Abstract
Recent findings indicate that glycosaminoglycans can play important roles in animal development. The genes sqv-3, -7, and -8, which are necessary for vulval morphogenesis in Caenorhabditis elegans, affect the biosynthesis of chondroitin and heparan sulfate glycosaminoglycans. We cloned sqv-1 and showed that the SQV-1 protein is a type II transmembrane protein that functions as a UDP-glucuronic acid decarboxylase. SQV-1 localizes to punctate cytoplasmic compartments and colocalizes with the SQV-7 nucleotide-sugar transporter, which probably acts in the Golgi apparatus. SQV-1 and SQV-7 are both expressed in the vulva and in oocytes, where they likely act in vulval morphogenesis and embryonic development, respectively. Progeny of sqv-7 and sqv-1 null mutants fail to initiate cytokinesis, possibly because they are unable to separate the plasma membrane from the eggshell, a defect analogous to that of incomplete vulval invagination.
Glycosaminoglycans (GAGs) are long unbranched carbohydrate polymers with specific repeating disaccharides; one sugar is usually a uronic acid (e.g., glucuronic acid) and the other is either N-acetylglucosamine or N-acetylgalactosamine. The importance of GAGs in animal development has become increasingly evident during the past few years. Mutations in the Drosophila melanogaster genes sugarless (1–3), sulfateless (4), and tout velu (5), which encode a UDP-glucose dehydrogenase, a heparan sulfate N-deacetylase/N-sulfotransferase and a heparan sulfate copolymerase, respectively, cause zygotic lethality with a segment polarity defect similar to that seen in wingless null mutants. Wingless is a member of the Wnt protein family and is involved in the control of cell differentiation and proliferation (reviewed in ref. 6). Heparan sulfate GAG has been implicated as a coreceptor for Wnt proteins (7). Mouse embryos deficient in the EXT-1 heparan sulfate copolymerase display embryonic lethality and fail to undergo normal gastrulation and to generate mesoderm (8). Mutants abnormal in the jekyll gene of the zebrafish are defective in UDP-glucose dehydrogenase and are deficient in the initiation of heart valve formation (9).
A screen for mutants defective in the morphogenesis of the vulva of Caenorhabditis elegans identified eight sqv (squashed vulva) genes (10). These mutants have reduced separation between the anterior and posterior halves of the vulva in the L4 larval stage. sqv-3, -7, and -8 have been molecularly identified and suggested to be involved in mediating glycosylation (11). SQV-3 and SQV-8 are glycosyltransferases necessary for the biosynthesis of a four-sugar linker attached to a protein core (serine-xylose-galactose-galactose-glucuronic acid), which is required for GAG biosynthesis (reviewed in ref. 12): SQV-3 is galactosyltransferase I, and SQV-8 is glucuronosyltransferase I (11, 13, 14). SQV-7 translocates three nucleotide-sugar substrates for GAG biosynthesis, UDP-glucuronic acid, UDP-N-acetylgalactosamine, and UDP-galactose, probably from the cytoplasm into the lumen of the endoplasmic reticulum and/or the Golgi apparatus, the site of GAG biosynthesis (15). In the accompanying paper (16), we show that sqv-4 encodes a UDP-glucose dehydrogenase, which synthesizes UDP-glucuronic acid.
Here we report the molecular identification and characterization of the sqv-1 gene and examine the expression of sqv-1 and sqv-7. Strong loss-of-function alleles of all sqv genes cause maternal-effect lethality (Mel). We identified a null allele of sqv-7 and show that the Mel phenotype is caused in part by the inability of the sqv mutants to initiate cytokinesis.
Materials and Methods
Strains and Genetics.
C. elegans strains were cultured as described by Brenner (17). N2 was the standard WT strain (17). Mutations used are described by Riddle et al. (18) except when noted: linkage group II, sqv-7(n3789) (this work) mnC1 [dpy-10(e128) unc-52(e444)] (19); linkage group IV, sqv-1(n2819, n2820, n2824, n2828, n2848, n2849, n3790, ku246) (ku246 was from Min Han; n3790 was from this work), unc-24(e138), dpy-20(e1282), eDf18, eDf19 (20), and nT1(n754) (21). The WT strain RW7000, which has many restriction fragment length polymorphisms when compared to the WT Bristol N2 strain, was also used (22, 23).
The deletion mutations sqv-1(n3790) and sqv-7(n3789) were isolated from a library of animals mutagenized with UV illumination and trimethylpsoralen, essentially as described by Jansen et al. (24) and Liu et al. (25), and backcrossed six times to N2. The sqv-1(n3790) deletion removes bases 2,981–5,690 of the cosmid D2096 and the entire sqv-1-coding sequence (Fig. 1A). The sqv-7(n3789) deletion removes bases 17,746–19,294 of the cosmid C52E12 and all but the first and part of the second exon of sqv-7 (for example, see Fig. 3F). sqv-7(n3789) also contains a tandem duplication of bases 19,295–19,316 of the cosmid C52E12.
Fig 1.
sqv-1 cloning and SQV-1 protein sequence comparisons. (A) Cloning of sqv-1. Genetic and physical maps of the sqv-1 region (Upper). The solid horizontal lines at the top indicate the extents of the chromosomal deletions eDf18 and eDf19; the dashed horizontal lines indicate uncertainty in their left end points. The short solid horizontal lines represent cosmid clones assayed in germline transformation experiments. The parallel vertical lines represent a gap in cosmid coverage of the C. elegans genome. The D2096 cosmid, which rescued the sqv-1 mutant phenotype, is shown in bold. (Lower) Subclones derived from the cosmid D2096 were tested for sqv-1 rescuing activity. Subclones that rescued the sqv-1 mutant phenotype are shown in bold. The rescue experiment results are shown as the number of transformed lines that rescued/total number of lines. The structure and location of the sqv-1 gene as deduced from the genomic and cDNA sequences relative to the minimal rescuing subclone are shown. Solid boxes indicate exons, and open boxes indicate untranslated sequences. The 5′-trans-spliced leader SL1 is indicated, and the arrow represents the 3′ poly(A) tail. The extent of the deletion in sqv-1(n3790) is indicated by a horizontal line. (B) Sequence alignment of SQV-1 and human and Drosophila homologs. The numbers on the right indicate amino acid positions. Residues identical in at least two proteins are shaded in black. Missense mutations in sqv-1 mutants are indicated. The addition of five amino acids (SFLLN) after amino acid 40 in an alternatively spliced form of the human protein is indicated. The putative transmembrane domains are underlined.
Fig 3.
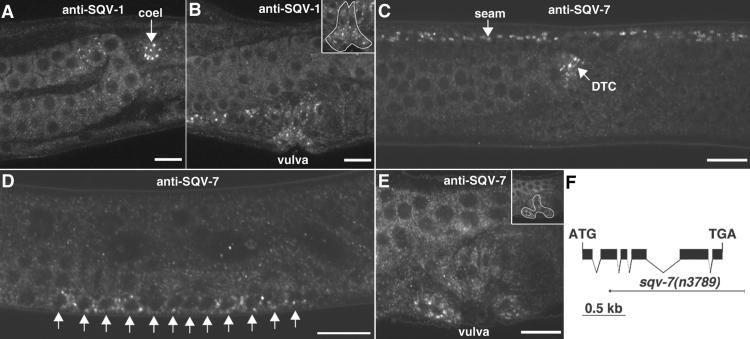
SQV-1 and SQV-7 localize to punctate cytoplasmic sites. (A and B) Whole-mount staining of WT animals by using anti-SQV-1-MBP rabbit polyclonal Abs. SQV-1 Abs localize to punctate cytoplasmic foci. (A) SQV-1 staining of a coelomocyte (coel) in an L4 larva is indicated by a white arrow. (B) SQV-1 staining of vulval cells during vulval morphogenesis. (Inset) The location of the vulval cells is indicated. (C–E) Whole-mount staining of WT animals by using anti-SQV-7 peptide Abs. SQV-7 Abs localize to punctate cytoplasmic foci. (C) SQV-7 staining in seam cells (seam) and the distal tip cell (DTC) in an L4 larva. Seam cells form a lateral line along the length of the worm from head to tail. The distal tip cell is located at the end of the developing gonad, which is visible to the left of the DTC. (D) SQV-7 staining in the 12 vulval precursor cells in an L3 larva. Ten of these 12 cells will divide once more to generate the 22 vulval descendants that form the vulva. (E) SQV-7 staining in vulval cells during vulval morphogenesis in an L4 larva. A subset of the 22 vulval cells is visible in this focal plane. (F) Deletion allele of sqv-7. The structure of the sqv-7 gene is shown by using solid boxes to indicate exons. The initiation and termination codons are indicated. The sqv-7(n3789) deletion is shown by using a thin solid line to depict the extent of the deletion and a short thick solid line to depict the extent of the duplication (see Materials and Methods). (Bars = 10 μm.)
Transgenic Animals.
We injected genomic DNA into unc-24(e138) sqv-1(n2819)/dpy-20(e1282) hermaphrodites at concentrations of 3–7 μg/ml with the dominant roller marker pRF4, as described by Mello et al. (26). Rol lines were established, and Unc Rol animals were examined for rescue of the sqv-1 mutant phenotype.
Molecular Biology.
Standard molecular biological techniques were used (27). The sequences of all PCR-amplified DNAs used for cloning were confirmed to exclude unintended mutations. The sequences of oligonucleotides used for DNA amplification or mutagenesis will be provided on request. The human cDNA clones 1875025, 1871770, 29917, 210962, 21151921, 32371, 208993, 2630577, and 54339, containing the human homolog of sqv-1 were provided to us by the Integrated Molecular Analysis of Genome and Their Expression consortium (28). The clones 1875025 and 1871770 have identical 5′-ends. The clone 29917 contains an alternative splice form, which is predicted to encode five additional amino acids not found in 1875025 and 1871770 (Fig. 1B).
sqv-1 cDNA.
A sqv-1 cDNA was isolated from an embryonic library (29) and found to contain an ORF identical to D2096.4, which is predicted to encode a protein of 467 aa. The D2096.4 ORF was cloned into pPD49.78 and pPD49.83 (from A. Fire), which are designed to express the transcriptionally fused ORF under the control of the C. elegans heat-shock promoters (30). We injected sqv-1(n2828)/nT1(n754) hermaphrodites with these vectors along with pRF4, and Rol lines were established. The expression of sqv-1 was induced by a 30-min heat-shock treatment at 33°C, and Rol animals were examined for rescue of the sqv-1 mutant phenotype.
UDP-Glucuronic Acid Decarboxylase Assay.
The sqv-1-coding sequence was cloned into pMAL-c2 and transformed into BL21 pLysS to generate a maltose-binding protein (MBP)-SQV-1 fusion protein. MBP-SQV-1 expression was induced by incubation with 1 mM isopropyl β-d-thiogalactoside at 20°C for 15 h. Escherichia coli were pelleted and resuspended in 0.1 M phosphate (pH 7), 1 mM glutathione, and 2 mM EDTA and lysed by using a French Pressure Cell (SLM Instruments, Rochester, NY). The soluble fraction was separated from insoluble inclusion bodies by centrifugation at 12,000 × g for 20 min. Soluble MBP-SQV-1 was purified further by binding to amylose resin and eluting with 10 mM maltose. Approximately 10 μg of purified MBP-SQV-1 was incubated with 2 mM UDP-glucuronic acid and 2 mM NAD+ for 1 h at 22–23°C. Nucleotide sugars were separated from protein by 20 min of centrifugation through a Microcon YM-10 (Millipore) equilibrated with water and methanol. The eluate was diluted 100-fold with water and mixed with an equal volume of methanol, injected into a capillary ion pair RP-HPLC (C18), and eluted with increasing concentrations of methanol. The HPLC column was coupled to an electrospray time-of-flight mass spectrometer (Mariner workstation, PerSeptive Biosystems, Framingham, MA) to detect the mass of nucleotide sugars present in the reaction sample (31).
Abs and Immunostaining.
A 26-aa [(C)RSKSTTISYKPLPMTMPIDVHKPRN] peptide corresponding to the carboxy-terminal end of SQV-7 was synthesized and injected into two rabbits (Zymed). Anti-SQV-7 antisera were affinity purified by binding to the SQV-7 peptide conjugated to SulfoLink coupling gel (Pierce) and elution with 100 mM glycine (pH 2.5), following the manufacturer's instructions.
The full-length sqv-1 ORF was cloned into the vectors pGEX-4T3 and pMAL-c2 to generate GST-SQV-1 and MBP-SQV-1 fusion proteins, respectively. The expression of both fusion proteins was induced by incubation with 1 mM IPTG at 37°C for 3 h, and both fusion proteins were purified by isolating the inclusion bodies followed by SDS/PAGE and electroelution of the fusion proteins. GST-SQV-1 was injected into two rabbits (Covance, Denver, PA). MBP-SQV-1 was injected into two rabbits and two rats (Covance). The anti-GST-SQV-1 Abs were affinity purified by incubation with MBP-SQV-1 fusion protein bound to Optitran (Schleicher & Schuell) strips and elution with 100 mM glycine, pH 2.5. Anti-MBP-SQV-1 Abs were affinity purified by incubation with GST-SQV-1 fusion proteins bound to Optitran strips and glycine elution. Whole worms were fixed and stained as described (16).
Results
Molecular Identification of sqv-1.
sqv-1 was mapped between unc-24 and dpy-20 on linkage group IV (10). We further mapped sqv-1 between the polymorphism bnP4 and the left endpoints of the deletions eDf18 and eDf19 (Fig. 1A). Thirteen cosmids in this interval were tested for the ability to rescue the sqv-1 mutant phenotype. A single cosmid, D2096, was able to rescue. A 3.6-kb Eco0109I–HindIII fragment of D2096, containing a single predicted gene D2096.4 (32), was sufficient for rescue (Fig. 1A).
We used this 3.6-kb minimal rescuing fragment as a probe to screen an embryonic cDNA library. One of the cDNAs obtained contains 1,771 bases of an ORF identical to that predicted for D2096.4. The expression of the D2096.4 ORF under the control of the C. elegans heat-shock promoters (30) rescued the vulval defect and maternal-effect lethality of sqv-1 mutants, indicating that the predicted coding sequence encodes a functional SQV-1 protein (data not shown).
We identified seven molecular lesions in D2096.4 in six of the seven sqv-1 alleles. Five alleles, n2820, n2824, n2828, n2848, and ku246, are missense mutations and one allele, n2819, contains two missense mutations (Fig. 1B). No molecular lesion of the seventh allele, n2849, has been identified. We obtained a deletion allele, n3790, which removes the entire coding sequence of sqv-1 (Fig. 1A), by PCR screening a library of mutagenized worms. Animals homozygous for n3790 show the same vulval and Mel phenotypes observed in the stronger missense mutants.
sqv-1 Encodes a Protein Similar to Nucleotide-Sugar Modifying Enzymes.
The SQV-1 protein is weakly similar to UDP-glucose epimerases and ribosylthymine 5′-diphosphate (TDP)-glucose dehydratases. For example, of the 467 aa of SQV-1, 67 (14%) are identical to the E. coli UDP-glucose epimerase and 78 (17%) are identical to E. coli TDP-glucose dehydratase (data not shown). We failed to detect either enzymatic activity by using recombinant SQV-1, and both of these E. coli genes expressed under the control of the C. elegans heat-shock promoters failed to rescue the sqv-1 mutant phenotype (data not shown). The C. elegans genome contains a UDP-glucose epimerase homolog, C47B2.6 (47% identity to the E. coli gene), and two TDP-glucose dehydratase homologs, F53B1.4 and C01F1.3 (35% and 30% identity, respectively, to the E. coli genes), all of which are more similar to their E. coli counterparts than to SQV-1 (data not shown). Thus, SQV-1 likely is neither a C. elegans UDP-glucose epimerase nor a TDP-glucose dehydratase.
SQV-1 contains a potential transmembrane domain near the amino terminus, suggesting it may be a type II transmembrane protein. SQV-1 is more similar to predicted proteins of as yet undefined function. Of 441 aa of the Drosophila melanogaster CG7979 gene product, 239 (54%) are identical to SQV-1 (Fig. 1B). We identified and determined the sequence of human cDNA clones (a National Cancer Institute EST project) that encode a protein closely related to SQV-1. Of 420 aa of the more common spliced form of this human protein, 236 (56%) are identical to SQV-1.
SQV-1 Has UDP-Glucuronic Acid Decarboxylase Activity.
Because previously cloned sqv genes are implicated in the biosynthesis of GAGs, we hypothesized that sqv-1 is also involved in GAG biosynthesis. Based on the sequence similarity to UDP-glucose epimerases and TDP-glucose dehydratases, we hypothesized that sqv-1 encodes an enzyme that modifies a nucleotide sugar. Many nucleotide-sugar modifying enzymes involved in GAG biosynthesis have been cloned and do not share a high degree of amino acid identity to SQV-1. Therefore, we tested SQV-1 for enzymatic activities for which corresponding genes had not yet been cloned in any species. One such enzymatic activity was that of UDP-glucuronic acid decarboxylase, which converts UDP-glucuronic acid to UDP-xylose. UDP-xylose is a donor substrate necessary for the initiation of the GAG-protein core linker region (reviewed in ref. 12). During the preparation of this manuscript, we noted that a fungal homolog of SQV-1 has been shown to have UDP-glucuronic acid decarboxylase activity (33).
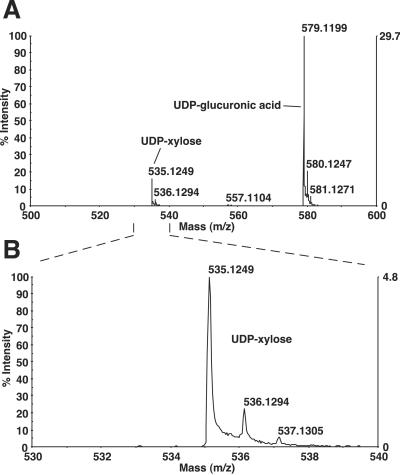
We tested the ability of a purified MBP-SQV-1 fusion protein to generate UDP-xylose from UDP-glucuronic acid. The reaction mixture was analyzed by using HPLC coupled to a mass spectrometry. Negative polarity mass spectra identified compounds with most abundant masses of 579 and 535 in the reaction sample (Fig. 2), corresponding to the molecular masses of UDP-glucuronic acid and UDP-xylose.
Fig 2.
Conversion of UDP-glucuronic acid to UDP-xylose by SQV-1. UDP-glucuronic acid and NAD+ were incubated with MBP-SQV-1, purified, and injected into a capillary ion pair RP-HPLC coupled to an electrospray time-of-flight mass spectrometer. (A) Negative polarity mass spectra of SQV-1 reaction sample from elution time of 8.02–35.02 min. The y axis indicates the intensities of the spectra relative to the most intense peak, and the x axis indicates the mass (m/z). Peaks of mass (m/z) of 500–600 are shown. (B) Magnification of mass spectra for mass (m/z) of 530–540. The highest peak at ≈535 represents the most abundant mass of UDP-xylose. The smaller peaks at ≈536 and ≈537 likely represent isotopic distributions of UDP-xylose.
SQV-1 Ab Stains Punctate Foci in the Cytoplasm.
We generated affinity-purified rabbit polyclonal Abs raised against GST-SQV-1 fusion protein and rabbit and rat polyclonal Abs raised against MBP-SQV-1 fusion protein. These Abs were found to stain punctate foci in the cytoplasm of many cells in WT worms (Figs. 3 A and B and 4A, data not shown). Staining was observed in the vulval and uterine cells (Fig. 3 A and B), and stronger staining was observed in oocytes and coelomocytes (Fig. 4A, data not shown). This punctate staining was not seen in animals homozygous for the sqv-1(n3790) null allele (data not shown). The presence of SQV-1 in the vulva is consistent with SQV-1s having a function in vulval morphogenesis. The presence of SQV-1 in oocytes is consistent with a cell-autonomous role of SQV-1 in normal embryonic development.
Fig 4.
SQV-1 and SQV-7 proteins colocalizes. (A) SQV-7 staining of oocytes. SQV-7 rabbit peptide Ab staining visualized by Texas red-conjugated secondary Abs. The three most mature oocytes (oocy), including the most proximal oocyte (prox oocy), are indicated. (B) SQV-1 staining of the same oocytes. SQV-1-MBP rat Ab staining visualized by FITC-conjugated secondary Abs. (C) Merged image of A and B, showing that SQV-1 and SQV-7 colocalize in oocytes. (Bar = 10 μm.)
SQV-7 Ab Stains Punctate Foci in the Cytoplasm.
We suspected that the punctate cytoplasmic staining by anti-SQV-1 Abs was caused by the localization of SQV-1 to a specific subcellular compartment, such as the Golgi apparatus. sqv-7 encodes a nucleotide-sugar transporter and translocates UDP-glucuronic acid, UDP-galactose, and UDP-N-acetylgalactosamine presumably from the cytosol to the lumen of the Golgi apparatus (15). We raised rabbit polyclonal antisera against a 26-aa peptide corresponding to the SQV-7 carboxy-terminus and affinity-purified the antisera against the same SQV-7 peptide. The anti-SQV-7 Abs stained punctate foci in the cytoplasm of several tissues in WT animals, including the vulva, seam cells, distal tip cells, and oocytes (Figs. 3 C–E and 4B, data not shown). By PCR screening, we obtained a sqv-7 null allele that deletes most of the sqv-7 ORF (Fig. 3F). SQV-7 Abs did not stain punctate foci in the vulval, seam cells, distal tip cells, or oocytes in animals homozygous for this sqv-7(n3789) null allele (data not shown).
SQV-1 Colocalizes with the SQV-7 Nucleotide-Sugar Transporter.
By using rat anti-SQV-1 Abs and rabbit anti-SQV-7 Abs, we found that SQV-1 and SQV-7 colocalized in oocytes (Fig. 4C). We also observed colocalization of the staining in vulval cells and pharynx (data not shown). We could not determine whether SQV-1 and SQV-7 colocalized in the seam cells or coelomocytes, because the Ab staining for one of the two antigens could not be resolved sufficiently above background. We speculate that significant amounts of SQV-1 and SQV-7 protein are present in the Golgi apparatus of several cells, including oocytes, and that lower concentrations are present in most if not all other cells.
The Maternal-Effect Lethality of sqv-7 Is Caused by a Defect in the Initiation of Cytokinesis.
Herman et al. (1998) reported that most progeny of mutants homozygous for stronger mutant alleles of sqv-1 to -7 arrest as one-cell stage embryos. We examined the embryonic arrest phenotype of sqv mutants further by comparing the development of WT embryos and embryos produced by animals homozygous for the sqv-7(n3789) null allele.
In the WT, fertilization triggers oocyte meiosis and the extrusion of polar bodies generated by meiosis. A separation between the plasma membrane and the eggshell becomes visible at around this time, and the space between the two membranes expands thereafter (Fig. 5 A–C). Shortly before fusion of the haploid maternal and paternal nuclei, the embryo undergoes a series of constrictions of the plasma membrane termed pseudocleavage (Fig. 5A). The maternal and paternal pronuclei, which are initially located at opposite ends of the embryo, meet and move to the middle of the embryo and rotate around each other (Fig. 5 B and C). Aster and mitotic spindle formation and nuclear division are followed by cytokinesis (Fig. 5 C–E).
Fig 5.
Early embryogenesis of WT and sqv-7(null) mutant animals. Nomarski photomicrographs of a WT embryo (A–E) and a sqv-7 embryo (F–J) from a sqv-7(n3789) homozygous hermaphrodite. (A and F) Pseudocleavage (the constriction of the plasma membrane as indicated by arrows) is observed in the WT embryo but not in the sqv-7 mutant. Maternal (m) and paternal (p) pronuclei are indicated. (B and G) Pronuclear meeting. The space between the plasma membrane and the eggshell (indicated by brackets) in the WT embryo is not evident in the mutant. The ends of the mutant egg along the A-P axis are indicated by short vertical lines. (C and H) The start of nuclear division along the A-P axis. (D and I) First mitosis. The initiation of cytokinesis is visible in the WT embryo but not in the mutant. The cleavage furrow is indicated by arrows. (E and J) After the first division. The two daughter cells (AB, P1) are labeled in the WT. The sqv-7 mutant embryo contains more than two nuclei (indicated by arrows) in a single cytoplasm. (Bar = 10 μm.)
We found that oocytes from the sqv-7 null mutants were fertilized. However, no signs of polar body extrusion, pseudocleavage, or separation of the plasma membrane and vitelline membrane were apparent in most progeny of sqv-7 null mutants (Fig. 5 F–J). The timing of migration, fusion, and rotation of the sperm and egg pronuclei and separation of postmitotic nuclei was similar to that observed in WT embryos (Fig. 5 F–I), but the nuclear division was not accompanied by the initiation of cytokinesis (Fig. 5 I and J). After the first nuclear division, at least three nucleus-like objects were found in the embryos instead of two (Fig. 5J). Similar defects were observed in the progeny of mothers homozygous for a strong loss-of-function mutation in any of the eight sqv genes (data not shown). The 4′,6-diamidino-2-phenylindole staining suggests the extra nucleus-like objects contain DNA (data not shown). We postulate that the extra nucleus-like objects are derived from the maternal DNA normally extruded in polar bodies. The nuclei continued to divide repeatedly without cytokinesis, resulting in a multinucleate embryo (data not shown).
Discussion
We have demonstrated that sqv-1 encodes a UDP-glucuronic acid decarboxylase that converts UDP-glucuronic acid to UDP-xylose. Mutations in all four other cloned sqv genes (sqv-3, -4, -7, and -8) cause the same developmental defects observed in sqv-1 mutants, and the proteins encoded by these four sqv genes have been implicated in the biosynthesis of chondroitin and heparan sulfate (10, 11, 15, 16, 34). UDP-xylose is used by xylosyltransferase to add xylose to specific serine residues in the protein core of proteoglycans and is required for biosynthesis of chondroitin and heparan sulfate GAGs (reviewed in ref. 12). We therefore propose that the abnormalities of the sqv-1 mutants are caused by a defect in the biosynthesis of chondroitin and/or heparan sulfate GAGs.
We found that SQV-1 protein is localized in punctate foci, presumably in a subcellular compartment within the cytoplasm. The SQV-7 transporter appears to localize to the same subcellular compartment. SQV-7 is a multipass transmembrane protein and is capable of transporting nucleotide sugars required for the biosynthesis of GAGs across the membranes of the Golgi apparatus (11, 15). SQV-1 has a putative transmembrane domain near its amino terminus and lacks a signal sequence, suggesting that it may be a type II transmembrane protein with all but its amino terminus in the lumen of the Golgi apparatus. Indeed, chicken UDP-glucuronic acid decarboxylase (a SQV-1 homolog) and xylosyltransferase localized to the endoplasmic reticulum and Golgi vesicles in chondrocytes (35). Furthermore, both the decarboxylase and xylosyltransferase are sensitized to trypsin by disruption of the endoplasmic reticulum and Golgi vesicles, suggesting that the catalytic domain of decarboxylase localizes to the luminal side. In chondrocytes, xylosylation of GAGs occurs in the vesicular regions of the endoplasmic reticulum and continues in the early Golgi apparatus (36). For these reasons, we propose that SQV-1 and SQV-7 act in the late endoplasmic reticulum and in the Golgi apparatus. In the discussion below, for simplicity, we focus on the localization of SQV-1 and SQV-7 to the Golgi apparatus.
Given that SQV-1 and the chicken UDP-glucuronic acid decarboxylases likely synthesize UDP-xylose in the lumen of the Golgi apparatus, we find some previous observations intriguing. Chickens have UDP-xylose transporter activities for transporting UDP-xylose from the cytosol to the lumen of the Golgi apparatus (35). The Drosophila homolog of SQV-7, Fringe connection, is a multisubstrate nucleotide sugar transporter and also may transport UDP-xylose (37). However, decarboxylation of UDP-glucuronic acid is the only reaction known to produce UDP-xylose, and in animals there appears to be no UDP-glucuronic acid decarboxylase in the cytosol, as the human and Drosophila SQV-1-like UDP-glucuronic acid decarboxylases are putative type II transmembrane proteins. The fungal homolog of SQV-1, by contrast, lacks a transmembrane domain and therefore likely resides in the cytosol (33). If UDP-xylose is synthesized only in the lumen of the Golgi apparatus in animals, what is the function of a UDP-xylose transporter? One possibility is that there is a mechanism of UDP-xylose production in the cytosol that is radically different from one involving a SQV-1-like protein. Another, not exclusive possibility, is raised by the observation that UDP-xylose is a strong inhibitor of some but not all UDP-glucose dehydrogenases (38, 39), which irreversibly convert UDP-glucose to UDP-glucuronic acid in the cytosol (reviewed in ref. 40). If the UDP-xylose transporters are permeases that allow bidirectional transport of UDP-xylose, then such a transporter may help in conservation of UDP-glucose (and glucose) in the cell.
The eggs of sqv-7 and sqv-1 null mutants are fertilized and initiate normal pronuclear migration, fusion, and postmitotic nuclear division. However, these embryos are unable to develop because of their inability to accompany nuclear division with cell division. The inability of these mutants to extrude excess maternal DNA in polar bodies, to undergo pseudocleavage, and to initiate cytokinesis suggest a defect in their ability to constrict the plasma membrane of the embryo.
Why might GAG biosynthesis affect cytokinesis? Perhaps the most visible defect accompanying the cytokinesis defect is the lack of separation between the plasma membrane and eggshell. In other organisms, such as sea urchins, fertilization of the egg by a sperm initiates cortical granule exocytosis, which releases proteases that cleave the proteins linking the vitelline envelope to the plasma membrane (41). At the same time, mucopolysaccharides released by the granules form an osmotic gradient, causing water to enter and swell the space between the vitelline envelope and the plasma membrane (reviewed in ref. 42). Therefore, one intriguing model is that the sqv mutants fail to release GAGs during cortical granule exocytosis, thereby reducing the osmotic gradient and the separation of the plasma membrane and eggshell.
Could the lack of separation between the plasma membrane and eggshell be the cause of the cytokinesis defect? Perhaps this lack of separation causes an abnormal adhesion between the eggshell and the plasma membrane, preventing constriction of the plasma membrane. Another possibility is that osmotic pressure is required in the space between the plasma membrane and eggshell for efficient constriction of the plasma membrane. It is also possible that because heparan sulfate GAGs can act as coreceptors for several signaling molecules, such as Wingless, Hedgehog, and FGF (reviewed in ref. 43), if the initiation of cytokinesis requires such a signaling molecule, the GAG biosynthetic defects of sqv mutants may prevent signaling. Because the vulval and embryonic defects of the sqv mutants both involve failures to form a fluid-filled extracellular space (between the anterior and posterior halves of the vulva and between the cell membrane and eggshell, respectively), we speculate that these defects arise through the same mechanism. An alteration in the properties of the extracellular matrix, possibly involving interaction with water, may well be responsible.
Mutations in the human homolog of sqv-3 are implicated as the cause of a progeroid variant of the connective-tissue disorder Ehlers-Danlos syndrome (14, 44). Ehlers-Danlos syndrome is a group of heritable disorders characterized by hyperelasticity of the skin and hypermobile joints. A skin culture from a patient with mutations in a sqv-3 homolog was reduced in galactosyltransferase activity and in glycosaminoglycan chains on a dermatan sulfate proteoglycan, decorin (45). sqv-1, as well as sqv-7 and sqv-8 also have close human counterparts and likely act in a corresponding pathway for the biosynthesis of GAGs. We propose that defects in the human counterparts of most and maybe all sqv genes can cause aging disorders and connective tissue diseases such as Ehlers-Danlos syndrome.
Acknowledgments
We thank Beth Castor for help with DNA sequencing, Andy Fire for heat-shock and GFP vectors, the Caenorhabditis Genetics Center for the mes-6(bn66) dpy-20(e1282)/nT1(n754) and fem-3(q20) strains, and Min Han for the unc-24(e138) sqv-1(ku246) strain. We are grateful to Lijuan Zhang, Mirek Lech, and Robert Rosenberg for the use of and assistance with the HPLC and mass spectrometer, which were maintained by National Institutes of Health Grants HL66105 and HL41484 (to R. Rosenberg). We thank Brad Hersh and Mike Hurwitz for critical reading of this manuscript. This work was supported by National Institutes of Health Grant GM24663. H.R.H. is an Investigator for the Howard Hughes Medical Institute.
Abbreviations
GAG, glycosaminoglycan
Mel, maternal-effect lethal
MBP, maltose-binding protein
TDP, ribosylthymine 5′-diphosphate
References
- 1.Binari R. C., Staveley, B. E., Johnson, W. A., Godavarti, R., Sasisekharan, R. & Manoukian, A. S. (1997) Development (Cambridge, U.K.) 124, 2623-2632. [DOI] [PubMed] [Google Scholar]
- 2.Haerry T. E., Heslip, T. R., Marsh, J. L. & O'Connor, M. B. (1997) Development (Cambridge, U.K.) 124, 3055-3064. [DOI] [PubMed] [Google Scholar]
- 3.Hacker U., Lin, X. & Perrimon, N. (1997) Development (Cambridge, U.K.) 124, 3565-3573. [DOI] [PubMed] [Google Scholar]
- 4.Lin X. & Perrimon, N. (1999) Nature 400, 281-284. [DOI] [PubMed] [Google Scholar]
- 5.Bellaiche Y., The, I. & Perrimon, N. (1998) Nature 394, 85-88. [DOI] [PubMed] [Google Scholar]
- 6.Nusse R. & Varmus, H. E. (1992) Cell 69, 1073-1087. [DOI] [PubMed] [Google Scholar]
- 7.Reichsman F., Smith, L. & Cumberledge, S. (1996) J. Cell Biol. 135, 819-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin X., Wei, G., Shi, Z., Dryer, L., Esko, J. D., Wells, D. E. & Matzuk, M. M. (2000) Dev. Biol. 224, 299-311. [DOI] [PubMed] [Google Scholar]
- 9.Walsh E. C. & Stainier, D. Y. (2001) Science 293, 1670-1673. [DOI] [PubMed] [Google Scholar]
- 10.Herman T., Hartwieg, E. & Horvitz, H. R. (1999) Proc. Natl. Acad. Sci. USA 96, 968-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herman T. & Horvitz, H. R. (1999) Proc. Natl. Acad. Sci. USA 96, 974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kjellen L. & Lindahl, U. (1991) Annu. Rev. Biochem. 60, 443-475. [DOI] [PubMed] [Google Scholar]
- 13.Okajima T., Yoshida, K., Kondo, T. & Furukawa, K. (1999) J. Biol. Chem. 274, 22915-22918. [DOI] [PubMed] [Google Scholar]
- 14.Almeida R., Levery, S. B., Mandel, U., Kresse, H., Schwientek, T., Bennett, E. P. & Clausen, H. (1999) J. Biol. Chem. 274, 26165-26171. [DOI] [PubMed] [Google Scholar]
- 15.Berninsone P., Hwang, H. Y., Zemtseva, I., Horvitz, H. R. & Hirschberg, C. B. (2001) Proc. Natl. Acad. Sci. USA 98, 3738-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang H.-Y. & Horvitz, H. R. (2002) Proc. Natl. Acad. Sci. USA 99, 14224-14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner S. (1974) Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riddle D. L., Blumenthal, T., Meyer, B. J. & Priess, J. R., (1997) C. elegans II (Cold Spring Harbor Lab. Press, Plainview, NY). [PubMed]
- 19.Herman R. K. (1978) Genetics 88, 49-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodgkin J., Doniach, T. & Shen, M. (1985) Cold Spring Harbor Symp. Quant. Biol. 50, 585-593. [DOI] [PubMed] [Google Scholar]
- 21.Ferguson E. L. & Horvitz, H. R. (1985) Genetics 110, 17-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emmons S. W., Yesner, L., Ruan, K. S. & Katzenberg, D. (1983) Cell 32, 55-65. [DOI] [PubMed] [Google Scholar]
- 23.Liao L. W., Rosenzweig, B. & Hirsh, D. (1983) Proc. Natl. Acad. Sci. USA 80, 3585-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen G., Hazendonk, E., Thijssen, K. L. & Plasterk, R. H. (1997) Nat. Genet. 17, 119-121. [DOI] [PubMed] [Google Scholar]
- 25.Liu L. X., Spoerke, J. M., Mulligan, E. L., Chen, J., Reardon, B., Westlund, B., Sun, L., Abel, K., Armstrong, B., Hardiman, G., et al. (1999) Genome Res. 9, 859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mello C. C., Kramer, J. M., Stinchcomb, D. & Ambros, V. (1991) EMBO J. 10, 3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J., Fritsch, E. F. & Maniatis, T., (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 28.Lennon G., Auffray, C., Polymeropoulos, M. & Soares, M. B. (1996) Genomics 33, 151-152. [DOI] [PubMed] [Google Scholar]
- 29.Okkema P. G. & Fire, A. (1994) Development (Cambridge, U.K.) 120, 2175-2186. [DOI] [PubMed] [Google Scholar]
- 30.Stringham E. G., Dixon, D. K., Jones, D. & Candido, E. P. (1992) Mol. Biol. Cell 3, 221-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuberan B., Lech, M., Zhang, L., Wu, Z. L., Beeler, D. L. & Rosenberg, R. D. (2002) J. Am. Chem. Soc. 124, 8707-8718. [DOI] [PubMed] [Google Scholar]
- 32.The C. elegans Sequencing Consortium (1998) Science 282, 2012-2018. [DOI] [PubMed] [Google Scholar]
- 33.Bar-Peled M., Griffith, C. L. & Doering, T. L. (2001) Proc. Natl. Acad. Sci. USA 98, 12003-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulik D. A., Wei, G., Toyoda, H., Kinoshita-Toyoda, A., Waldrip, W. R., Esko, J. D., Robbins, P. W. & Selleck, S. B. (2000) Proc. Natl. Acad. Sci. USA 97, 10838-10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kearns A. E., Vertel, B. M. & Schwartz, N. B. (1993) J. Biol. Chem. 268, 11097-11104. [PubMed] [Google Scholar]
- 36.Vertel B. M., Walters, L. M., Flay, N., Kearns, A. E. & Schwartz, N. B. (1993) J. Biol. Chem. 268, 11105-11112. [PubMed] [Google Scholar]
- 37.Selva E. M., Hong, K., Baeg, G. H., Beverley, S. M., Turco, S. J., Perrimon, N. & Hacker, U. (2001) Nat. Cell Biol. 3, 809-815. [DOI] [PubMed] [Google Scholar]
- 38.Schiller J. G., Bowser, A. M. & Feingold, D. S. (1973) Biochim. Biophys. Acta 293, 1-10. [DOI] [PubMed] [Google Scholar]
- 39.Campbell R. E., Sala, R. F., van de Rijn, I. & Tanner, M. E. (1997) J. Biol. Chem. 272, 3416-3422. [DOI] [PubMed] [Google Scholar]
- 40.Perozich J., Leksana, A. & Hempel, J. (1995) Adv. Exp. Med. Biol. 372, 79-84. [PubMed] [Google Scholar]
- 41.Vacquier V. D., Tegner, M. J. & Epel, D. (1973) Exp. Cell Res. 80, 111-119. [DOI] [PubMed] [Google Scholar]
- 42.Austin C. R., (1965) Fertilization (Prentice–Hall, Englewood Cliffs, NJ).
- 43.Perrimon N. & Bernfield, M. (2000) Nature 404, 725-728. [DOI] [PubMed] [Google Scholar]
- 44.Okajima T., Fukumoto, S., Furukawa, K. & Urano, T. (1999) J. Biol. Chem. 274, 28841-28844. [DOI] [PubMed] [Google Scholar]
- 45.Quentin E., Gladen, A., Roden, L. & Kresse, H. (1990) Proc. Natl. Acad. Sci. USA 87, 1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]