Abstract
Activation of the retinoblastoma (RB) protein through dephosphorylation arises in cells upon exit from M phase and in response to environmental stresses, including DNA damage. We provide here for the first time evidence that these responses are co-ordinately affected in a subset of tumor derived cell lines. We find that RB dephosphorylation is not apparent in these cells during progression into G1. Importantly these cells also do not respond with RB activation after DNA damage during S phase. Moreover and as a consequence they display phenotypes classically associated with RB− cells, showing accelerated apoptosis after DNA damage and DNA re-replication after spindle-checkpoint activation. A large body of literature provides evidence that controls governing inactivation of RB are lost in tumors. The results presented here indicate that the reverse reaction, namely the activation of RB from an inactive precursor, may also be compromised. Our findings indicate that this type of defect may be coupled with hypersensitivity to DNA damage and an increase in genomic instability in response to spindle-checkpoint activation thus bearing potentially important medical implications.
Keywords: retinoblastoma protein, DNA damage, spindle checkpoint, cell cycle, M Phase exit
The tumor suppressor product of the retinoblastoma (RB) susceptibility gene is regarded as a key regulator of the cell-cycle progression and fate determination (for review, see ref. 1). RB exerts its growth-suppressive effect through its ability to bind and interact with a variety of cellular proteins (for review, see ref. 2), and its protein interacting ability is controlled by phosphorylation (for review, see refs. 3 and 4).
Hypophosphorylated RB binds and sequesters cellular proteins, most notably transcription factors of the E2F/DP family, thereby regulating the transcription of genes (for review, see ref. 5). Its protein-binding function is abrogated when RB undergoes phosphorylation, which occurs in actively cycling cells during late G1 and is mediated by the concerted action of cyclin D/CDK complexes and cyclin E/CDK2. Reversal of phosphorylation arises after completion of mitosis and is executed by a multimeric complex of protein phosphatase type 1 (PP1); through this reversal, RB is returned to its active, growth-suppressive state (for review, see ref. 6). More recent results suggest that RB activation is also seen at other positions in the cell cycle and in response to stresses, including hypoxia, DNA damage, and long-term challenge of the spindle checkpoint, indicating that this may be key to cell survival and the preservation of genetic stability under these circumstances (7, 8).
The enzymatic machinery and regulation associated with RB inactivation has been extensively studied, including the recognition that deregulation of the RB kinases results in extension of lifespan, cooperating toward cell immortalization (for review, see refs. 9 and 10). The processes and circumstances by which RB becomes activated have received disproportionately less attention and, thus far, no evidence has been provided that these processes are affected in tumor cells.
We report here the existence of tumor cell lines where M phase exit does not coincide with accumulation of active RB. We further demonstrate that these tumor cells also do not respond with RB activation after DNA damage, and that, despite the expression of principally functional RB protein, these cells behave like RB− cells in several respects.
Materials and Methods
Cell Culture and Associated Procedures.
Cell lines were grown in DMEM or American Type Culture Collection Eagle's minimal essential medium (MEM) supplemented with 10% (vol/vol) FCS at 37°C in 5% CO2. Synchronization of cells in metaphase was achieved after sequential arrest in thymidine (2 mM) followed by release into 400 nM nocodazole for 12 h. Mitotic cells were enriched by shake off and either harvested directly or reseeded for mitotic release. Spindle-checkpoint challenge of cells was achieved by long-term culture in 50 nM nocodazole. U2OS with tetracycline-regulated constitutively active RB (11) were maintained in DMEM/10% (vol/vol) FCS supplemented with 2 μg/ml tetracycline/300 μg/ml G418/0.5 μg/ml puromycin. The RB transgene was induced through culture in tetracycline-free medium. To monitor the cell-cycle distribution, aliquots of the cells were routinely analyzed for DNA content by fluorescence activated cell sorter (FACS). Treatment and analysis of cells with cisplatin were performed essentially as described (12). Cell lysates were prepared in HB lysis buffer (50 mM Hepes, pH 7.4/150 mM NaCl/20 mM EDTA/1 mM DTT/2 mM PMSF/1% aprotinin/2.5 μg/ml leupeptin/10 mM NaF/10 mM β-glycerophosphate/1 mM NaVanadate/0.5% Triton X-100).
Cell-Cycle Analysis.
Cell-cycle analysis was performed essentially as described (11). Detection of BrdUrd incorporation used an FITC-coupled anti-BrdUrd monoclonal antibody (Becton Dickinson) and followed the procedures given by the manufacturer. BrdUrd labeling was conducted by adding BrdUrd to the tissue culture medium before harvest for a period of 4 h.
Immunoblot Analysis.
Immunoblot analysis followed standard procedures. For the Western blots, the following antibodies were used: PharMingen 14001A (pan RB), PharMingen 14441A (nonphosphorylated Ser-608; ref. 13); rabbit antiserum with specificity for phosphorylated Ser-608 was raised by using a 10-mer phosphopeptide spanning Ser-608 of RB coupled to purified protein derivative (PPD). Antiserum with specificity for the glutathione transferase tag was raised in rabbits. Antibodies for human p16, cyclins D1/D2, D3, and cyclin E were purchased from Santa Cruz Biotechnology. Antibodies for different subunits of PP1 were kindly provided by E. Villa-Moruzzi (University of Pisa, Pisa, Italy), and PP1 PhosphoT320 was purchased from Cell Signaling Technology (Beverly, MA).
GST Pull-Down.
Unfused GST tag or GST-fused full-length E2F-1 were produced and purified as described (13). Purified recombinant proteins adsorbed to glutathione Sepharose 4B beads were incubated with cell lysates for 2 h at 4°C. Subsequently, the beads were washed extensively, and bound proteins were eluted in SDS containing gel-loading buffer. Lysates used for treatment with λ-phosphatase were produced with buffer lacking phosphatase inhibitors. Lysates equivalent to 4 × 105 cells were diluted fourfold with λ-phosphatase buffer (50 mM Tris, pH 7.6/5 mM DTT/2 mM MnCL2/100 ng/ml BSA) and treated with 400 units λ-phosphatase for 60 min at 30°C. Controls included omission of phosphatase and addition of phosphatase inhibitors (10 mM β-glycerophosphate/10 mM NaF/2 mM NaVO3) concurrent with the phosphatase enzyme.
CDK Activity Assays.
Cells were scraped into HB lysis buffer, and aliquots normalized for equal amounts of cells were precipitated with anti-CDK2 (06-505) and anti-CDC2 (06-966), both from Upstate Biotechnology (Lake Placid, NY), or with anti-cyclin D1 (MS-211), D2 (214), and D3 (216), all from Neomarkers (Fremont, CA). Immunocomplexes were collected by using protein A/G Sepharose beads and washed three times with lysis buffer and once in kinase buffer. Reactions for CDK2 and cyclin D-associated kinase activity were performed at 30°C in 40 μl of kinase buffer A (50 mM Hepes-KOH, pH 7.4/1 mM MnCl2/10 mM MgCl2/10 mM β-glycerophosphate/1 mM DTT/0.1 μM protein kinase A inhibitor/2.5 μg/ml leupeptin/1% aprotinin/1 mM PMSF), whereas reactions for CDC2-associated activity were performed at 27°C in 40 μl of kinase buffer B (50 mM Hepes-KOH, pH 7.4/10 mM MgCl2/25 mM β-glycerophosphate/12 mM NaF/75 mM NaCl/0.1% Triton X-100/1 mM DTT/0.1 μM protein kinase A inhibitor/2.5 μg/ml leupeptin/1% aprotinin/1 mM PMSF). Precisely 0.5 μg of substrate (GST-Rb 763–928 for CDK 2 and CDK4/6 or histone H1 for CDC2) was used, and reactions contained 10 μM cold ATP and 0.2 μCi/μl [γ-32P]ATP (1 Ci = 37 GBq). Reactions were run for 20 min and stopped by adding 10 μl of 6 times Laemmli SDS-sample buffer and by boiling for 3 min. Samples were loaded in SDS/12% polyacrylamide gel and exposed to x-ray film or quantified with a Storm 860 phosphorimager.
Results
Lack of RB Activation in U2OS Osteosarcoma Cells upon G1 Entry.
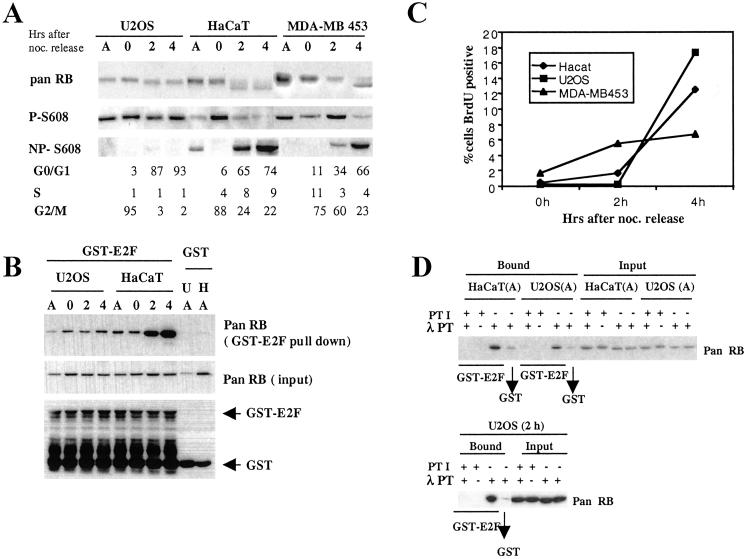
To characterize changes in phosphorylation associated with RB activation, we studied the process during M phase exit in U2OS osteosarcoma, MDA-MB 453 breast cancer, and epithelial-derived HaCaT cells. In doing so, we noticed a striking difference between these cell lines. Although fast migrating, underphosphorylated RB accumulated in both HaCaT and MDA-MB 453, this form did not become apparent in U2OS cells (Fig. 1A). Results consistent with these were obtained with antibodies specific for the phosphorylated and nonphosphorylated state of Ser-608 in RB. In both HaCaT and MDA-MB 453 cells, the level of RB phosphorylated on Ser-608 clearly decreased during release from M phase, whereas the amount of RB underphosphorylated at this site increased, indicating a relative loss of Ser-608 phosphorylated RB in both of these cell lines after M phase exit. In contrast, U2OS cells treated in the same way did not show significant changes in phosphorylation, as assessed by these two antibodies. This result suggests the existence of a defect in U2OS, affecting the accumulation of underphosphorylated RB after progression from mitosis into G1.
Fig 1.
Lack of RB activation in U2OS osteosarcoma cells. (A) RB phosphorylation analysis. Cell lysates were prepared from asynchronously growing cells (A) or metaphase cells released into G1 for 0, 2, and 4 h and analyzed by using antiretinoblastoma antibodies as follows: pan RB, recognizing all forms of RB; P-Ser-608, recognizing RB phosphorylated on Ser-608; NP-Ser-608, recognizing RB unphosphorylated on Ser-608. A portion of cells was subjected to FACS-based cell-cycle analysis for DNA content. Analysis of three different cell lines (U2OS, HaCaT, and MDA-MB 453) is presented. (B) E2F-binding analysis. Cell lysates were adsorbed to either GST-E2F1 or unfused GST. Bound proteins were analyzed by immunoblot by using pan RB antibody. GST protein equal loading in the experiment is shown by Western blot with an anti-GST antibody. (C) G1 progression. Metaphase cells were released into medium containing BrdUrd. Cells were harvested as indicated, and the percentage of BrdUrd+ cells was determined by FACS. (D) Activation of E2F binding by phosphatase treatment in vitro. Cell lysates from either asynchronous cells or cells released from metaphase for 2 h (2h) were treated with λ-phosphatase (λ PT) in the presence or absence of phosphatase inhibitors (PTI). Lysate was subjected to binding selection (bound) by using GST-E2F1, GST, or loaded directly (input).
The lack of production of underphosphorylated RB after release from mitotic arrest was not caused by poor or delayed exit of U2OS from M phase. Parallel cell-cycle analysis revealed that U2OS cells exited M phase with an efficiency similar to or greater than HaCaT or MDA-MB 453 (Fig. 1A). Furthermore, although there was a difference in the speed and efficacy of G1 transit between the different cell lines, as indicated by the timing of BrdUrd incorporation after release from the mitotic block (Fig. 1C), this did not correlate with the ability to accumulate underphosphorylated RB.
To investigate whether the lack of Ser-608 dephosphorylation signifies a lack of generation of active RB species, we assessed the ability of the RB species present in the different lysates to bind to recombinant E2F-1 in vitro. We observed a clear increase in E2F binding RB species in HaCaT cells after their release into G1 occurring concurrent with the appearance of the fast migrating form of RB and a decrease in Ser-608 phosphorylation (Fig. 1B). However, no change in E2F binding was detected in the U2OS cells after mitotic exit, suggesting a deficiency in these cells to generate RB with E2F-1 binding ability.
However, RB with E2F-1-binding ability was generated when U2OS cell lysate from either nonsynchronized cells or cells released from mitosis was treated with λ-phosphatase (Fig. 1D). This result indicates that the observed lack of E2F-1 binding with untreated lysates does not reflect a principal disability of the RB in these cells to bind these transcription factors. Together, these results suggest the existence of an alteration in U2OS that prevents the accumulation of underphosphorylated, active RB after M phase exit.
Defective Accumulation of Underphosphorylated RB in Other Tumor Cell Lines.
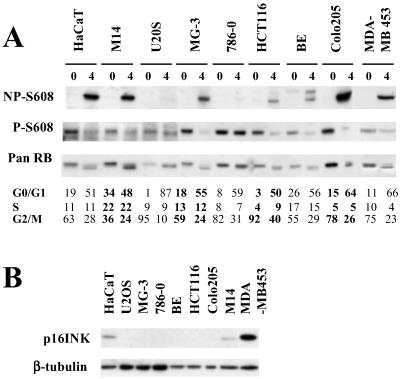
To examine whether a similar defect exists in other tumor cells, we examined another six cell lines derived from different tumor types and randomly selected from the cell lines of the National Cancer Institute (NCI) Anticancer Drug Screen Panel. All cell lines were treated by a regime identical to that used in the previous experiments, and cell-cycle analysis confirmed that all of them blocked and exited from M phase with an efficacy comparable to HaCaT, U2OS, and MDA-MB 453. Analysis of cell lysates by either pan- or phosphorylation state-specific RB antibodies indicated defective accumulation of dephosphorylated RB in one of these six cell lines, the renal carcinoma-derived 786-0 (Fig. 2A). Thus, defective RB activation after M Phase exit may not be frequent but clearly can be identified in other tumor cell lines apart from U2OS. To investigate possible mechanisms for the absence of RB activation, we examined expression of upstream regulation of RB. Lack of RB activation was not correlated with loss of p16 expression, as indicated by Western blotting (Fig. 2B). Absence of p16 expression was observed in both activation-defective cell lines and cell lines with RB activation competence. Furthermore, the expression of the main catalytic PP1 subunits, PP1α, -δ, and -γ, previously implicated in RB dephosphorylation (14), were indistinguishable between activation-competent and -incompetent cells. In addition, the expression levels and regulation of the three different D type cyclins, the CDK inhibitors, p21CIP or p27KIP, and the activity associated with CDC2 and CDK2, were indistinguishable between activation positive and negative cell lines. We noted a three- to fivefold higher cyclin D-associated activity in 786-0 cells. However, the activity of this kinase was not increased in U2OS (Figs. 6 and 7, which are published as supporting information on the PNAS web site, www.pnas.org). These results indicate that defective RB activation is not associated with a consistent and obvious change in the RB kinase regulation or the loss of catalytic PP1 core units.
Fig 2.
Changes in RB phosphorylation after metaphase release in selected tumor cell lines. (A) Cell lines randomly selected from the NCI tumor cell panel were treated and analyzed as described for Fig. 1A. Cell lines and their origins were as follows. HCT116, BE, Colo205 (colorectal cancer derived), 786-0 (renal cancer derived), MG-3 and U2OS (osteosarcoma derived) and M14 (melanoma derived). HaCaT and MDA-MB 453 were taken along as RB activation competent controls. (B) Lysate from asynchronously growing tumor cell lines was probed with antibody to human p16INK4a. Each lane contained lysate equivalent to 1 × 105 cells.
DNA Re-Replication After Spindle Disruption in Cells with RB Activation Defect.
The above experiments suggest that certain tumor cells, although expressing principally functional RB, fail to activate RB in a cell cycle-dependent manner. This suggestion raises the possibility that such cell lines may share features and behaviors with bona fide RB− tumor cells. One such phenotype is an unlicensed DNA replication in response to G2-checkpoint challenge through spindle disruption or DNA damage (15, 16). RB+ cells will refrain from DNA synthesis under these conditions, whereas cells with loss of RB function enter unlicensed rounds of DNA synthesis, resulting in increased ploidy and death.
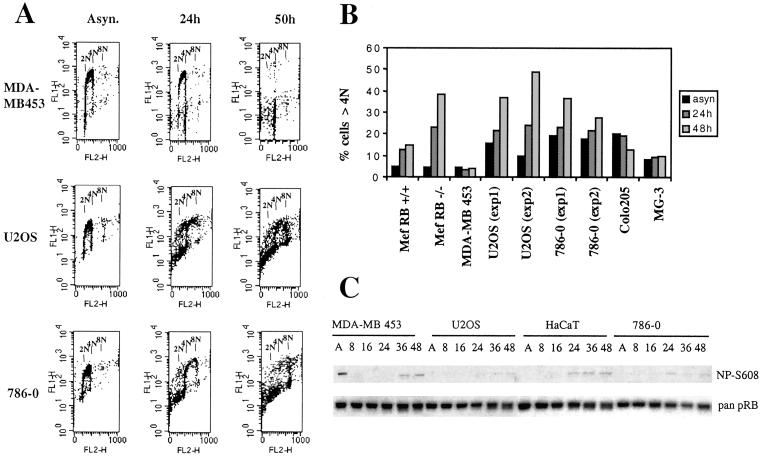
Therefore, we examined DNA replication after spindle-checkpoint challenge by using low concentrations of nocodazole in RB activation deficient U2OS and 786-0 alongside with activation competent MDA-MB 453 cells (Fig. 3A). Whereas DNA synthesis ceased in MDA-MB 453 during the 50 h observation period and the majority of the cells arrested with a DNA content of 4 N, this was not the case in U2OS and 786-0. Both, U2OS and 786-0, continued to incorporate BrdUrd and by 50 h a significant percentage of the cells featured a DNA content of 8 N, indicating that these cells, despite their inability to complete mitosis, went through an additional round of DNA synthesis (Fig. 3A). The increase in ploidy was in degree on par with that seen in RB−/− MEFs examined in parallel and consistently occurred in U2OS and 786-0 but not in RB+/+ MEFs or two other RB activation competent tumor cells, Colo-205 and MG-3, that we randomly chose from our selection (Fig. 3B). Concurrent with the nocodazole treatment the appearance of RB lacking Ser-608 phosphorylation was seen in MDA-MB 453 cells and in HaCaT cells and this response was markedly reduced in both U2OS and 786-0 (Fig. 3C). We conclude therefore that, like RB gene loss, deficient RB activation renders cells susceptible to increased ploidy and associated genome instability in response to checkpoint challenge in G2.
Fig 3.
DNA re-replication upon spindle-checkpoint challenge in RB activation-defective cells. As indicated, cells were cultured in the presence of 50 nM nocodazole for the time period indicated and analyzed for incorporation of BrdUrd and DNA content by FACS. (A) Raw BrdUrd incorporation data in cells as indicated. (B) Numerical representation of FACS-based analysis. Mouse embryo fibroblasts (MEF) RB−/− and MEF RB+/+ represent early passage mouse embryo fibroblasts derived from RB knockout and RB (WT) littermates, respectively. (C) Cell lysates obtained at different time points after nocodazole exposure were analyzed by using antibody for Ser-608 unphosphorylated (NP-Ser-608) or pan RB. Lysate amounts comprising equivalent numbers of cells were loaded in each lane.
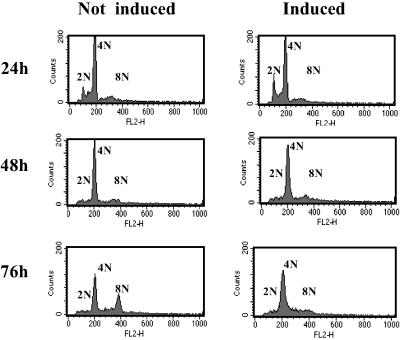
To exclude the possibility that a downstream defect in the RB pathway rather than the lack of RB activation was responsible for the unlicensed re-replication, we repeated our analysis by using a U2OS cell clone that expresses a constitutively active version of RB under the control of tetracycline (11). To achieve G2/M phase specific expression of the RB transgene we synchronized these cells in S phase by using thymidine and induced the transgene after their release into nocodazole containing medium. These conditions were chosen to minimize the effect of the retinoblastoma transgene before G2/M. As seen in Fig. 4 this treatment allowed cells to accumulate with a 4 N DNA content irrespective of whether they were induced for transgene expression or not. However, in the absence of transgene expression, these cells, like the parental U2OS, re-replicated their DNA. This is indicated by the significant rise in the number of cells with an 8 N DNA content after culture in nocodazole. However, in cells in which RB activity was supplied through transgene expression this response was completely suppressed. This demonstrates that the expression of active RB restores mitotic-checkpoint control in U2OS cells, excluding genetic or epigenetic changes in RB effector functions as an explanation for the lack of replication control after spindle disruption in these cells.
Fig 4.
Expression of constitutively active RB inhibits DNA re-replication after spindle-checkpoint activation. U2OS expressing a constitutively active form of RB under the control of tetracycline were synchronized in S phase by culture in thymidine for 16 h. Cells were subsequently released into medium containing low levels (50 nM) of the spindle inhibitor nocodazole in either the presence (not induced) or absence (induced) of tetracycline. Cells were processed for FACS analysis.
Lack of RB Activation in Response to DNA Damage.
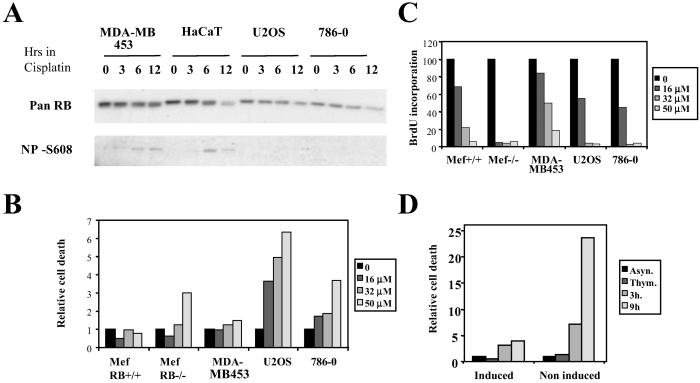
Previous work has indicated that conversion of inactive RB into an underphosphorylated active form also occurs during S phase in response to chemically induced DNA damage (12). To assess whether cell lines with a defect in cell cycle-dependent RB activation may also be defective for RB activation in response to challenge of this S phase checkpoint, we asked whether RB activation can be seen after treatment of S phase cells with cisplatin. We find that under these circumstances RB activation becomes apparent in both MDA-BD 453 and HaCaT cells within 3 and 6 h after addition of the damaging agent but is not seen in either 786-0 or U2OS, which are defective for RB activation after M phase release (Fig. 5A).
Fig 5.
RB activation and cell death after cisplatin challenge during S phase. (A) RB activation. RB activation-competent (MDA-MB453 and HaCaT) and -incompetent (U2OS and 786-0) cells were enriched for S phase by culture for 24 h in thymidine-containing medium. Cells were subsequently treated with 50 μM cisplatin for different lengths of time. Samples marked with 0 did not receive cisplatin. Lysates were generated and probed with either a pan RB antibody or antibody specific for RB unmodified on Ser-608. (B) Cell death after cisplatin treatment. S phase-enriched cells were treated with increasing doses of cisplatin for 16 h. Cells were subsequently washed and cultured in growth medium for an additional 3 h and processed for FACS analysis. Death response was assessed by scoring the percentage of cells with a sub-G1 DNA content. (C) Inhibition of S-phase progression after cisplatin treatment. S phase-enriched cells were treated with increasing doses of cisplatin as described in B. They were subsequently washed and cultured in the presence of BrdUrd for 4 h. Cells then were collected, fixed, and analyzed for DNA content and BrdUrd incorporation. (D) U2OS with tetracycline-regulated expression of constitutively active RB were exposed to thymidine for 24 h in the presence or absence of tetracycline; cisplatin at 50 μM was subsequently added to the culture for a further 16 h. Cells then were released into thymidine- and cisplatin-free growth medium for 3 or 9 h and analyzed for death response as in B.
Significantly, 786-0 and U2OS also show a significantly enhanced death response as assessed by the appearance of sub G1 cells 16 h post addition of cisplatin (Fig. 5B), consistent with the notion that cells lacking RB function have increased propensity to death after challenge of this checkpoint (12). The reduced death response seen in RB activation competent cells is not due to a decreased reactivity to the DNA damaging agent. This is indicated by the fact that BrdUrd incorporation is inhibited to a similar degree in all cells used (Fig. 5C).
Importantly, induction of constitutively active RB in U2OS concurrent with DNA damage resulted in a complete rescue of the death response, indicating that the increased death response is directly linked to the absence of RB activity (Fig. 5D).
Together these results indicate that tumor cells with defective cell-cycle-dependent activation of RB may also be defective for activation of RB in response to other triggers and after such triggers display a phenotypic behavior characteristically seen in cells with mutational loss of the RB gene.
Discussion
We present evidence here for the existence of cells in which conversion of inactive RB into an active form does not arise. Although it has been speculated that defective RB activation might represent an oncogenic event equivalent to mutational inactivation of this tumor suppressor, experimental evidence that this type of defect exists has not been presented. Here we show that cell lines with this defect can be found with a frequency of approximately two in eight, which closely resembles the frequency of mutational RB loss detected across the NCI panel. By extension, therefore, lack of RB activation may arise in tumors at a frequency on par with loss of RB function via gene mutation.
Our results indicate lack of dephosphorylation of Serine 608 of RB in the activation defective cells. However, a lack of dephosphorylation can also be seen at a number of other sites (data not shown) and coincides with a lack of E2F binding competent RB species, which together indicates that functional activation of RB truly does not arise in such cells.
The mechanistic reason(s) underlying deficiency for RB activation in the instances described is currently unknown. Theoretically, two different possibilities exist, a constitutive and/or untimely activation of RB inactivating kinases or the loss of RB phosphatase activity. A large body of literature provides evidence for deregulation and inappropriate regulation of kinases responsible for RB inactivation during tumor development. Production of cyclin D1, the activator of CDK4/CDK6, is increased as a consequence of Ras activation and its stability is increased in cells with defective integrin signaling. Furthermore p16INK4a, a specific inhibitor of CDK4/6, is frequently lost in tumor cells. However we have not found any consistent differences in the steady state levels of either cyclin D or cyclin E, the CDK2 activator which co-operates with cyclin D toward RB inactivation in late G1, or their associated activities. The defective RB activation is also not linked to loss of expression of the p16INK4a. It is noteworthy in this context that both RB activation defective cell lines identified indeed respond to p16INK4a overexpression with G1 arrest (data not shown), indicating that the defect observed does not confer resistance to INK mediated inhibition of cyclin D kinases. It is conceivable that cyclin D activated kinase activity in these cells is required for the inactivation of RB de novo synthesized during G1. The defect appears further not linked to p53 loss, as four p53− tumor cell lines (M14 melanoma, MG-3 osteosarcoma, Colo-205 rectal carcinoma and MDA-MB 453 breast carcinoma) are fully competent for RB dephosphorylation after G1 entry. Moreover, the p53− MDA-MB 453 cells activate RB in response to cisplatin treatment during S phase, indicating that this response is independent of p53.
Members of the PP1 are implicated in RB dephosphorylation (17). Recent evidence suggests that distinct PP1 catalytic core units have varied preference for the dephosphorylation of individual sites, suggesting that differently phosphorylated forms of RB may be generated in response to these various enzymes (18). However, Ser-608 is dephosphorylated by all of them, suggesting that, as far as this site is concerned, function of the different isoenzymes is redundant.
Our results do not provide evidence for defective or deregulated expression of catalytic PP1 cores, which precludes simple loss of catalytic core units as an explanation for the phenotype observed. This fact is, perhaps, not surprising, because loss of PP1 catalytic core enzymes in invertebrates has severe phenotypic consequences, revealing nonredundant functions for different cores (19–21). However, the PP1 enzymes are multimeric structures associated with regulatory subunits that function to direct substrate specificity, subcellular localization, and catalytic activity (for review, see refs. 22 and 23); alterations affecting the function of these subunits may lead to selective loss of target dephosphorylation and phenotypes more compatible with cell growth. The subunit composition of the enzyme(s) involved in dephosphorylation of RB has not been resolved so far.
Our results demonstrate that RB activation is affected not only upon entry into G1 in the cell lines described but, in addition, that these same lines do not respond with RB underphosphorylation upon DNA damage. This strongly suggests that the biochemical defect(s) inherent in these cell lines might affect RB activation in response to different triggers, thus affecting a variety of biological responses elicited through RB.
Importantly, our results indicate that loss of RB activation is linked to phenotypes classically seen in RB− cells. More specifically, these phenotypes affect the response to spindle-checkpoint activation and DNA damage. Spindle poisons including taxol, vinblastine, and vicristine, all of which are used in clinical applications, may cause increased ploidy and associated genetic instability in tumors with defective RB activation. In contrast, DNA-modifying drugs that induce damage responses may be more effective in leading to cell-death responses in such tumors. Thus, the phenotype described in this article has potentially important clinical implications.
Supplementary Material
Acknowledgments
We thank Georgina Platt for critical comments. This work was supported by Cancer Research United Kingdom. S.W. was the recipient of an Institute of Cancer Research studentship.
Abbreviations
RB, retinoblastoma
PP1, protein phosphatase type 1
FACS, fluorescence activated cell sorter
References
- 1.Zheng L. & Lee, W. H. (2001) Exp. Cell Res. 264, 2-18. [DOI] [PubMed] [Google Scholar]
- 2.Morris E. J. & Dyson, N. J. (2001) Adv. Cancer Res. 82, 1-54. [DOI] [PubMed] [Google Scholar]
- 3.Mittnacht S. (1998) Curr. Opin. Genet. Dev. 8, 21-27. [DOI] [PubMed] [Google Scholar]
- 4.Adams P. D. (2001) Biochim. Biophys. Acta 1471, M123-M133. [DOI] [PubMed] [Google Scholar]
- 5.Harbour J. W. & Dean, D. C. (2000) Genes Dev. 14, 2393-2409. [DOI] [PubMed] [Google Scholar]
- 6.Tamrakar S., Rubin, E. & Ludlow, J. W. (2000) Front. Biosci. 5, D121-D137. [DOI] [PubMed] [Google Scholar]
- 7.Ludlow J. W. & Nelson, D. A. (1995) Semin. Cancer Biol. 6, 195-202. [DOI] [PubMed] [Google Scholar]
- 8.Amellem O., Sandvik, J. A., Stokke, T. & Pettersen, E. O. (1998) Br. J. Cancer 77, 862-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee M. H. & Yang, H. Y. (2001) Cell Mol. Life Sci. 58, 1907-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmero I. & Peters, G. (1996) Cancer Surv. 27, 351-367. [PubMed] [Google Scholar]
- 11.Chew Y. P., Ellis, M., Wilkie, S. & Mittnacht, S. (1998) Oncogene 17, 2177-2186. [DOI] [PubMed] [Google Scholar]
- 12.Knudsen K. E., Booth, D., Naderi, S., Sever-Chroneos, Z., Fribourg, A. F., Hunton, I. C., Feramisco, J. R., Wang, J. Y. & Knudsen, E. S. (2000) Mol. Cell. Biol. 20, 7751-7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarkowska T., U, S., Harlow, E. & Mittnacht, S. (1997) Oncogene 14, 249-254. [DOI] [PubMed] [Google Scholar]
- 14.Nelson D. A., Krucher, N. A. & Ludlow, J. W. (1997) J. Biol. Chem. 272, 4528-4535. [DOI] [PubMed] [Google Scholar]
- 15.Khan S. H. & Wahl, G. M. (1998) Cancer Res. 58, 396-401. [PubMed] [Google Scholar]
- 16.Harrington E. A., Bruce, J. L., Harlow, E. & Dyson, N. (1998) Proc. Natl. Acad. Sci. USA 95, 11945-11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludlow J. W., Glendening, C. L., Livingston, D. M. & DeCarprio, J. A. (1993) Mol. Cell. Biol. 13, 367-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubin E., Mittnacht, S., Villa-Moruzzi, E. & Ludlow, J. W. (2001) Oncogene 20, 3776-3785. [DOI] [PubMed] [Google Scholar]
- 19.Axton J. M., Dombradi, V., Cohen, P. T. & Glover, D. M. (1990) Cell 63, 33-46. [DOI] [PubMed] [Google Scholar]
- 20.Doonan J. H., MacKintosh, C., Osmani, S., Cohen, P., Bai, G., Lee, E. Y. & Morris, N. R. (1991) J. Biol. Chem. 266, 18889-18894. [PubMed] [Google Scholar]
- 21.Ohkura H., Kinoshita, N., Miyatani, S., Toda, T. & Yanagida, M. (1989) Cell 57, 997-1007. [DOI] [PubMed] [Google Scholar]
- 22.Bollen M. (2001) Trends Biochem. Sci. 26, 426-431. [DOI] [PubMed] [Google Scholar]
- 23.Aggen J. B., Nairn, A. C. & Chamberlin, R. (2000) Chem. Biol. 7, R13-R23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.