we show that Kap β2B (transportin-2) (31) is a nuclear export factor and mediates mRNA export in cooperation with TAP.
Experimental Procedures
RanGAP Assay.
Kap β2A was purified as described (33). Full-length Kap β2B amplified by PCR from human brain cDNA (CLONTECH) was sequenced (GenBank accession no. AF007748), cloned in the BamHI/EcoRI sites of pGEX 4T-3(Tev) vector (Amersham Pharmacia), and expressed in Escherichia coli BL21(DE3) (Novagen). The GST moiety was cleaved off with Tev protease (Amersham Pharmacia). RanGAP1 (Schizosaccharomyces pombe Rna1p) and Ran were purified as described (34, 35). Wild-type Ran was loaded with [γ-32P]GTP (36) in the presence of 10 mM EDTA/20 mM Hepes-KOH, pH 7.3/110 mM potassium acetate/2 mM magnesium acetate/1 mM EGTA/2 mM DTT. RanGAP assay was conducted as described (36). Reaction mixtures containing 100 pM Ran[γ-32P]GTP were preincubated with 0–1,000 nM Kaps for 20 min on ice. The GTPase reactions were started by addition of Rna1p (40 nM) and immediately placed at 25°C for 15 min. Reactions were stopped by addition of 1 ml of charcoal suspension and centrifuged. The released γ-32P was measured by scintillation counting.
Coimmunoprecipitation.
HeLa cells were transfected by using Lipofectamine with the pCMV-Tag plasmid (Stratagene) from which the FLAG and myc sequences were removed, and the hemagglutinin (HA) tag fused to the N terminal of Kap β2B was introduced between the XhoI/SmaI sites. Cell lysates were prepared 48 h later and subjected to immunoprecipitation as described (37). In brief, the cells were lysed by using 1% Nonidet P-40, and Ran loaded with guanylyl-5′-imidodiphosphate (GMP-PNP) was added together with GMP-PNP to final concentrations of 2 μM and 1 mM, respectively. After 1 h of incubation at 4°C, the samples were incubated with protein G beads for 1 h at 4°C, and the retained proteins were analyzed by SDS/PAGE followed by immunoblotting with anti-HA antibodies (Santa Cruz Biotechnology) (1:200) or rabbit anti-TAP antibodies (1:500), followed by horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG, respectively (1:3,000), detected by enhanced chemiluminiscence. Where required, RNases A (300 μg/ml) and T1 (10 units/ml) were added.
Nuclear Export Assay.
TAP-GFP and GFP-TAP constructs were generated by cloning full-length TAP into the EcoRI/BamHI sites of pEGFP-N1 or pEGFP-C1 plasmids (CLONTECH), respectively. TAPmutNES-GFP construct was made by substituting a segment excised by the PpuM1 enzyme with a segment generated by PCR with a reverse primer that contained R→A mutations of residues 97, 98, 100, and 105 (38). GFP-TAPmutC construct was made by substituting a BspE1–BglII segment of TAP with a segment generated by PCR using a reverse primer coding for the mutation S585→P (38). HeLa cells were transiently transfected with 0.4 μg of plasmid DNA by using Lipofectamine Plus (Life Technologies, Grand Island, NY). Twenty-four hours after transfection, the cells were permeabilized with 35 μg/ml digitonin for 5 min on ice. Cells were incubated with final concentrations of 5 μM RanGTP (to compensate for the loss of endogenous RanGTP), including or excluding 5 μM Kap β2A or β2B for 30 min at 22°C. Nuclear export of TAP was observed by using fluorescence microscopy. Digital images of the same cells were taken at 0 and 30 min. The nuclear fluorescence intensities were measured for at least 20 nuclei for each experimental point by using the software IMAGEJ (http://rsb.info.nih.gov/ij/).
Kap β2B Detection in Poly(A)+ Complexes.
HeLa nuclei were isolated as described (39). Nuclei were lysed by incubating for 30 min at 22°C in 100 mM Tris⋅HCl, pH 8/100 mM NaCl/0.5% Triton X-100/1 mM DTT/10 mM EDTA/RanGMP-PNP as described. The lysate was centrifuged, and the supernatant was incubated with oligo(dT) cellulose (Monomer Sciences, New Market, AL) for 1 h at 22°C. Where required, RNases A (300 μg/ml) and T1 (10 units/ml) were added. The column was washed, and mRNA–protein complexes were eluted with 0.1 M NaOH, precipitated with trichloroacetic acid, and analyzed by SDS/PAGE. For immunoblots the complexes were transferred to nitrocellulose and probed with anti-Kap β2A or β2B antibodies visualized by enhanced chemiluminiscence. The anti-Kap β2A antibodies were raised against the full-length protein expressed in E. coli. The Kap β2B peptide 344–359, which has little similarity to Kap β2A (33), was used as an immunogen for anti-Kap β2B antibodies. The antibodies do not crossreact, as tested against recombinant proteins.
RNA Interference.
The short interfering RNA (siRNA) duplexes specific for Kap β2B (nucleotides 1041–1063) r(GAGGCUGAGCGGCCUGAUGGCU)d(TT) and r(AGCCAUCAGGCCGCUCAGCCUC)d(TT) and control inverted duplexes r(UCGGUAGUCCGGCGAGUCGGAG)d(TT) and r(CUCCGACUCGCCGGACUACCGA)d(TT) were synthesized by Xeragon (Huntsville, AL). HeLa cells were transfected by using Oligofectamine (Invitrogen) as described (www.mpibpc.gwdg.de/abteilungen/100/105/sirna.html). Inhibition of Kap β2B gene expression was monitored for up to 96 h after transfection by RT-PCR of fragments 10–1351 of Kap β2A and 1–1049 of Kap β2B. At various time points, poly(A)+ RNA was visualized as described (40) by using biotinylated oligo(dT)50 detected by Fluorescein Avidin DN (Vector Laboratories). Cyclin B1 was detected by immunofluorescence (41) by using anti-cyclin B1 antibodies (Sigma). 28S RNA was detected by using a biotinylated oligonucleotide corresponding to positions 2316–2350 of the 28S human ribosomal gene (GenBank accession no. M11167), followed by Fluorescein Avidin DN. Digoxigenin-labeled β-actin (Roche Diagnostics) and GAPDH (BIO/CAN, Mississauga, ON, Canada) probes were detected by using anti-digoxigenin-rhodamine Fab (1/500, Roche) (42).
Results
Kap β2B Is Associated with TAP in the Presence of RanGTP.
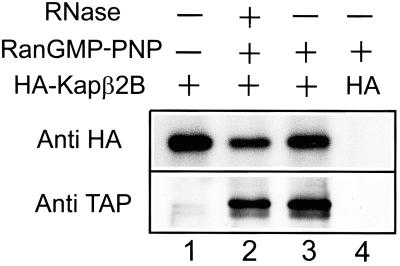
HA-tagged Kap β2B was expressed in HeLa cells, and cell extracts were subjected to immunoprecipitation with anti-HA antibodies. TAP was present in the immunoprecipitated material, and the interaction strictly depended on the presence of RanGTP, as expected for complexes between nucleocytoplasmic export factors and their cargoes (Fig. 1). RNase treatment did not have any effect, excluding the possibility that the two partners coprecipitate because they bind independently to RNA.
Fig 1.
Kap β2B and TAP form complexes in the presence of RanGTP. HA-Kap β2B (lanes 1–3) or a control construct (HA only, lane 4) were expressed in HeLa cells immunoprecipitated from nuclear extracts by using anti-HA antibodies in the presence (lanes 2–4) or absence (lane 1) of Ran loaded with a nonhydrolyzable GTP analog. The complexes were analyzed by immunoblotting using anti-HA and anti-TAP antibodies. TAP binding to Kap β2B is not abolished by treatment with RNases A1 and T1 (lane 2).
The Affinity of Kap β2B for RanGTP Is in the Range Typical for Export Factors.
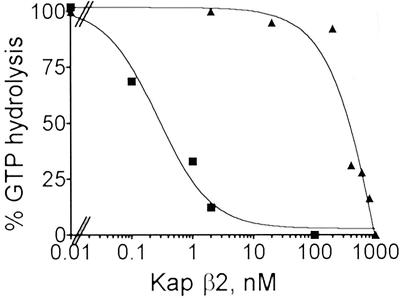
RanGAP1 stimulates the GTPase activity of Ran (43), and binding of Kaps to RanGTP strongly reduces the stimulation by RanGAP1. The dissociation constants of the complexes formed by the Kaps with RanGTP can be estimated from the dose dependence of this effect (44–47). This assay revealed that the Kap β2B/RanGTP complex has an apparent dissociation constant (Kd) of ≈300 nM, whereas the corresponding value for Kap β2A is ≈0.3 nM (Fig. 2). It is known that in the absence of their cargoes the nucleocytoplasmic export factors have high Kd values for RanGTP, whereas the Kd of import factors is in the low nanomolar range (44–47). The Kd of the Kap β2B/RanGTP complex is in the range typical for export factors (>1 μM for CRM1 and CAS, ≈1 μM for exportin-t, and ≈40 nM for exportin-4).
Fig 2.
The dissociation constant (Kd) of the Kap β2B/RanGTP complex is typical for a nucleocytoplasmic export factor. Ran complexed with [γ-32P]GTP was preincubated with the concentrations of Kap β2A or Kap β2B indicated on the horizontal axis, and the hydrolysis of Ran-bound GTP was determined after 15 min of incubation with RanGAP1. Kap β2A, ▪; Kap β2B, ▴. Based on the curves, a Kd of ≈300 nM was computed for the Kap β2B/RanGTP complex, and 0.3 nM for Kap β2A/RanGTP. The Kd values obtained from two independent experiments differed by <5%.
Kap β2B Mediates TAP Export from Nuclei.
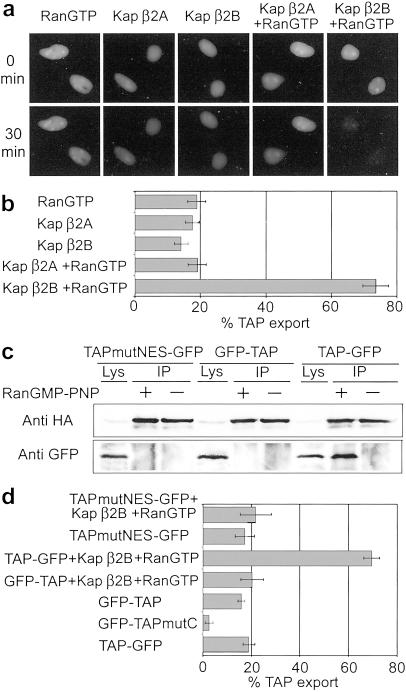
HeLa cells were transfected with a plasmid coding for TAP tagged C terminally with GFP. At 24 h after transfection the fusion protein was visible in the nuclei of most cells in agreement with previous reports (30, 48). At this time point the cells were treated with digitonin, which selectively permeabilizes plasma membranes (49). The cells were then incubated with Kaps and an energy-regenerating system in the presence or absence of RanGTP. After 30 min of incubation small reductions of TAP-GFP were detected in the nuclei of permeabilized cells if RanGTP, Kap β2A, Kap β2B, or Kap β2A and RanGTP were added to the incubation medium (Fig. 3 a and b). However, when Kap β2B was added in combination with RanGTP, the majority of TAP-GFP disappeared from the nuclei (Fig. 3 a and b), indicating that Kap β2B mediates TAP export. (The exported TAP is not visible because it diffuses through the permeabilized plasma membranes.)
Fig 3.
Kap β2B mediates RanGTP-dependent nuclear export of TAP in a permeabilized cells assay. (a) HeLa cells expressing TAP-GFP were permeabilized with digitonin then incubated with purified Kaps in the presence or absence of RanGTP. After 30 min of incubation with RanGTP and Kap β2B, the majority of TAP-GFP is exported from the nuclei. (b) Quantification of GFP-TAP export, measured as decrease of nuclear fluorescence intensity at 30 vs. 0 min. A highly significant increase in export (P = 5 × 10−8, t test, two-tailed) is observed when cells are incubated with RanGTP and Kap β2B. (c) Immunoblots of material immunoprecipitated by using anti-HA antibodies from cells coexpressing HA-Kap β2B and mutant TAP (TAPmutNES-GFP), or GFP-TAP, or TAP-GFP. Lys, cell lysate; IP, immunoprecipitated material. (d) Export of TAPmutNES-GFP or GFP-TAP is not stimulated by Kap β2B and RanGTP (the four upper bars). A mutation that abolishes the interaction of TAP with nucleoporins reveals the contribution of Kap β2B-independent export (GFP-TAPmutC vs. GFP-TAP, P = 4 × 10−10). The difference between TAP-GFP and GFP-TAP is not statistically significant. (Bars = SEM.)
A nuclear export signal (NES) domain was identified in the N-terminal region of TAP (amino acid residues 83–110), but the mechanism underlying its export activity remained unknown (38). We tested the ability of this NES to bind to and be exported by Kap β2B by using a construct containing four simultaneous point mutations shown to inactivate the NES (38). Wild-type TAP-GFP or mutant TAP-GFP (TAPmutNES-GFP) and HA-tagged Kap β2B were coexpressed in HeLa cells and subjected to coimmunoprecipitation with anti-HA antibodies (Fig. 3c). The mutant NES does not bind Kap β2B, indicating that this N-terminal NES is the TAP domain responsible for binding to Kap β2B. As expected, Kap β2B has no effect on the export of the mutant NES in the permeabilized cells assay (Fig. 3d). [A nuclear localization sequence partially overlaps with the NES, but the four mutations do not abolish the nuclear import activity (38) and do not prevent the predominantly nuclear localization of the mutant.] The position of TAP NES suggests that Kap β2B does not participate in the TAP-mediated export of CTE-containing viral RNAs, because the TAP-binding sites for the two ligands overlap (30). If GFP is fused to the N terminus of wild-type TAP, Kap β2B is unable to bind and stimulate export of the TAP construct (Fig. 3 c and d).
Previous studies have shown that TAP can be exported from the nuclei by direct interaction with nucleoporins (24–30). To compare this pathway with Kap β2B-mediated export, we used a TAP C-terminal point mutant (GFP-TAPmutC) that does not associate with nucleoporins (38). As expected, this mutant displays impaired export (Fig. 3d). Comparisons of the export rates of both TAP mutants and wild-type TAP reveal, however, that the direct binding of TAP to nucleoporins has a significantly lower contribution than the Kap β2B-mediated export (Fig. 3d).
Kap β2B Is Associated with Nuclear Poly(A)+ RNA.
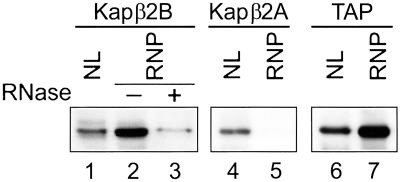
We investigated whether a physical association occurs in the nuclei between the putative carrier Kap β2B and its mRNA cargo. Poly(A)+ RNAs complexed with their associated proteins were isolated from HeLa nuclear lysate by using oligo(dT) cellulose, and the associated proteins were subjected to immunoblotting. Both Kap β2B and TAP are present and enriched in the poly(A)+ RNA complexes, and Kap β2A is absent (Fig. 4).
Fig 4.
Kap β2B and TAP, but not Kap β2A, are present in poly(A)+ RNA complexes isolated from HeLa cells nuclei. Nuclear lysates (NL) were incubated with oligo(dT) cellulose, and the bound ribonucleoprotein complexes (RNP) were eluted and analyzed by immunoblotting using anti-Kap β2B, anti-Kap β2A, or anti-TAP antibodies. Equal amounts of total protein were applied on gel in each lane except lane 3, which contains the same volume eluted from the oligo(dT) cellulose as lane 2. The Kap β2B signal is strongly reduced on treatment with RNases A1 and T1 (lane 3).
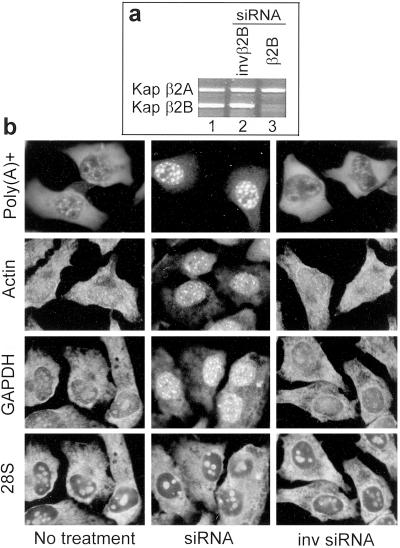
Down-Regulation of Kap β2B Induces Nuclear Accumulation of Poly(A)+ mRNA.
Kap β2B expression was inhibited in HeLa cells by transfection with duplex siRNA. Kap β2B mRNA starts to decline at 72 h after transfection and becomes virtually absent at 96 h (Fig. 5a). The amount of Kap β2A mRNA is not affected, although the siRNA used is 42% identical to the Kap β2A sequence. A strong accumulation of poly(A)+ RNA is visible in the nuclei of all cells treated with Kap β2B siRNA (Fig. 5b), indicating that the export of at least a large proportion, if not the vast majority, of the mRNA species is mediated by Kap β2B. To verify that the increased poly(A)+ signal in the nuclei reflects reduced RNA export and is not due to a perturbed polyadenylation process, the export of two specific mRNA species, β-actin and GAPDH, was monitored. Nuclear accumulations of these mRNAs occur in cells treated with Kap β2B siRNA (Fig. 5b). The speckled pattern of mRNA accumulated in the nuclei is similar to the pattern generated by TAP down-regulation or use of nonfunctional TAP mutants in Drosophila and yeast (50–53).
Fig 5.
Kap β2B siRNA produces nuclear accumulation of poly(A)+ RNA in HeLa cells. (a) Kap β2B RNA is strongly reduced by Kap β2B siRNA, as detected by RT-PCR (lane 3). Kap β2B RNA in cells treated with the inverted siRNA (lane 2) is the same as in the untreated cells (lane 1). Kap β2A RNA is not affected. (b) Accumulations of poly(A)+, β-actin, and GAPDH RNAs are visible in the nuclei of cells transfected with Kap β2B siRNA (Middle), in comparison with cells treated with inverted siRNA (Right) or untreated cells (Left). 28S RNA export is not affected (double staining for 28S and GAPDH RNAs is shown).
The export of 28S RNA is not affected, ruling out a general blocking of nucleocytoplasmic traffic. In addition, the predominantly cytoplasmic distribution of cyclin B1 is not visibly affected (data not shown). Cyclin B1 shuttles between the nucleus and the cytoplasm (41), and its unchanged localization indicates that protein import by Kap β1 and protein export by CRM1 are not affected by the siRNA.
Discussion
The data presented support the conclusion that Kap β2B is a major carrier for export of cellular mRNA and TAP connects Kap β2B to the mRNAs to be exported, whereas the direct interaction of TAP with nucleoporins allows lower rates of mRNA export.
Kap β2B has 84% amino acid identity and 92% homology with Kap β2A, which was shown to function in nuclear import of heterogeneous ribonucleoproteins (31–33). The nonidentical residues are scattered all along the sequences, and the longest segment of contiguous nonidentical residues has a length of only nine residues. The apparently minor differences in the primary structure produce dramatic differences in the interaction of the two Kaps with RanGTP and the cargoes; whereas RanGTP dissociates TAP from Kap β2A (30), Kap β2B binds TAP only in the presence of RanGTP (Figs. 1 and 3).
An explanation for the difference can be sought on the basis of the x-ray structure of the complex between Kap β2A and RanGTP (54). Surprisingly, the majority of the 36 residues of Kap β2A that are in direct contact with RanGTP are conserved in Kap β2B (only three conservative and one nonconservative substitution or deletion occur, depending on how the alignment is made). A major difference, however (only 2 identical residues out of 15), occurs in an acidic region of Kap β2A (residues 346–360), which does not contain, but is flanked by, residues in direct contact with RanGTP. Although this segment still contains mostly acidic residues in Kap β2B, its charge is lowered from −10 to −5, and it includes a lysine not present in Kap β2A. This segment is placed in the center of the loop L7 of Kap β2A (54), which was recently shown to couple physically the Ran and cargo-binding sites of Kap β2A (55), explaining the release of the cargo induced by RanGTP binding. It is probable that the different loop composition of Kap β2B explains the stable binding rather than cargo release induced by Ran-GTP.
We confirmed a previous study that reported the presence of an NES proximal to the N terminus of TAP (38). This NES is, however, ineffective in a TAP construct that has GFP fused to the N terminus, probably because of steric hindrance. This observation may explain why previous studies that used large protein domains fused to TAP N terminus did not detect this export domain and reached the conclusion that TAP export cannot occur in the absence of direct binding of TAP to nucleoporins (27, 56–58).
Most nucleocytoplasmic transport pathways are conserved from yeast to mammals. However, no obvious orthologue of Kap β2B exists in yeast, which is nevertheless not a unique situation among proteins of the Kap β family. Approximately 22 structurally related Kap β proteins occur in mammalian cells, but only 14 in Saccharomyces cerevisiae (6–10), indicating that some nucleocytoplasmic transport pathways exist in higher eukaryotes but not in yeast. Among the Kaps shown to function in export, exportin-4 lacks also an obvious yeast orthologue (45). A possible explanation for the difference in mRNA export between yeast and higher eukaryotes is that the export rate allowed by direct interaction of TAP with nucleoporins is sufficient in yeast, but not in higher eukaryotes. Apparently, in the latter case evolution added a more efficient pathway, probably to accommodate the largely increased number of mRNA species, the extensive splicing, the larger size of nuclei, and the increased complexity of nuclear pore complexes. Other systems besides yeast, e.g., Xenopus oocytes, may be endowed exclusively with Ran-independent mRNA export pathways (59, 60). Alternatively, Ran-dependent and independent pathways could coexist in Xenopus oocytes, but the high degree of redundancy could require complex approaches to clarify their relative contributions.
The data indicate that in mammalian cells, Kap β2B is responsible for the export of a high proportion or possibly the majority of the mRNA species. Because mRNA represents a complex group of cargo molecules, it is probable that besides bulk export, specialized pathways for certain mRNA subsets also exist. Other proteins besides TAP or related proteins may function in these specialized pathways, independently or in conjunction with Kap β2B. For instance, it was recently shown that c-fos mRNA is exported by the protein HuR in cooperation with Kap β2B, or by HuR in cooperation with pp32, APRIL, and the protein export factor CRM1, but neither pathway is involved in the export of poly(A)+ RNA in general (43).
Variations in the rates of mRNA export are expected to affect, at least transiently, cellular protein levels. The detailed knowledge of the components and mechanisms of the mRNA export system may lead to discovery of new mechanisms of regulation of gene expression, based on global or differential modulation of mRNA export.
Acknowledgments
We thank Drs. Elias Coutavas for kindly providing Rna1p and Ran clones and Elisa Izaurralde for the very generous support with anti-TAP antibodies and constructs. This work was supported by National Institutes of Health Grant R01 GM57569 (to A.R.).
Abbreviations
Kap, karyopherin
siRNA, short interfering RNA
NES, nuclear export signal
HA, hemagglutinin
References
- 1.Rout M. P. & Aitchison, J. D. (2001) J. Biol. Chem. 276, 16593-16596. [DOI] [PubMed] [Google Scholar]
- 2.Vasu S. K. & Forbes, D. J. (2001) Curr. Opin. Cell Biol. 13, 363-375. [DOI] [PubMed] [Google Scholar]
- 3.Allen T. D., Cronshaw, J. M., Bagley, S., Kiseleva, E. & Goldberg, M. W. (2000) J. Cell Sci. 113, 1651-1659. [DOI] [PubMed] [Google Scholar]
- 4.Ryan K. J. & Wente, S. R. (2000) Curr. Opin. Cell Biol. 12, 361-371. [DOI] [PubMed] [Google Scholar]
- 5.Stoffler D., Fahrenkrog, B. & Aebi, U. (1999) Curr. Opin. Cell Biol. 11, 391-401. [DOI] [PubMed] [Google Scholar]
- 6.Komeili A. & O'Shea, E. K. (2001) Annu. Rev. Genet. 35, 341-364. [DOI] [PubMed] [Google Scholar]
- 7.Macara I. G. (2001) Microbiol. Mol. Biol. Rev. 65, 570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quimby B. B. & Corbett, A. H. (2001) Cell Mol. Life Sci. 58, 1766-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayliss R., Corbett, A. H. & Stewart, M. (2000) Traffic 1, 448-456. [DOI] [PubMed] [Google Scholar]
- 10.Sweitzer T. D., Love, D. C. & Hanover, J. A. (2000) Curr. Top. Cell Regul. 36, 77-94. [DOI] [PubMed] [Google Scholar]
- 11.Azuma Y. & Dasso, M. (2000) Curr. Opin. Cell Biol. 12, 302-307. [DOI] [PubMed] [Google Scholar]
- 12.Kuersten S. M., Ohno, M. & Mattaj, I. W. (2001) Trends Cell Biol. 11, 497-503. [DOI] [PubMed] [Google Scholar]
- 13.Strawn L. A., Shen, T. & Wente, S. R. (2001) J. Biol. Chem. 276, 6445-6452. [DOI] [PubMed] [Google Scholar]
- 14.Zenklusen D., Vinciguerra, P., Strahm, Y. & Stutz, F. (2001) Mol. Cell. Biol. 21, 4219-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon J. H., Love, D. C., Guhathakurta, A., Hanover, J. A. & Dhar, R. (2000) Mol. Cell. Biol. 20, 8767-8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei E. P., Krebber, H. & Silver, P. A. (2001) Genes Dev. 15, 1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strasser K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A. G., Aguilera, A., Struhl, K., Reed, R. & Hurt, E. (2002) Nature 417, 304-308. [DOI] [PubMed] [Google Scholar]
- 18.Jensen T. H., Boulay, J., Rosbash, M. & Libri, D. (2001) Curr. Biol. 11, 1711-1715. [DOI] [PubMed] [Google Scholar]
- 19.Zhao J., Jin, S. B., Bjorkroth, B., Wieslander, L. & Daneholt, B. (2002) EMBO J. 21, 1177-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodge C. A., Colot, H. V., Stafford, P. & Cole, C. N. (1999) EMBO J. 18, 5778-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watkins J. L., Murphy, R., Emtage, J. L. & Wente, S. R. (1998) Proc. Natl. Acad. Sci. USA 95, 6779-6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.York J. D., Odom, A. R., Murphy, R., Ives, E. B. & Wente, S. R. (1999) Science 285, 96-100. [DOI] [PubMed] [Google Scholar]
- 23.Brodsky A. S. & Silver, P. A. (2000) RNA 6, 1737-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braun I. C., Herold, A., Rode, M. & Izaurralde, E. (2000) Mol. Cell. Biol. 22, 5405-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant R. P., Hurt, E., Neuhaus, D. & Stewart, M. (2002) Nat. Struct. Biol. 9, 247-251. [DOI] [PubMed] [Google Scholar]
- 26.Katahira J., Straesser, K., Saiwaki, T., Yoneda, Y. & Hurt, E. (2002) J. Biol. Chem. 277, 9242-9246. [DOI] [PubMed] [Google Scholar]
- 27.Wiegand H. L., Coburn, G. A., Zeng, Y., Kang, Y., Bogerd, H. P. & Cullen, B. R. (2002) Mol. Cell. Biol. 22, 245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt I. & Gerace, L. (2001) J. Biol. Chem. 276, 42355-42363. [DOI] [PubMed] [Google Scholar]
- 29.Kang Y., Bogerd, H. P. & Cullen, B. R. (2000) J. Virol. 74, 5863-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachi A., Braun, I. C., Rodrigues, J. P., Pante, N., Ribbeck, K., von Kobbe, C., Kutay, U., Wilm, M., Gorlich, D., Carmo-Fonseca, M. & Izaurralde, E. (2000) RNA 6, 136-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siomi M. C., Eder, P. S., Kataoka, N., Wan, L., Liu, Q. & Dreyfuss, G. (1997) J. Cell Biol. 138, 1181-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollard V. W., Michael, W. M., Nakielny, S., Siomi, M. C., Wang, F. & Dreyfuss, G. (1996) Cell 86, 985-994. [DOI] [PubMed] [Google Scholar]
- 33.Bonifaci N., Moroianu, J., Radu, A. & Blobel, G. (1997) Proc. Natl. Acad. Sci. USA 94, 5055-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matunis M. J., Coutavas, E. & Blobel, G. (1996) J. Cell Biol. 135, 1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coutavas E., Ren, M., Oppenheim, J. D., D'Eustachio, P. & Rush, M. G. (1993) Nature 366, 585-587. [DOI] [PubMed] [Google Scholar]
- 36.Askjaer P., Bachi, A., Wilm, M., Bischoff, F. R., Weeks, D. L., Ogniewski, V., Ohno, M., Niehrs, C., Kjems, J., Mattaj, I. W. & Fornerod, M. (1999) Mol. Cell. Biol. 19, 6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brownawell A. M. & Macara, I. G. (2002) J. Cell Biol. 156, 53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bear J., Zolotukhin, A. S., Tabernero, C., Hudson, E. A. & Felber, B. K. (1999) Mol. Cell. Biol. 19, 6306-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dignam J. D. (1990) Methods Enzymol. 182, 194-203. [DOI] [PubMed] [Google Scholar]
- 40.Huang S., Deerinck, T. J., Ellisman, M. H. & Spector, D. L. (1994) J. Cell Biol. 126, 877-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J., Bardes, E. S., Moore, J. D., Brennan, J., Powers, M. A. & Kornbluth, S. (1998) Genes Dev. 12, 2131-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallouzi I. E. & Steitz, J. A. (2001) Science 294, 1895-1901. [DOI] [PubMed] [Google Scholar]
- 43.Bischoff F. R., Klebe, C., Kretschmer, J., Wittinghofer, A. & Ponstingl, H. (1994) Proc. Natl. Acad. Sci. USA. 91, 2587-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bischoff F. R. & Görlich, D. (1997) FEBS Lett. 419, 249-254. [DOI] [PubMed] [Google Scholar]
- 45.Lipowsky G., Bischoff, F. R., Schwarzmaier, P., Kraft, R., Kostka, S., Hartmann, E., Kutay, U. & Görlich, D. (2000) EMBO J. 19, 4362-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kutay U., Lipowsky, G., Izaurralde, E., Bischoff, F. R., Schwarzmaier, P., Hartmann, E. & Görlich, D. (1998) Mol. Cell 1, 359-369. [DOI] [PubMed] [Google Scholar]
- 47.Kutay U., Hartmann, E., Treichel, N., Calado, A., Carmo-Fonseca, M., Prehn, S., Kraft, R., Görlich, D. & Bischoff, F. R. (2000) J. Biol. Chem. 275, 40163-40168. [DOI] [PubMed] [Google Scholar]
- 48.Herold A., Suyama, M., Rodrigues, J. P., Braun, I. C., Kutay, U., Carmo-Fonseca, M., Bork, P. & Izaurralde, E. (2000) Mol. Cell. Biol. 20, 8996-9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adam S. A., Sterne-Marr, R. & Gerace, L. (1992) Methods Enzymol. 219, 97-110. [DOI] [PubMed] [Google Scholar]
- 50.Herold A., Klymenko, T. & Izaurralde, E. (2001) RNA 7, 1768-1780. [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkie G. S., Zimyanin, V., Kirby, R., Korey, C., Francis-Lang, H., Van Vactor, D. & Davis, I. (2001) RNA 7, 1781-1792. [PMC free article] [PubMed] [Google Scholar]
- 52.Hurt E., Strasser, K., Segref, A., Bailer, S., Schlaich, N., Presutti, C., Tollervey, D. & Jansen, R. (2000) J. Biol. Chem. 275, 8361-8368. [DOI] [PubMed] [Google Scholar]
- 53.Segref A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Lührmann, R. & Hurt, E. (1997) EMBO J. 16, 3256-32571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chook Y. M. & Blobel, G. (1999) Nature 399, 230-237. [DOI] [PubMed] [Google Scholar]
- 55.Chook Y. M., Jung, A., Rosen, M. K. & Blobel, G. (2002) Biochemistry 41, 6955-6966. [DOI] [PubMed] [Google Scholar]
- 56.Braun I. C., Herold, A., Rode, M., Conti, E. & Izaurralde, E. (2001) J. Biol. Chem. 276, 20536-20543. [DOI] [PubMed] [Google Scholar]
- 57.Lévesque L., Guzik, B., Guan, T., Coyle, J., Black, B. E., Rekosh, D., Hammarskjöld, M. L. & Paschal, B. M. (2001) J. Biol. Chem. 276, 44953-44962. [DOI] [PubMed] [Google Scholar]
- 58.Kang Y. & Cullen, B. R. (1999) Genes Dev. 13, 1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Izaurralde E., Kutay, U., von Kobbe, C., Mattaj, I. W. & Görlich, D. (1997) EMBO J. 16, 6535-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clouse K. N., Luo, M. J., Zhou, Z. & Reed, R. (2001) Nat. Cell Biol. 3, 97-99. [DOI] [PubMed] [Google Scholar]