Abstract
A large number of hDAF transgenic pigs to be used for xenotransplantation research were generated by using sperm-mediated gene transfer (SMGT). The efficiency of transgenesis obtained with SMGT was much greater than with any other method. In the experiments reported, up to 80% of pigs had the transgene integrated into the genome. Most of the pigs carrying the hDAF gene transcribed it in a stable manner (64%). The great majority of pigs that transcribed the gene expressed the protein (83%). The hDAF gene was transmitted to progeny. Expression was stable and found in caveolae as it is in human cells. The expressed gene was functional based on in vitro experiments performed on peripheral blood mononuclear cells. These results show that our SMGT approach to transgenesis provides an efficient procedure for studies involving large animal models.
Keywords: SMGT, hyperacute rejection, swine
Transgenic animals have for the most part been produced by using microinjection of exogenous DNA into male pronuclei of the zygote (1). This technique, although highly successful in mice, is not as efficient in farm animals, limiting its general utility (2, 3).
We have pursued an alternative procedure for producing transgenic animals: sperm-mediated gene transfer (SMGT) (4). We show that sperm cells bind and internalize exogenous DNA, giving them the dual function of acting as a vector for transmitting not only their own but also exogenous DNA (5–8). Sperm cells harboring the gene of interest can be used by in vitro fertilization or by artificial insemination to yield transgenic animals (4, 9, 10).
Because of our initial description of the technique in mice, successful SMGT experiments have been reported in several species, ranging from echinoids to mammals, although large animal studies have been limited to demonstrating the presence of the gene of interest in the DNA of the transgenic animals (2, 11, 12).
Given our interest in xenotransplantation and the possibility that a donor animal will need to express several transgenes (13, 14), we have used SMGT to produce pigs transgenic for human decay accelerating factor (hDAF), which has been shown to help overcome hyperacute rejection of a pig organ by nonhuman primates (15). The hDAF transgenic pigs can now be used to create transgenic pigs with multiple transgenes, each of which contributes to overcoming different aspects of the rejection responses.
We show here that SMGT is highly efficient for generating transgenic pigs, and that the transgene is not only integrated into the DNA but is also transcribed, translated, functional, and transferred to the next generation. We have developed an efficient procedure that, when applied to farm animals, has two additional advantages: low cost and ease of use.
The ability to create transgenic large animals by SMGT with high yields of positive founders at relatively low cost compared with microinjection is important when one needs to create a multitransgene animal. The method will also be of benefit to those whose work requires the use of transgenic large animal models in medicine and animal biotechnology.
Materials and Methods
Animals.
Animal use met local, national, and European Economic Community guidelines. Tissue harvest was performed under general anesthesia. Prepubertal synchronized [1,250 units of eCG; 60 h later, 750 units of hCG (Intervet, Holland)] gilts (Large White or Large White × Landrace) were inseminated 43 h after hCG injection by using 1–1.5 × 109 DNA-treated sperm cells per gilt from selected, trained boars (Landrace) that had abstained for 4–5 days.
Preparation of Sperm.
After evaluation, semen from the selected pig donors was collected (33). Seminal fluid was removed by washing sperm in swine fertilization medium [SFM; 1 liter contains 11.25 g glucose, 10 g sodium citrate (2⋅H2O), 4.7 g EDTA (2⋅H2O), 3.25 g citric acid (H2O), 6.5 g Trizma adjusted to pH 7.4] supplemented with 6 mg/ml BSA prewarmed to 37°C. After this treatment, the medium was kept at room temperature (RT). Semen was incubated for 5 min, transferred to 50-ml tubes (SFM/BSA), spun at 800 × g for 10 min (25°C), and supernatants were aspirated, sperm was suspended and spun again at 800 × g for 10 min (17°C) and discarded. Sperm cells were resuspended and counted by using a hemocytometric chamber.
Sperm/DNA Uptake and Artificial Insemination.
Washed sperm cells (109) were diluted to 120 ml SFM/BSA (17°C). XhoI linearized plasmid DNA (0.4 μg per × 106 sperm) was added for 2 h (17°C). The flask was inverted every 20 min to prevent sedimentation of sperm. The final 20 min of incubation was at RT with heating (37°C) for 1 min before artificial insemination (performed by standard procedures).
DNA and RNA Extraction and Blotting.
Genomic DNA for Southern blots and RNA from snap-frozen tissues for Northern blots were studied according to standard protocols. The probe used was the entire hDAF minigene. Under our high stringency experimental conditions, we did not observe any hybridization in DNA samples from control animals.
PCR and RT-PCR Analysis.
One microgram of genomic DNA was amplified for both the coding and the regulatory regions. Primers (GIBCO/BRL): hDAF, 5′-CTGCTGCTGGTGCTGTTGTG-3′ (forward), 5′-TAGCGTCCCAAGCAAACCTG-3′ (reverse) amplify the hDAF minigene (1,575-bp fragment); LTR-RSV, 5′-TTCGCGGATGTACGGGCCAGA-3′ (forward), 5′-TTGGAGGTGCACACCAATGT-3′ (reverse) amplify a 402-bp fragment; PCR was driven by r-Tth TaqXL and Amplitaq Gold DNA polymerase (Perkin–Elmer). PCR reactions were conducted in triplicate. Of the total RNA, 3 μg was reverse transcribed by using Superscript II RT (GIBCO/BRL). Of the cDNA obtained, 1/10 was amplified by using the hDAF primers. PCR products were analyzed on agarose gels and by Southern blotting. The RT-PCR experiments were subjected to the routine controls. The primers used amplified fragments of 1,575 and 1,059 bp from the hDAF minigene and the cDNA, respectively, and discriminated between amplification of cDNA or of transgenic DNA on the basis of expected size. The risk of contaminating genomic DNA coamplification was also ruled out by running the PCR reactions without prior reverse transcription. Furthermore, as an additional control, a PCR reaction used the primers that amplified the LTR-RSV. All of the controls gave negative results.
Fluorescence in Situ Hybridization (FISH).
In situ hybridization was performed on metaphase chromosomes prepared from PHA-activated pig lymphocytes (16, 17). A biotin-labeled hDAF probe (2 kb) was generated from the RSV-hDAF plasmid (PCR). Primers: 5′-ACTAGTGGATCTTTGTTCTAACC-3′ (forward), 5′-GGAATTCTAGATCCTTGGCTAAG-3′ (reverse) (GIBCO/BRL). The negative control used metaphase plates from lymphocytes of control animals, and the positive control used the All Human Centromeres (α-satellite) probe digoxigenin-labeled (P5095-DG.5, Oncor).
hDAF Immunohistochemistry.
Immunohistochemistry was performed on frozen tissues (7-μm thick) fixed with 2% (wt/vol) paraformaldehyde/1% (vol/vol) acetic acid in PBS for 10 min at RT. Seven anti-hDAF mAbs were used at 1:50 dilution: IA10 (PharMingen), Bric110, Bric216, Bric220, Bric230 (International Blood Group Reference Laboratory, Bristol, UK), IC-6 (Wako Chemicals, Richmond, VA) and IF-7 (Upstate Biotechnology, Lake Placid, NY). Isotype-matched IgG mAbs (Sigma) were used as controls. Detection was by a two-step protocol with a LSAB-2 kit. Staining was revealed by using 3-amino-9-ethylcarbazole (DAKO) for 15 min. Aqueous hematoxylin/eosin (H&E) was used for counterstaining. Staining intensity was measured by an imaging analyzer (Image Pro-Plus). Results are given as the mean percentage of cells showing positivity ± SD.
Low-Density Triton-Insoluble (LDTI) Fraction Recovery and Western Blotting.
Heart biopsies (0.5 μg) were frozen in liquid nitrogen. Tissues were ground for 5 min at medium frequency in a Spex/6700 Freezer Mill apparatus (Glen Creston, U.K.), resuspended in 3 ml of MBS-Triton, and a discontinuous MBS-sucrose gradient was generated, as described (18). After ultracentrifugation, twelve 375-μl fractions were sequentially recovered, and the LDTI material was included in fractions IV to VI, which correspond to the 5–30% interface of the gradient. Protein content was determined by Folin's method. Fractions were electrophoresed and Western blotted (8). The mAbs used were IA10 anti-hDAF (1:500) and anti-caveolin 3 (1:5,000, Transduction Laboratories, Lexington, KY). Blots were analyzed by chemiluminescence (Amersham Pharmacia).
hDAF Protein Expression in Red Blood Cells.
Pig erythrocyte suspension from heparin-treated pig blood (500 μl) was resuspended in 1:2 (vol/vol) MBS-Triton and processed with 15 strokes in a tight-fitting Dounce homogenizer (Wheaton Scientific) on ice. Protein lysates were analyzed by Western blots (8). An identical amount of protein, as determined by Folin's method, was loaded onto each lane; protein transfer was controlled by Ponceau red staining.
Macrophage Resistance to Challenge with Human Serum in Vitro.
Peripheral blood mononuclear cells obtained by lympholyte-H separation (Cedarlane Laboratories) from heparin-treated blood were suspended at 4 × 106 cells per ml in RPMI medium 1640 plus 10% (vol/vol) FCS (GIBCO/BRL) and allowed to adhere to glass slides for 1 day. Human serum (25% fresh or heat-inactivated) was added to the adherent cells. The control was 10% (vol/vol) FCS RPMI medium 1640. The number of viable adherent cells was evaluated by Trypan blue dye exclusion at times 0, 10, 30, 60, and 120 min, in triplicate for each time point.
Results
Sperm–DNA Interaction.
Sperm–DNA interaction is the key step for the outcome of the SMGT procedure. The amount of exogenous DNA incorporated by sperm cells varies in different animals and under different experimental conditions, and therefore selection of sperm donors and optimization of DNA uptake was performed (M.L., M.F., M.L.B., C.D.S., V. Varzi, H.J.W., and E.S., unpublished work). We found a direct correlation between the quality of sperm, evaluated by standard procedures (19), and the outcome of the sperm–DNA interaction. In this study, 2 of 20 boars studied were used as sperm donors.
Production of hDAF Transgenic Pigs by SMGT.
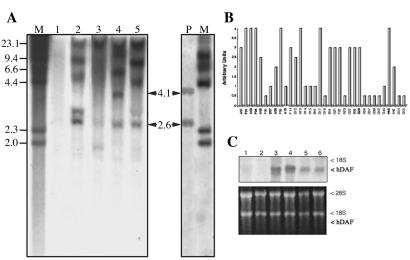
Sperm cells from a selected boar were used as vectors for transferring a 6.8-kb construct containing an hDAF minigene into eggs by artificial insemination (Fig. 1; ref. 20). Eight separate fertilizations performed over 18 months in 15 gilts generated 93 piglets that were studied by Southern blotting, PCR, RT-PCR, and Northern blot analyses. DNA from tail and ear tissues from 53 of the 93 animals (57%) contained hDAF sequences. In six fertilizations, 46–88% of pigs were positive for hDAF; two fertilizations gave negative results. PCR was performed separately for hDAF and RSV regulatory regions. There was 100% concordance between the presence of hDAF and of RSV promoter sequences in two repeat experiments performed on the same biopsies with different DNA extractions. Fig. 2A shows Southern blots of HindIII-digested DNA from one control, four transgenic animals, and, as a positive control, DNA from a nontransgenic pig mixed with the RSVhDAF plasmid. HindIII cuts twice in the RSVhDAF plasmid, giving rise to two bands of 2,655 bp and 4,125 bp. Lane 2 shows bands of >23, 9.1, 5.2, 3.1, and 2.65 kb; lane 3 shows bands of >23, 2.65, and 1.9 kb; lane 4 shows bands of >23, 12, 6.3, 4.1, and 2.65 kb; and lane 5 shows bands of >23, 8.25, 4.4, and 2.65 kb. No bands are visible in lane 1. The 2.65-kb band corresponds to the internal band of the RSVhDAF construct. The other bands represent fragments generated by HindIII digestion of transgenic pig DNA; their number and sizes are consistent with the bands predicted, considering all of the patterns (head to tail, head to head, or tail to tail) generated by transgene integration in single or multiple sites. Northern blots and RT-PCR were performed on RNA samples from pigs positive for DNA. The hDAF transgene was transcribed in all tissues tested in 34/53 (64%) of the animals. Tissues found positive at birth were still positive 6–12 months later. hDAF mRNA expression varied considerably between animals (Fig. 2 B and C) and in different tissues of the same animal (21), as is also seen with microinjection. The highest expressing pigs at birth had eightfold more hDAF mRNA on average than the lowest expressing pigs. Northern blots of nontransgenic pig tissues prepared in an identical manner were negative.
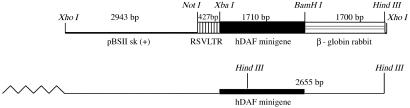
Fig 1.
Map of the RSVhDAF construct. RSVhDAF is 6,780 bp long, including the hDAF minigene (1,710 bp, containing the first exon, first intron, second exon, followed by the cDNA-encoding exons III–XI, with the exception of exon X, which contains Alu sequences), the LTR-RSV regulatory region, the rabbit β globin, and the pBSII sk(+) vector. Cloning sites are shown above the schematic. The construct was linearized with XhoI. HindIII digestion of RSVhDAF yields two bands, 2,655 bp and 4,125 bp.
Fig 2.
DNA and RNA analysis of hDAF genetically modified pigs. (A) Southern blots of DNA from the tail of four transgenic pigs: D12, H36, F04, and D07 (lanes 2–5). DNA from a control pig (lanes 1) and DNA from control pig plus RSVhDAF plasmid (line P). Fifteen micrograms of DNA were digested with HindIII, run on 1% agarose gels, and blotted and probed with the hDAF minigene. The two bands generated by HindIII are in lane P. After HindIII digestion of the transgenic pig genomic DNA, the following pattern can be predicted to occur: (i) 1 integration site, 1 copy: an internal 2,655-bp band and one band of >4,125 bp; (ii) 1 integration site, 2 copies, head-to-tail orientation: two copies of the 2,655-bp band plus one 4,125-bp band and one band of >4,125 bp; (iii) 1 integration site, 2 copies, head-to-head orientation: two copies of the 2,655-bp band and one 8,250-bp band; (iv) 1 integration site, 2 copies, tail-to-tail orientation: two copies of the 2,655-bp band and two bands of >4,125 bp; (v) more than one integration site, 1 copy in each: multiple copies of the 2,655-bp band and multiple bands of >4,125 bp of different sizes. Combinations of the above cases are also possible. A further degree of complexity can result from rearrangements of DNA after integration. Lane 1: no bands are visible in the negative control. Lane 2: pattern is compatible with array 5 or with a combination of patterns 4 and 5, as described above, and also presents a rearranged band (3.1 kb). Lane 3: pattern is compatible with array 1 plus a rearranged band (1.9 kb). Lane 4: pattern is compatible with a combination of 2 and 5. Lane 5: pattern is compatible with a combination of patterns 3 and 4 or 3 and 5. M: HindIII-digested λ. (B) The histogram shows RNA analysis by Northern blot in the genetically modified pigs using the hDAF minigene as a probe. The intensities of the hDAF transcript bands were quantified with a Molecular Image FX Documentation System (Bio-Rad). The mean of three analyses (one liver and two ear samples) was plotted as arbitrary units standardized on the 28S and 18S RNA from each sample. Only ear sections were analyzed in pigs D54 and D55. Labels in bold correspond to animals selected for further breeding. (C) Northern blots from two representative animals: A01, a high expresser (lanes 3 and 4) and D18, a low expresser (lanes 5 and 6) and a control pig (lanes 1 and 2). Lanes 1, 3, and 5 are from ear samples; lanes 2, 4, and 6 are from the liver. Ten micrograms of RNA per lane were run onto a 1% formaldehyde agarose gel, blotted, and hybridized with the hDAF minigene. The positions of hDAF RNA and the ribosomal RNA are shown. The size of the hDAF transcript was evaluated with respect to 18S and 28S rRNA and to a 0.24- to 9.5-kb RNA ladder (not shown). Ethidium bromide staining of 28S and 18S rRNA is shown to document equal loading.
The Transgene Is Integrated and Transmitted to the F1 Generation.
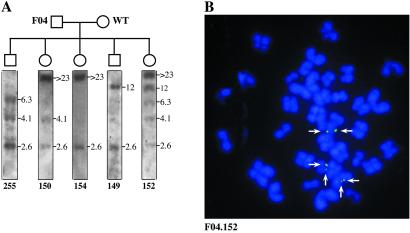
Ten founders expressing different levels of hDAF mRNA (bold in Fig. 2B) generated 205 F1 piglets in 20 litters. All 10 founders produced offspring, and all gave rise to transgenics with frequencies ranging from 20% to 50% (22). Fig. 3A displays the results of germ-line transmission of the RSVhDAF transgene from a founder boar (Fig. 2A, lane 4, F04) to five F1 piglets. The bands in the F1 can be interpreted as representing one or more integrations also present in the founder. F04 gave rise to 38 progeny (25/38 transgenics), of which 5 are shown. Each of the others presented one of the banding patterns shown in the figure. The patterns of piglets 154 and 149 are consistent with one integration site, one copy each; piglets 255 and 150 are consistent with one integration site, two copies with head-to-tail orientation; piglet 152 is consistent with three integration sites, each of which is present in the other piglets.
Fig 3.
F1 transmission and FISH analysis. (A) Germline transmission of the RSVhDAF transgene from a founder boar (Fig. 2A, lane 4, F04) to 5 of 38 F1 piglets of its progeny. DNA was extracted, digested, and Southern blotted as in Fig. 2A. The size of the bands is indicated. (B) FISH analysis on metaphase chromosomes of lymphocytes from F1 transgenic pig 152 (shown in A). Chromosomes clearly show three copies of the integrated transgene. The DNA is counterstained in blue (DAPI); the probe in green (FITC) appears light blue after merging, indicating stable integrations. Two signals per chromosome, one for each sister chromatid, are indicated (arrows).
FISH analysis on F1 piglet 152 is shown in Fig. 3B. Three integration events in different chromosomes are visible. The presence of one spot per sister chromatid, i.e., two adjacent fluorescent signals per chromosome, testify that the transgene is not only stably inserted in the genome but replicates in S phase, thus appearing on FISH as a normal endogenous gene. Integration of the transgene was confirmed by FISH on four founders and nine F1 pigs.
The hDAF Transgene Is Expressed as DAF Protein.
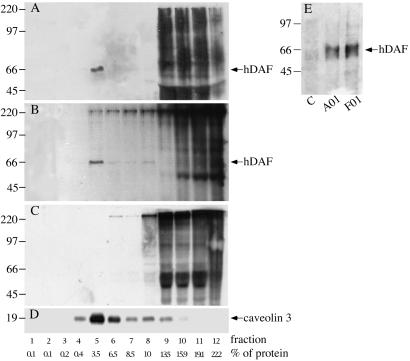
DAF in human cells is located in caveolae, the low-density triton-insoluble (LDTI) plasma-membrane domains, on the cell surface (23). We found that the hDAF transgenic protein was similarly localized in pig cells. Western blots on heart lysates from seven transgenic and two nontransgenic pigs and from a human heart were tested for expression of hDAF protein and for caveolin 3, a marker of caveolae domains, by using mAbs to evaluate whether hDAF was expressed and present in the same protein fractions as caveolin. hDAF was present and migrated similarly (66 kDa) in fraction 5 of heart lysates from transgenic pig B16 (a low-RNA expresser) and from the human heart, but hDAF was not present in nontransgenic heart extracts (Fig. 4 A–C). Caveolin 3 was seen in fraction 5 of human, transgenic, and nontransgenic pig heart, consistent with other reports studying human tissues (19, 23). Thus, hDAF mRNA is translated in pig cells, and the hDAF protein is located on pig membranes similarly to DAF on human cells. The several bands visible throughout fractions 9–12 from human, transgenic, and nontransgenic pig lysates are nonspecific and caused by the high protein content (80–90% of the total protein cell lysate). Red blood cells from two founders analyzed (Fig. 4C) were positive for expression of hDAF, and samples from nontransgenic pigs were negative. Similar results were obtained with samples from platelet-rich plasma from six founders analyzed (data not shown).
Fig 4.
hDAF protein expression in transgenic pigs. Transgenic hDAF protein is present on the plasma-cell membrane and localizes in the caveolin-rich fractions. The low-density triton-insoluble (LDTI) caveolin-rich domains were isolated from human (A), transgenic pig B16 (B and D), and nontransgenic pig (C) hearts. Tissue lysates were subjected to sucrose density centrifugation, and 125-μl aliquots of the 12 gradient fractions under nonreducing conditions were boiled, separated on SDS/10% (hDAF analysis) or 12% (caveolin 3 analysis) PAGE, and probed in a Western blot assay with IA10 anti-hDAF (A–C) and anti-caveolin 3 (D) mAbs. The percentage of total protein present in the fractions is shown. Fraction 5, the richest in caveolin 3, clearly contains the bulk of the hDAF (66-kDa band) in the human and in the transgenic pig hearts. No specific bands are revealed by the antibody in the nontransgenic pig, indicating that the antibody does not cross-react with pig DAF. The mAb IA10 does not cross-react with endogenous porcine DAF. Western blot analysis of lysates from erythrocytes (E). Lane 1: sample from a control pig. Lanes 2 and 3: from red blood cells of transgenic pigs A01 and F01. The mAb used was IA10 anti-hDAF.
The hDAF Transgene Is Expressed in Tissues of Transgenic Pigs.
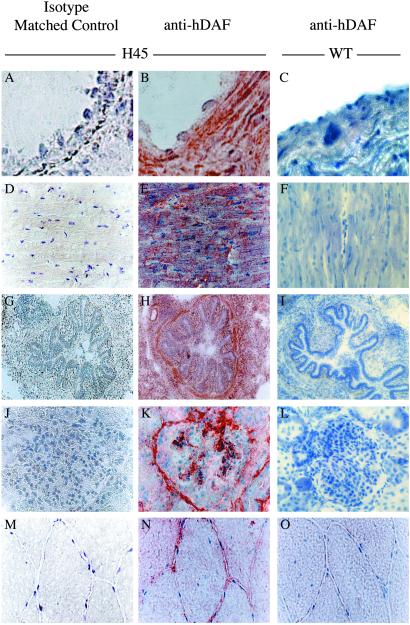
hDAF expression was examined by immunohistochemistry on different organs by using multiple anti-hDAF mAbs, each recognizing a different hDAF protein subdomain SCR1–4 (24); results of one are shown in Fig. 5. All tissues analyzed were positive for hDAF. The strongest expression was observed on endothelial cell plasma membranes (90 ± 5%). hDAF was differentially expressed in various tissues but was expressed homogeneously on the endothelial lamina of all vessels in all organs examined. Evaluation of staining intensity was performed independently and blindly by three pathologists. Of 18 transgenic founder pigs showing different levels of RNA expression, 15 were positive for hDAF by immunohistochemistry. Experiments on 10 positive pigs are reported in Table 1. The pluses indicate the score given by two or three pathologists; in no case did any two pathologists differ by more than a single plus.
Fig 5.
Immunostaining for hDAF on frozen tissue sections from transgenic pigs. Serial sections of aorta (A and B), heart (D and E), lung (G and H), kidney (J and K), and muscle (M and N) from transgenic pig H45 were stained with IF7 anti-hDAF mAb (B, E, H, K, and N) or an isotype matched control (A, D, G, J, and M). The aortic wall shows strong positivity mostly in the extracellular matrix. The heart shows hDAF expression mostly in the plasmalemma of the muscle fibers. In the lung, the protein is expressed at a very high level in all tissues. The vascular tuft of the glomeruli and the vascular spaces of the Bowman capsule of the kidney show strong positivity. In skeletal muscle, hDAF was expressed in the sarcoplasm and on the sarcolemma of fibers in a peripheral location. Sections of aorta, heart, lung, kidney, and muscle from a control pig were stained with the same anti-hDAF mAb (C, F, I, L, and O). Staining was with the ABC method with hematoxylin counterstain. (A–C, ×1,000; D–O, ×400.)
Table 1.
Expression of hDAF in transgenic pig tissues
| Pig | Muscle | Aorta | Lung | Heart | Kidney | Liver |
|---|---|---|---|---|---|---|
| A01 | +++ | +++ | +++ | ++++ | +++ | ++ |
| A03 | +++ | ++ | +++ | +++ | ++ | +++ |
| A05 | ++ | + | ++ | + | + | ++ |
| B16 | ++ | + | + | ++ | + | ++++ |
| B28 | ++++ | ++++ | ++++ | ++++ | ++++ | ++ |
| D17 | ++ | ++ | ++ | + | + | ++ |
| D18 | ++ | + | + | + | + | +++ |
| F01 | ++++ | ++++ | ++++ | +++ | ++++ | ++ |
| F06 | + | ND | + | + | + | + |
| H45 | ++++ | ++++ | ++++ | +++ | +++ | ++++ |
Frozen sections of muscle, aorta, lung, heart, kidney, and liver from 10 transgenic pigs were stained by the immunoperoxidase technique using a panel of anti-hDAF mAbs that recognize different epitopes of the protein. The results are reported as staining intensity. The evaluation was done subjectively and blindly by three different observers using the following criteria: +, weakly positive (vessels, muscle cell membranes); ++, medium positive (vessels, muscle cell membranes); +++, strongly positive (vessels, muscle cell membranes); ++++, very strongly positive (vessels, muscle cell membranes parenchimas and extracellular matrix). ND, not determined.
Function of the hDAF Gene Product.
Pig adherent cells from peripheral blood mononuclear cells obtained from 15 transgenic founders (representative of the varying levels of hDAF RNA expression described in Fig. 2B) and 6 nontransgenic pigs were assessed for their resistance to lysis by antibodies and complement in fresh human serum (HS). The extent of lysis based on the number of surviving cells found at different times after the addition of antibodies and complement in one representative experiment is reported in Table 2. Macrophages of transgenic pig A03 were markedly more resistant to lysis than those of the nontransgenic pig, demonstrating the known function of hDAF. Heat inactivation of serum (HIHS) prevented lysis of the WT pig cells. If results are expressed as the percentage of the number of surviving cells after treatment with HS divided by the number of surviving cells after treatment with HIHS, the results can be summarized as follows. In the cells of the 6 control pigs tested, survival in all cases was <10%, whereas the minimum survival of cells from the 15 transgenic pigs was in all cases >75%, and usually close to 100%. The results obtained with the other 14 transgenic and 5 control animals were essentially identical to those reported in Table 2, notwithstanding the inclusion of cells from animals expressing low levels of hDAF. Consistent with the data presented here are results (unpublished data) on aortic endothelial cells from (i) hDAF transgenic pigs, (ii) control pigs transfected with the hDAF minigene, and (iii) control pigs. Cells in categories i and ii were resistant to lysis by human serum, whereas the cells in category iii were readily lysed.
Table 2.
hDAF confers resistance to complement mediated cell lysis
| Pig
|
Treatment
|
Surviving cells per mm2 culture slides (% of surviving cells) | ||
|---|---|---|---|---|
| 30 min | 60 min | 120 min | ||
| Control | None | 182 ± 13 (99.5) | 118 ± 9 (94.5) | 134 ± 11 (83.2) |
| HS | 2 ± 1 (1.2) | 4 ± 2 (2.4) | 2 ± 1 (1.2) | |
| HIHS | 176 ± 11 (98.3) | 132 ± 12 (93.7) | 135 ± 8 (75.4) | |
| A03 | None | 119 ± 19 (98.1) | 131 ± 16 (97.0) | 100 ± 12 (84.1) |
| HS | 100 ± 11 (84.6) | 132 ± 11 (98.5) | 96 ± 10 (81.6) | |
| HIHS | 122 ± 6 (97.6) | 124 ± 15 (99.2) | 100 ± 11 (80.0) | |
Macrophages from transgenic pigs show increased resistance to challenge with human serum in vitro. Macrophages from a representative animal obtained from 4 × 106 per ml PBMCs cultured in RPMI medium 1640 10% FCS o/n were caused to adhere to tissue culture-treated glass slides and exposed for different lengths of time with RPMI medium 1640 10% FCS (None), RPMI medium 1640 plus 25% fresh human serum (HS) as source of antibodies and active complement, or RPMI medium 1640 plus 25% heat inactivated human serum (HIHS) as toxicity control. Each experimental point was performed in triplicate. After the challenges, the cultures were fixed with acetone and stained with H&E. At least 10 HPF per triplicate were counted with the help of a 1-mm2 grid. The values are reported as a mean ± SD of the viable macrophage number per mm2; the percentage of surviving cells is indicated in parentheses.
Discussion
We describe herein the production of hDAF transgenic pigs that express hDAF by using SMGT. The key feature of this study is that, by this method, we have obtained a large number of pigs that have the hDAF minigene integrated in the genome and that correctly express the hDAF protein on the cell membranes of the desired tissues. The efficiency of transgenesis obtained by SMGT is greatly superior to that obtained by any other method (2, 4, 11, 25).
To date, the production of transgenic animals has relied almost exclusively on the microinjection technique in which the gene of interest is introduced into the male pronucleus of the fertilized eggs. However, that procedure is relatively complex, inefficient, and expensive (3, 25). Cloning technology is at a very early stage and presents difficulties that must be solved before it can be used routinely for large animal transgenesis (2, 26). Several years ago, we described SMGT as an alternative procedure for the production of transgenic animals (4). In this method, the gene of interest is taken up by sperm cells in vitro and subsequently integrated into the sperm DNA (5–8). Successful transgenesis occurs after fertilization by admixture of the DNA-loaded sperm cells with the eggs in vitro or simply by in vivo artificial insemination (4, 9, 10). These studies have been confirmed in a number of laboratories that have reported successful SMGT in fish, honeybee, sea urchin, Xenopus, mouse, chicken, and sheep (11) and, recently, by a technique not significantly different from ours that makes use of intracytoplasmic sperm injection (27).
The pig is the most likely donor animal for xenotransplantation of organs, but may well require multiple transgenes to be a satisfactory donor for humans (28, 29). Given the high efficiency of transgenesis, SMGT could greatly facilitate the production of such pigs.
Given the importance of hDAF in overcoming the first rejection barrier to pig-to-primate transplants, we elected to generate pigs expressing hDAF by SMGT as our starting point for studies on xenotransplantation. The several hDAF transgenic founders and F1 we derived expressed hDAF at the mRNA and protein levels in all tissues examined. The expressed protein confers resistance against the action of human complement. The efficiency of creating transgenic pigs by SMGT is significantly greater than by using microinjection. In eight experiments, 53 of 93 pigs generated were transgenic (57%). This result contrasts with reported efficiencies of 0.5–4% in pigs using microinjection (25), i.e., a 25-fold improvement in efficiency. That the gene detected was hDAF, and not swine DAF, was based on the high stringency used with an hDAF probe, on the absence of any sequence detected in nontransgenic pigs, and was supported by the observation that fragments of different sizes were present in the transgenic founder animals on Southern blots. Atypical banding patterns appear to be a common feature of SMGT, and multiple integrations were the most frequent occurrence in our transgenics. Analysis of the integration sites of transgenic DNA is beyond the scope of this paper; however, it has been noted that, with SMGT in mice, the DNA patterns can indeed be identical in different animals, leading to the suggestion that integration can occur in preferential sites. Many studies have shown that binding, internalization, and the possible integration of exogenous DNA into the sperm are not random events. Specific factors seem to be involved in these processes, supplying an explanation for integration at specific sites (5, 8, 10, 30). Instances in which no transgenics were produced and the variation in efficiency in the successful experiments may be ascribed to problems that arise during the preparation of the sperm, in which damage to the sperm membrane may occur, or during the sperm–DNA interaction step, where residual traces of the protein IF-1 from seminal fluid can be inhibitory (7, 33). The conditions described in Materials and Methods must be followed with great care. It is essential to use fresh sperm because frozen-thawed sperm undergo accelerated capacitation and manifest other functional changes (31).
Not all pigs (34/53) that carried hDAF DNA transcribed the gene, a finding consistent with other reports (32). Protein expression occurred in 15/18 (83%) of those founder pigs examined in which mRNA was detected. hDAF protein was localized in caveolae, as is described for normally expressed DAF protein in humans; i.e., hDAF colocalized with caveolin 3, a marker for caveolae (18, 23). Thus, the hDAF transgene is processed in the pig cells to localize in a manner not different from normal DAF. Moreover, the level of expression of hDAF protein in our transgenic pigs was comparable to the level in human heart, even though the transgenic pig studied in Fig. 4A exhibited low-RNA expression.
Endothelial cells in all tissues examined by immunohistochemistry expressed high levels of hDAF. The expressed hDAF was functional in that hDAF-expressing pig macrophages were resistant to human antibodies and complement (human serum), whereas macrophages from nontransgenic animals were lysed after only 30 min. Also, hearts from transgenic pigs functioned longer and better and with less damage compared with nontransgenic hearts in ex vivo perfusion experiments (M.L. and M. Yacoub, unpublished work).
The results presented here describe the SMGT procedure for creating transgenic large animals, a method we have previously described only for mice (others who confirmed our findings have also primarily used small animals). The possibility of generating transgenic pigs efficiently and reproducibly will, hopefully, not only be an advantage for creating multitransgene pig donor animals, but also enable strategies to fulfill many of the promises originally expected from the introduction of transgenic livestock.
Acknowledgments
We thank Maria Rosaria Rossini for the generous gift of plasmid RSVhDAF; Vincenzo Varzi, Bianca Moioli, Pierino Mattoni, Valerio Faeti, Enrico Poletti, Gianni Marchetto, Alvaro Tattini, and Delia Mandelli for assistance and care of animals; Raffaello Cortesini for support and encouragement; and Pierluigi Donini for helpful discussions and critical reading of the manuscript. This work was supported by Ministero per le Politiche Agricole Grants DM 581/7240/96, 564/7240/97, and 404/7240/99 (to M.L.), Ministero per la Ricerca Scientifica Grant DD 21.09.99, n462 ric. (to M.L.), and by the Consiglio Nazionale delle Ricerche Biotech Target Project. This work is dedicated to the memory of Prof. Dario Alfani, a dear friend and colleague whom we miss enormously.
Abbreviations
SMGT, sperm-mediated gene transfer
RT, room temperature
hDAF, human decay accelerating factor
FISH, fluorescence in situ hybridization
References
- 1.Wall R. J. (2001) Cloning Stem Cells 3, 209-220. [DOI] [PubMed] [Google Scholar]
- 2.Chan A. W. S. (1999) Cloning 1, 25-46. [DOI] [PubMed] [Google Scholar]
- 3.Wolf E., Schernthaner, W., Zakhartchenko, V., Prelle, K., Stojkovic, M. & Brem, G. (2000) Exp. Physiol. 85, 615-625. [PubMed] [Google Scholar]
- 4.Lavitrano M., Camaioni, A., Fazio, V. M., Dolci, S., Farace, M. G. & Spadafora, C. (1989) Cell 57, 717-723. [DOI] [PubMed] [Google Scholar]
- 5.Lavitrano M., French, D., Zani, M., Frati, L. & Spadafora, C. (1992) Mol. Reprod. Dev. 31, 161-169. [DOI] [PubMed] [Google Scholar]
- 6.Francolini M., Lavitrano, M., Lamia, C. L., French, D., Frati, L., Cotelli, F. & Spadafora, C. (1993) Mol. Reprod. Dev. 34, 133-139. [DOI] [PubMed] [Google Scholar]
- 7.Zani M., Lavitrano, M., French, D., Lulli, V., Maione, B., Sperandio, S. & Spadafora, C. (1995) Exp. Cell Res. 217, 57-64. [DOI] [PubMed] [Google Scholar]
- 8.Lavitrano M., Maione, B., Forte, E., Francolini, M., Sperandio, S., Testi, R. & Spadafora, C. (1997) Exp. Cell Res. 233, 56-62. [DOI] [PubMed] [Google Scholar]
- 9.Sperandio S., Lulli, V., Bacci, M. L., Forni, M., Maione, B., Spadafora, C. & Lavitrano, M. (1996) Anim. Biotech. 7, 59-68. [Google Scholar]
- 10.Maione B., Lavitrano, M., Spadafora, C. & Kiessling, A. A. (1998) Mol. Reprod. Dev. 50, 406-409. [DOI] [PubMed] [Google Scholar]
- 11.Wall R. J. (1999) Transgenic Res. 8, 313-315. [DOI] [PubMed] [Google Scholar]
- 12.Rieth A., Pothier, F. & Sirard, M. A. (2000) Mol. Reprod. Dev. 57, 338-345. [DOI] [PubMed] [Google Scholar]
- 13.Osman N., McKenzie, I. F., Ostenried, K., Ioannou, Y. A., Desnick, R. J. & Sandrin, M. S. (1997) Proc. Natl. Acad. Sci. USA 94, 14677-14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowan P. J., Aminian, A., Barlow, H., Brown, A. A., Chen, C. G., Fisicaro, N., Francis, D. M., Goodman, D. J., Han, W., Kurek, M., et al. (2000) Transplantation 69, 2504-2515. [DOI] [PubMed] [Google Scholar]
- 15.Cozzi E., Bhatti, F., Schmoeckel, M., Chavez, G., Smith, K. G., Zaidi, A., Bradley, J. R., Thiru, S., Goddard, M., Vial, C., et al. (2000) Transplantation 70, 15-21. [PubMed] [Google Scholar]
- 16.Renzi L., Pacchierotti, F. & Russo, A. (1996) Mutagenesis 11, 133-138. [DOI] [PubMed] [Google Scholar]
- 17.Trask B. & Pinkel, D. (1990) Methods Cell Biol. 33, 383-400. [DOI] [PubMed] [Google Scholar]
- 18.Lisanti M. P., Scherer, P. E., Vidugiriene, J., Tang, Z., HermanowskiVosatka, A., Tu, Y. H., Cook, R. F. & Sargiacomo, M. (1994) J. Cell Biol. 126, 111-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holt C., Holt, W. V., Moore, H. D., Reed, H. C. & Curnock, R. M. (1997) J. Androl. 18, 312-323. [PubMed] [Google Scholar]
- 20.Mulder L. C., Mora, M., Ciccopiedi, E., Melli, C., Nuti, S., Marinucci, G., Bruzzone, P., Lazzeri, M., Lorenzini, R., Alfani, D., et al. (1995) Transplant. Proc. 27, 333-335. [PubMed] [Google Scholar]
- 21.Cappello F., Stassi, G., Lazzereschi, D., Renzi, L., Di Stefano, C., Marfe, G., Giancotti, P., Wang, H. J., Stoppacciaro, A., Forni, M., et al. (2000) Transplant. Proc. 32, 895-896. [DOI] [PubMed] [Google Scholar]
- 22.Lazzereschi D., Forni, M., Cappello, F., Bacci, M. L., Di Stefano, C., Marfe, G., Giancotti, P., Renzi, L., Wang, H. J., Rossi, M., et al. (2000) Transplant. Proc. 32, 892-894. [DOI] [PubMed] [Google Scholar]
- 23.Parolini I., Sargiacomo, M., Lisanti, M. P. & Peschle, C. (1996) Blood 87, 3783-3794. [PubMed] [Google Scholar]
- 24.Coyne K. E., Hall, S. E., Thompson, S., Arce, M. A., Kinoshita, T., Fujita, T., Anstee, D. J., Rosse, W. & Lublin, D. M. (1992) J. Immunol. 149, 2906-2913. [PubMed] [Google Scholar]
- 25.Niemann H. & Kues, W. A. (2000) Anim. Reprod. Sci. 60–61, 277-293. [DOI] [PubMed] [Google Scholar]
- 26.Dai Y., Vaught, T. D., Boone, J., Chen, S., Phelps, C. J., Ball, S., Monahan, J. A., Jobst, P. M., McCreath, K. J., Lamborn, A. E., et al. (2002) Nat. Biotechnol. 20, 251-255. [DOI] [PubMed] [Google Scholar]
- 27.Perry A. C., Wakayama, T., Kishikawa, H., Kasai, T., Okabe, M., Toyoda, Y. & Yanagimachi, R. (1999) Science 284, 1180-1183. [DOI] [PubMed] [Google Scholar]
- 28.Cozzi E. & White, D. J. (1995) Nat. Med. 1, 964-966. [DOI] [PubMed] [Google Scholar]
- 29.Robson S. C., Schulte, A. M., Esch, J. & Bach, F. H. (1999) Ann. N.Y. Acad. Sci. 875, 261-276. [DOI] [PubMed] [Google Scholar]
- 30.Zoraqi G. & Spadafora, C. (1997) DNA Cell Biol. 16, 291-300. [DOI] [PubMed] [Google Scholar]
- 31.Curry M. R. (2000) Rev. Reprod. 5, 46-52. [DOI] [PubMed] [Google Scholar]
- 32.Cozzi E., Tucker, A. W., Langford, G. A., Pino-Chavez, G., Wright, L., O'Connell, M. J., Young, V. J., Lancaster, R., McLaughlin, M., Hunt, K., et al. (1997) Transplantation 64, 1383-1392. [DOI] [PubMed] [Google Scholar]
- 33.Lavitrano, M., Forni, M., Bacci, M. L., Di Stefano, C., Varzi, V., Wang, H. J. & Seren, E. (2002) Mol. Reprod. Dev., in press. [DOI] [PubMed]