Abstract
RNA interference (RNAi) using double-stranded RNA has been used for the systematic analysis of gene function in invertebrate organisms. Here we have explored the use of short interfering RNA (siRNA) to knock down gene expression during the development of mammalian postimplantation embryos. The developing CNS system of embryonic day 10 mouse embryos was used as a model tissue. siRNA prepared by endoribonuclease digestion (esiRNA) was injected into the lumen of the neural tube at specific regions and delivered into neuroepithelial cells by directed electroporation. Injected and electroporated embryos were grown for 1 day in whole-embryo culture and the effects of RNAi were examined. esiRNA directed against β-galactosidase (β-gal), coelectroporated into neuroepithelial cells together with reporter plasmids expressing GFP and β-gal, abolished expression of β-gal but not GFP, showing the specificity of the esiRNA-mediated RNAi. To demonstrate RNAi of endogenous gene expression, we used heterozygous embryos of a knock-in mouse line expressing GFP from the Tis21 locus, a gene turned on in neuroepithelial cells that switch from proliferation to neurogenesis. GFP-directed esiRNA electroporated into neuroepithelial cells of such embryos blocked the GFP expression normally occurring on the onset of neurogenesis. Taken together, our data indicate that esiRNA delivered in a tissue-specific manner by topical injection followed by directed electroporation can efficiently silence endogenous gene expression in mammalian postimplantation embryos.
Understanding the function of genes requires methods that allow manipulation of gene expression. The discovery that double-stranded RNA (dsRNA) can be used for RNA interference (RNAi) in certain invertebrates and plants has allowed the systematic analysis of gene function in these organisms (1–6). In most mammalian cells, however, dsRNA triggers the IFN response, which leads to general shutdown of gene expression and/or cell death (7). dsRNA is therefore not useful for gene function analysis in most mammalian cells. In contrast, short interfering RNA (siRNA), which can either be added to cells exogenously or produced intracellularly from DNA templates expressing short hairpin RNAs (8–10), does not trigger the IFN response, but it is an efficient mediator of posttranscriptional gene silencing in mammalian cell lines (8, 11–13).
To understand gene function in a physiological context, however, methods are required that allow their investigation in complex systems such as the whole animal. In the mouse, gene function can be studied through gene targeting by using homologous recombination in embryonic stem cells. However, the generation of gene knockout mice is cost, time, and labor intensive. We therefore explored the possibility of using exogenously added siRNA to trigger RNAi in mice, focusing on mouse postimplantation embryos.
Whole-embryo culture supports the normal development of mouse postimplantation embryos for up to 2 days in vitro (14). In addition, whole-embryo culture can conveniently be combined with various methods of introducing foreign DNA into cells of the developing embryo, including electroporation (15–17). By using the neuroepithelium of embryonic day (E) 10 mouse embryos as a model target tissue, we investigated the use of electroporation as a possible means of introducing siRNA, specifically endoribonuclease-prepared siRNA (esiRNA), into neuroepithelial cells in defined regions of the developing CNS, to achieve tissue- and region-specific RNAi during subsequent development in whole-embryo culture.
Methods
Preparation of esiRNA.
esiRNA was prepared as described (13). In brief, ssRNA was obtained from PCR-derived templates carrying T7 and T3 promoters by using the MEGAscript kit from Ambion (Austin, TX). The following primers were used: T7-β-galactosidase (β-gal), 5′-TAATACGACTCACTATAGGGAGAATCGTAATCACCCGAGTGTGA; T3-β-gal, 5′-AATTACCCTCACTAAAGGGAGCCCTAATCCGAGCCAGTTTA; T7-EGFP, 5′-TTAATACGACTCACTATAGGTGAGCAAGGGCGAGGA; and T3-EGFP, 5′-TAATTAACCCTCACTAAAGGGTACAGCTCGTCCATGCCGA.
Annealed dsRNA (100 μg) was digested with 0.2 μg RNase III for 1 h at 20°C. The sample was mixed with 5 vol of PN buffer (Qiagen, Hilden, Germany) and loaded onto a QIAquick column (Qiagen). The flowthrough, containing dsRNA of 15–40 bp, was precipitated with 0.7 vol of 2-propanol. The pellet was washed with 750 μl of 70% ethanol and dissolved in 1 μM EDTA, 10 μM Tris⋅HCl (pH 8.0) to an RNA concentration of 0.5 μg/μl.
Whole-Embryo Electroporation and Culture.
Manipulation of mouse embryos was performed at room temperature in Dulbecco's PBS containing 10% of heat-inactivated FCS and 100 units/ml penicillin/streptomycin. E10 embryos were dissected from the uterine walls and freed of decidua capsularis and Reichert's membrane, and the yolk sac was opened. For injection and electroporation (16), embryos were immobilized in a mould of agarose. By using a glass capillary controlled by a standard micromanipulator (Narishige MN-153, Tokyo) and connected to a pneumatic PicoPump (PV820, WPI Instruments, Waltham, MA), 0.3–0.6 μl (corresponding to ≈1/10 of the luminal volume of the anterior neural tube) of PBS containing 1–4 μg/μl plasmid DNA [pSVpaXΔ expressing β-gal under the control of the early simian virus 40 promoter (18), pEGFP-N2 expressing GFP under the control of the cytomegalovirus promoter (CLONTECH)] and esiRNAs (0.1–0.5 μg/μl) as indicated in the figure legends were injected into different regions of the neural tube or other cavities of the embryo. Immediately after injection, five square electrical pulses of 30 V, 50 ms each at 1-s intervals, were delivered through platinum electrodes (7 mm diameter, 1 cm distance) mounted on plastic tweezers by using a BTX (San Diego) ECM830 electroporator. The orientation of the electric field was used to direct the uptake of the nucleic acids to specific regions of the neural tube or other organs. After electroporation, embryos were placed in a whole-embryo culture incubator (Ikemoto, Tokyo) and allowed to continue their normal development for 24 h in 0.5 ml per embryo of a 2:1 mixture of immediately centrifuged, heat-inactivated mouse serum (Harlan Sera-Lab, Crawley Down, U.K.) and DMEM in a continuous-flow atmosphere of 60% O2, 5% CO2, 35% N2 (50 ml per min) (14).
Light Microscopy.
Embryos were fixed overnight at 4°C in 4% paraformaldehyde in 120 mM phosphate buffer, pH 7.4, equilibrated in 30% sucrose in PBS and embedded in Tissue-Tek. Cryosections (10 μm) were prepared, permeabilized with 0.3% Triton X-100 in PBS, quenched with 10 mM NH4Cl, and subjected to immunohistochemistry according to standard procedures by using mouse monoclonal β-gal antibody (Sigma) and anti-mouse rhodamine-conjugated secondary antibody (Jackson ImmunoResearch) in PBS containing 5% BSA, 5% FCS, and 0.1% Triton X-100. In some experiments, unfixed embryos were embedded in low-melting agarose, and vibratome sections (150 μm) were prepared, analyzed by fluorescence microscopy, fixed, and processed for fluorescence microscopy of cryosections as above. Images were collected by using IPLAB 3.5.1 software, and fluorescence of defined regions was quantified by using IMAGE 1.62.
Results and Discussion
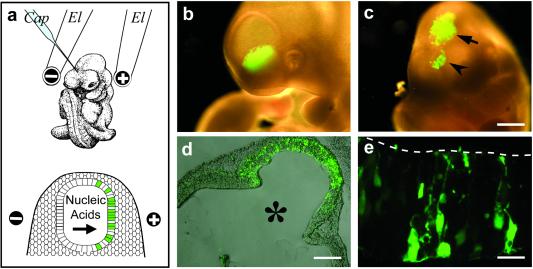
By monitoring GFP expression from a plasmid with a constitutive promoter (pEGFP-N2) as an indicator of cellular nucleic acid uptake, we first examined the organ- and region-specific delivery of nucleic acid upon injection into the lumen of the neural tube at defined positions, followed by electroporation with the electric field in a desired orientation (Fig. 1a). Indeed, analysis of E10 mouse embryos developing for a further 24 h in whole-embryo culture after the injection and electroporation showed that, depending on the segment of the neural tube injected and on the orientation of the electroporation electrodes, GFP expression could be directed to specific regions of the neuroepithelium (Fig. 1 b–e).
Fig 1.
Approach used for tissue-specific RNAi in postimplantation mouse embryos. (a) Cartoon illustrating the region-specific injection (Cap, capillary) of siRNA into the lumen of the neural tube of an E10 mouse embryo, with the electroporation electrodes (El) in the lateral, cathode-right/anode-left orientation (Upper), and the uptake of siRNA into the left side of the neuroepithelium on electroporation (Lower). (b–e) Directed uptake of nucleic acids into neuroepithelial cells on region-specific injection into the lumen of the anterior neural tube of E10 mouse embryos followed by electroporation, exemplified by the use of pEGFP-N2 and expression of GFP on subsequent whole-embryo culture for 24 h. (b and c) Anterior region of whole unfixed embryos showing GFP expression in the left ventral telencephalon on injection into the telencephalic neural tube and electroporation by using the lateral, cathode-right/anode-left orientation (b), and in the dorsal telencephalon (c, arrowhead) and dorsal mesencephalon (c, arrow) on injection into the mesencephalic neural tube and electroporation by using a cathode-caudal/anode-rostral orientation. (Bar = 500 μm.) (d) Coronal-horizontal cryosection through the mesencephalon of the embryo in c showing GFP expression in the neuroepithelium (combined phase-contrast and fluorescence microscopy). The lumen of the neural tube is indicated by *. (Bar = 200 μm.) (e) Higher magnification fluorescence micrograph of a horizontal cryosection through the hindbrain of another embryo on injection into the rhombencephalic neural tube and electroporation by using a cathode-dorsal/anode-ventral orientation, showing GFP expression in individual neuroepithelial cells and neurons derived therefrom. The luminal surface of the neural tube is indicated by the dashed line. (Bar = 10 μm.)
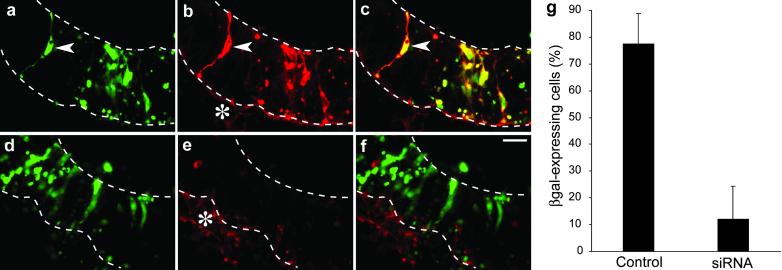
We then investigated the use of siRNA to achieve RNAi on transfected DNA. For this purpose, two viral promoter-based expression vectors (see Methods) were used in combination, one (pEGFP-N2) driving GFP expression as a positive control and the other (pSVpaXΔ) driving β-gal expression as the target of RNAi. The two plasmids were coinjected into the lumen of the telencephalic neural tube of E10 mouse embryos and coelectroporated into the neuroepithelium, and the embryos were allowed to continue normal development in whole-embryo culture for a further 24 h. Analysis of GFP fluorescence and β-gal immunoreactivity showed that the coelectroporation of the two reporter genes was highly efficient because almost all neuroepithelial cells expressing GFP also expressed β-gal (Fig. 2 a–c and g).
Fig 2.
Specificity of esiRNA-mediated RNAi in the neuroepithelium of postimplantation mouse embryos developing in whole-embryo culture. E10 mouse embryos were injected into the lumen of the telencephalic neural tube with the GFP-expressing plasmid pEGFP-N2 plus the β-gal-expressing plasmid pSVpaXΔ either without (a–c and g, Control) or with (d–f and g, siRNA) β-gal-directed esiRNAs, followed by directed electroporation (lateral, cathode-right/anode-left orientation) and whole-embryo culture for 24 h. (a–f) Horizontal cryosections through the left telencenphalon were analyzed by double fluorescence for expression of GFP (green; a and d) and β-gal immunoreactivity (red; b and e). Neuroepithelial cells expressing both GFP and β-gal (arrowheads) appear yellow in the merge (c and f). Note the lack of β-gal expression in neuroepithelial cells in the presence of β-gal-directed esiRNAs. Upper and lower dashed lines indicate the luminal (apical) surface and basal border of the neuroepithelium, respectively. Asterisks in b and e indicate the basal lamina and underlying mesenchymal cells, which cross-react with the secondary antibody used to detect β-gal immunoreactivity. (Bar in f = 20 μm.) (g) Quantitation of the percentage of GFP-expressing neuroepithelial cells that also express β-gal without (Control) or with (siRNA) application of β-gal-directed esiRNAs. Data are the mean of three embryos analyzed as in a–f. (Bars indicate SD.)
We generated endoribonuclease-prepared siRNA (13) covering a 1,365-nt stretch of the β-gal mRNA. In comparison to chemically synthesized siRNAs, esiRNAs have the advantage that screening for effective mediators of mRNA silencing is not necessary because the mixture of different siRNAs usually contains effective molecules. In addition, the use of a mixture of different siRNAs targeted against the same mRNA seems to increase the silencing efficiency (13).
When β-gal-directed esiRNA was included together with the GFP- and β-gal-expression plasmids in the injection and electroporation of the neural tube of E10 mouse embryos, β-gal expression as observed after 24 h of whole-embryo culture was abolished in the vast majority of GFP-positive neuroepithelial cells (Fig. 2 d–g). These results indicate the efficient delivery of esiRNA into neuroepithelial cells and the selective silencing of the targeted mRNA.
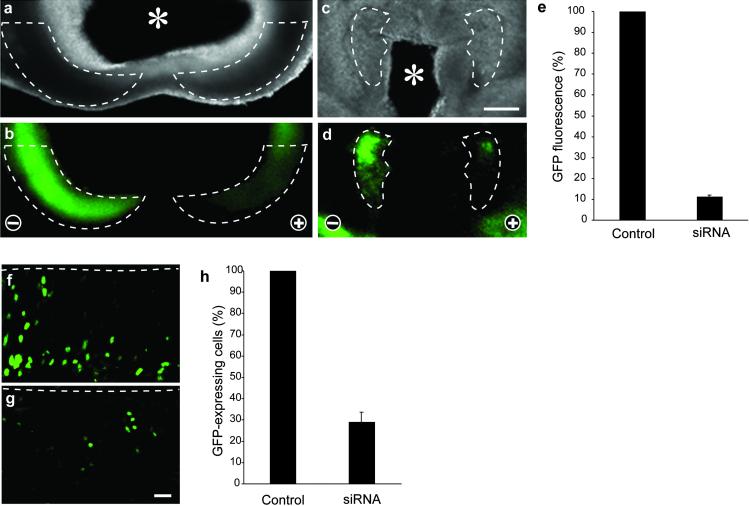
It was important to determine whether esiRNA and the present method of delivery to cells can be used to silence endogenous gene expression. For this purpose, we used (phenotypically WT) heterozygous embryos of a mouse knock-in line expressing GFP (carrying a nuclear localization signal) from the Tis21 locus (Tis21+/tm2(Gfp)Wbh; W.H. and W.B.H, unpublished work). Tis21 is an antiproliferative gene that, during the development of the CNS, is specifically expressed in neuron-generating neuroepithelial cells (19), with an onset of expression in the mouse telencephalon at E10. Hence, to study mRNA silencing, Tis21 is superior to a gene constitutively expressed in the neural tube, whose translation product would be present already before RNAi and might persist, making an evaluation of the effects of siRNA difficult. We therefore investigated the potential use of esiRNA for silencing endogenous gene expression during mouse development by monitoring the appearance of GFP fluorescence in the neuroepithelium of E10 Tis21+/tm2(Gfp)Wbh mouse embryos.
We generated GFP-esiRNA covering virtually the entire coding sequence (712 nt) of GFP (excluding the nuclear localization signal) in the mRNA transcribed from the knocked-in Tis21 locus. This GFP-esiRNA was directed into neuroepithelial cells in specific regions of the developing E10 mouse brain by topical injection into the lumen of the anterior neural tube and directed electroporation as above. The orientation of the electrodes was such (cathode-right/anode-left) that the esiRNA would be electroporated selectively into neuroepithelial cells of the left half of the brain, with the right half serving as an internal positive control for Tis21 locus-driven GFP expression. Analysis of GFP fluorescence after 24 h of whole-embryo culture showed that, indeed, endogenous gene expression was prevented in those segments of the neuroepithelium targeted by the site of esiRNA injection (telencephalon or diencephalon), and that the silencing was selective for the left, anode-facing half of the brain (Fig. 3 a–d). As judged from the GFP fluorescence per area, silencing in the most affected regions was almost complete (Fig. 3e) and even for the entire telencephalon or diencephalon was at least 70% efficient (data not shown). Because the total fluorescence in a given area is the product of the fluorescence per cell times the number of fluorescent cells, the latter number was also quantitated. This quantitation revealed a dramatic decrease in the number of fluorescent neuroepithelial cells in the affected region compared with the corresponding region on the contralateral side (Fig. 3 f–h). However, we also observed that the esiRNA-triggered decrease in the number of fluorescent neuroepithelial cells (Fig. 3h) was slightly less than the reduction in total fluorescence per area (Fig. 3e). Consistent with this observation, the few neuroepithelial cells in a silenced region that still expressed GFP seemed to show, on average, a lower level of fluorescence than the control GFP-expressing neuroepithelial cells, that is, those in the corresponding region on the contralateral side.
Fig 3.
esiRNA-mediated RNAi of a gene endogenously expressed during the development of postimplantation mouse embryos. Heterozygous E10 Tis21+/tm2(Gfp)Wbh mouse embryos were injected with GFP-directed esiRNAs into the lumen of the telencephalic (a and b) or diencephalic (c, d, f, and g) neural tube, followed by directed electroporation (lateral, cathode-right/anode-left orientation) and whole-embryo culture for 24 h. (a–d) Low-power dark-field (a and c) and fluorescence (b and d) micrographs of horizontal vibratome sections through the telencenphalon (a and b) and diencephalon (c and d). The left half of the brain is on the right side of the panels. Note the silencing of GFP expression in defined regions of the neuroepithelium (dashed lines) in the left, anode-facing half of the embryo. Asterisks in a and c indicate the lumen of the neural tube. (Bar in c = 200 μm.) (f and g) Higher-magnification fluorescence micrographs of horizontal cryosections through the right (f, cathode-facing) and left (g, anode-facing) half of the diencephalon at boundary to the telencephalon, prepared from the vibratome section shown in d. Dashed lines indicate the luminal (apical) surface of the neuroepithelium. (Bar in g = 20 μm.) (e and h) Quantitation of GFP fluorescence (e) and GFP-expressing cells (h) in the right (cathode-facing) and left (anode-facing) half of the neuroepithelium. Data in e are the mean of three distinct regions of the neuroepithelium, each from an independent embryo, two of which are the regions shown in b and d. Data in h are the mean of two cryosections prepared from the vibratome section shown in d; the region counted was the entire diencephalic neuroepithelium to the boundary to the telencephalon, including the fields shown in f and g. For each embryo/determination, values obtained for the left (anode-facing) half of the neuroepithelium (siRNA) were expressed as percentage of that for the right (cathode-facing) half (Control); the latter was arbitrarily set to 100. [Bars indicate SD (e) or the variation of the duplicates from the mean (h).]
The percentage of silenced neuroepithelial cells observed on esiRNA-triggered RNAi of endogenous gene expression (Fig. 3h) seemed to be greater than expected from the proportion of neuroepithelial cells transfected on electroporation of the reporter plasmids, which was not more than half of the neuroepithelial cells in a given area (Fig. 2 a and b). This finding may reflect a greater electroporation efficiency because the small esiRNAs may enter cells more easily than large plasmid DNA. Alternatively, the high proportion of silenced cells raises the possibility that some kind of cell-to-cell propagation mechanism of RNAi may exist in mammals, as is known to be the case for systemic RNAi in Caenorhabditis elegans (1, 20).
Our results establish the use of esiRNA for specific mRNA silencing in the mouse embryo and document an approach to perform siRNA-triggered RNAi in a tissue-specific manner. They also extend recent reports (21, 22) showing siRNA-triggered RNAi in adult mice that appeared while the present work was being prepared for publication. Long dsRNA, which trigger the IFN response in most mammalian cells (4), have successfully been used for RNAi only in mouse oocytes and preimplantation embryos (23–25) due to the absence of the IFN response at these early developmental stages. The present demonstration of tissue-specific siRNA-triggered RNAi in postimplantation mouse embryos suggests that this powerful method can be used for investigating the function of individual genes in mammalian development. Our data showing that the knocking down of the protein of interest in the targeted cell population (the neuroepithelial cells) is dramatic (≈90% on average, with the majority of cells showing no detectable expression), but not complete, as would be the case in a knockout mouse, does not invalidate this conclusion because numerous examples exist in which this degree of reduction in gene product has led to a loss-of-function phenotype (26). It will be worthwhile to investigate whether even more efficient knocking down can be achieved by further optimizing the present experimental parameters.
Several aspects of the present experimental approach deserve comment. First, it should be noted that our combination of topical injection of esiRNA, directed electroporation, and whole-embryo culture as an approach to trigger and observe the effects of tissue-specific RNAi is not restricted to the developing CNS but can, in principle, be applied to various organs of the mouse embryo including those lacking internal cavities. In a separate set of experiments, we injected pEGFP-N2 into the cavity of the developing heart of E10 mouse embryos, without or with GFP-directed esiRNA, followed by electroporation and whole-embryo culture for 24 h. This procedure did not have any obvious effects on heart development and functionality. The presence of GFP-directed esiRNA prevented the GFP expression observed in the control heart (data not shown). In addition, nucleic acids have successfully been delivered into target cells by injection directly into the tissue (e.g., muscle) rather than into a cavity surrounded by tissue, followed by electroporation (27).
Second, the present experimental approach is applicable at various developmental stages, because the start of whole-embryo culture of mouse embryos can be any time point between E7 and E12 (14). Moreover, the current limitation of the whole-embryo culture period to about 2 days does not preclude the use of electroporation for tissue-specific delivery of siRNA in long-term studies of mouse development, because electroporation of mouse embryos can also be performed in utero (28, 29). In this context, although the duration of esiRNA-triggered RNAi in the mouse embryo remains to be determined, this process lasted for up to 10 days in mammalian cell lines (13), and in adult mice efficient silencing was seen for at least 4 days after siRNA administration (22). In addition, it may be worth exploring the use of DNA templates expressing short hairpin RNAs (8–10).
Third, we find it likely that mixtures of esiRNAs, targeting the mRNAs of various proteins, can be used at the same time. It is therefore conceivable that with the present approach, genomewide functional screens, such as those initiated in C. elegans by using long dsRNA (2, 3), will now be possible in the mouse to systematically investigate not only neurogenesis but also other aspects of mammalian development.
Acknowledgments
We thank Patricia Vegh for excellent technical assistance. W.B.H. was supported by grants from the Deutsche Forschungsgemeinschaft (SPP 1109, Hu 275/7-1; SPP 1111, Hu 275/8-1) and the Fonds der Chemischen Industrie.
Abbreviations
RNAi, RNA interference
siRNA, short interfering RNA
esiRNA, endoribonuclease-prepared siRNA
β-gal, β-galactosidase
dsRNA, double-stranded RNA
E, embryonic day
References
- 1.Fire A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- 2.Fraser A. G., Kamath, R. S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M. & Ahringer, J. (2000) Nature 408, 325-330. [DOI] [PubMed] [Google Scholar]
- 3.Gönczy P., Echeverri, C., Oegema, K., Coulson, A., Jones, S. J., Copley, R. R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. (2000) Nature 408, 331-336. [DOI] [PubMed] [Google Scholar]
- 4.Hannon G. J. (2002) Nature 418, 244-251. [DOI] [PubMed] [Google Scholar]
- 5.Schmid A., Schindelholz, B. & Zinn, K. (2002) Trends Neurosci. 25, 71-74. [DOI] [PubMed] [Google Scholar]
- 6.Wang M. B. & Waterhouse, P. M. (2002) Curr. Opin. Plant Biol. 5, 146-150. [DOI] [PubMed] [Google Scholar]
- 7.Stark G. R., Kerr, I. M., Williams, B. R., Silverman, R. H. & Schreiber, R. D. (1998) Annu. Rev. Biochem. 67, 227-264. [DOI] [PubMed] [Google Scholar]
- 8.Brummelkamp T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- 9.Paddison P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. (2002) Genes Dev. 16, 948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuschl T. (2002) Nat. Biotechnol. 20, 446-448. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 12.Elbashir S. M., Harborth, J., Weber, K. & Tuschl, T. (2002) Methods 26, 199-213. [DOI] [PubMed] [Google Scholar]
- 13.Yang D., Buchholz, F., Huang, Z., Goga, A., Chen, C. Y., Brodsky, F. M. & Bishop, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 9942-9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cockroft D. L. (1990) in Postimplantation Mammalian Embryos, eds. Copp, A. J. & Cockroft, D. L. (IRL, Oxford), pp. 15–40.
- 15.Oback B., Cid-Arregui, A. & Huttner, W. B. (2000) in Viral Vectors: Basic Science and Gene Therapy, ed. García-Carrancá, A. C.-A. A. (Eaton Publishing, Natick, MA), pp. 277–293.
- 16.Osumi N. & Inoue, T. (2001) Methods 24, 35-42. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi M., Sato, K., Nomura, T. & Osumi, N. (2002) Differentiation (Berlin) 70, 155-162. [DOI] [PubMed] [Google Scholar]
- 18.Buchholz F., Ringrose, L., Angrand, P. O., Rossi, F. & Stewart, A. F. (1996) Nucleic Acids Res. 24, 4256-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iacopetti P., Michelini, M., Stuckmann, I., Oback, B., Aaku-Saraste, E. & Huttner, W. B. (1999) Proc. Natl. Acad. Sci. USA 96, 4639-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winston W. M., Molodowitch, C. & Hunter, C. P. (2002) Science 295, 2456-2459. [DOI] [PubMed] [Google Scholar]
- 21.McCaffrey A. P., Meuse, L., Pham, T. T., Conklin, D. S., Hannon, G. J. & Kay, M. A. (2002) Nature 418, 38-39. [DOI] [PubMed] [Google Scholar]
- 22.Lewis D. L., Hagstrom, J. E., Loomis, A. G., Wolff, J. A. & Herweijer, H. (2002) Nat. Genet. 32, 107-108. [DOI] [PubMed] [Google Scholar]
- 23.Wianny F. & Zernicka-Goetz, M. (2000) Nat. Cell Biol. 2, 70-75. [DOI] [PubMed] [Google Scholar]
- 24.Zernicka-Goetz M. (2000) Nature 405, 733. [DOI] [PubMed] [Google Scholar]
- 25.Svoboda P., Stein, P., Hayashi, H. & Schultz, R. M. (2000) Development (Cambridge, U.K.) 127, 4147-4156. [DOI] [PubMed] [Google Scholar]
- 26.Bargmann C. I. (2001) Genome Biol. 2, 10051-10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aihara H. & Miyazaki, J. (1998) Nat. Biotechnol. 16, 867-870. [DOI] [PubMed] [Google Scholar]
- 28.Tabata H. & Nakajima, K. (2001) Neuroscience 103, 865-872. [DOI] [PubMed] [Google Scholar]
- 29.Saito T. & Nakatsuji, N. (2001) Dev. Biol. 240, 237-246. [DOI] [PubMed] [Google Scholar]