Abstract
Modularity in animal development is thought to have facilitated morphological diversification, but independent change of those traits integrated within a module might be restricted. Correlations among traits describe potential developmental constraints on evolution. These have often been postulated to explain patterns of morphological variation and have been examined theoretically but seldom analyzed experimentally. Here, we use artificial selection to explore the modular organization of butterfly wing patterns and the extent to which their evolution is constrained by the genetic correlations among repeated pattern elements. We show that, in Bicyclus anynana butterflies, despite the evidence that all eyespots are developmentally coupled, the response to selection for increased size of one individual eyespot can proceed in a manner largely independent from selection imposed on another eyespot. We argue that among-eyespot correlations are unlikely to have constrained the evolutionary diversification of butterfly wing patterns but might be important when only limited time is available for adaptive evolution to occur. The ease with which we have been able to produce independent responses to artificial selection on different eyespots may be linked to a legacy of natural selection favoring individuality. Our results are discussed within the context of the evolution of modularity and individuality of serially repeated morphological traits.
The idea that in animals groups of traits are developmentally integrated within modules has been receiving much attention in evolutionary developmental biology (1–5). The modularity of developing organisms has supposedly facilitated independent evolution of groups of traits belonging to different modules, but it also may have led to the concerted evolution of traits within one module (1, 2). Genetic correlations have been extensively documented for many morphological (and other) traits (examples in ref. 6). It is generally accepted that such patterns of covariance represent potential developmental constraints that can limit independent evolutionary change of coupled traits (7–9). Studies of evolutionary constraints arising from developmental coupling have concentrated primarily on the description of the genetic correlations between traits (10, 11) and on theoretical models predicting their effects on evolutionary change (8, 12, 13). Few experimental data exist to directly test such predictions, especially in the context of exploring the underlying developmental mechanisms. Evolutionary developmental biology is beginning to provide an approach to analyzing how development might introduce biases into the production of those phenotypes that become available for natural selection, and thus, for adaptive evolution (14–16). One related issue to which evo-devo can contribute is the study of how serially repeated elements (e.g., vertebrate teeth, arthropod body segments) acquire characteristic properties and differentiate from each other during evolution. The patterns of color on butterfly wings provide ideal material to study morphological integration and the evolution of developmental independence (i.e., individuality) of serially repeated traits.
Butterfly wing patterns are made up of different types of discrete pattern elements often repeated along the anterior-posterior axes of the wings (17). Serially homologous elements show positive correlations in different species, whereas different types of pattern elements seem largely independent (10, 18, 19). Eyespots are common pattern elements composed of concentric rings of different colors. They often have a clear adaptive function and are amenable to detailed developmental characterization (20, 21). Bicyclus anynana butterflies have a series of marginal eyespots on different wing surfaces, each centered in an individualized wing area bordered by veins. It has been shown that high additive genetic variance exists for several features of eyespot morphology in this species (e.g., size and color-composition; refs. 19 and 22). Artificial selection on a single eyespot has consistently produced rapid changes not only for the target eyespot but also for other eyespots, especially on the same wing surface (19, 22). Single mutations also generally affect all eyespots in concert (23). Such positive genetic correlations presumably reflect the shared developmental basis of the different eyespots (19, 21). Indeed, all butterfly eyespots are formed around groups of central organizing cells that show a characteristic expression of several wing patterning genes (21, 24–26). The whole pattern, rather than each individual eyespot, thus seems to constitute a semi-independent developmental module (27), leading to predictions about constraints on the evolution of butterfly wing patterns (23).
How independently can the evolution of individual eyespots proceed? The descriptions of eyespot development and the patterns of genetic variances and covariances within B. anynana raise the prediction that response to selection on one eyespot will be highly dependent on the selection imposed on other eyespots (23). Here, we test this prediction for the size of the two eyespots on the dorsal forewing of these butterflies. These eyespots are characterized by a conserved pattern of relative size (a smaller anterior, and a larger posterior eyespot) and by a positive genetic correlation between the two (19). We use artificial selection as a tool to explore the evolutionary potential available in our outbred stock of B. anynana for independent changes of eyespot size. We compare how readily one eyespot can respond to selection for increased size when the other is (i) also selected for increased size, (ii) under stabilizing selection for size, or (iii) selected for reduced size. Because of the genetic coupling between the two eyespots, we expect to be able to produce concerted changes across eyespots (in i) more readily than when selecting in opposite directions (iii), or when only one is free to change (ii).
Materials and Methods
Artificial Selection on Dorsal Eyespot Size.
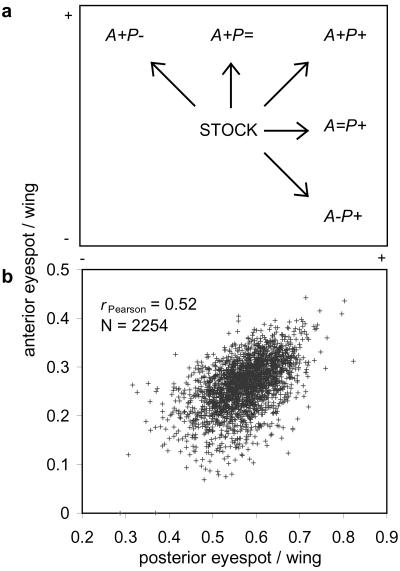
Artificial selection targeted the size of the anterior eyespot (A) and the posterior eyespot (P) on the dorsal forewing of Bicyclus anynana butterflies. We derived different lines selected for increased size of one eyespot but with varying modes of selection imposed on the other: (i) parallel selection for increased size (“+” direction); (ii) stabilizing selection on size (“=” directions); and (iii) antagonistic selection for reduced size (“−” directions; Fig. 1a). We established two replicate lines for each mode of directional selection and three unselected control (UC) lines starting from the same outbred stock population used in previous selection experiments (19, 22, 23, 28, 29). Females (2,254) were measured at G0 (generation 0; Fig. 1b), and 45 of these were randomly selected to produce the next generation of one UC replicate line. The remaining butterflies were split randomly into two groups from which the two sets of replicates for all other directions were derived (first the additional UCs and then the directional selection lines). In subsequent generations, 150–200 females were measured per line. Selected females were mated with about 50 males chosen randomly and allowed to lay eggs. To increase selection intensity, the number of parents was progressively reduced in the course of the experiment (no indication of inbreeding depression on egg-hatching success; ref. 30). From G1 to G5, we selected 40 females per line; from G5 to G8, 35 females were selected; and from G8 to G10, 30 females were selected. Selection was continued for 10 generations, with closely similar intensities across all lines.
Fig 1.
Relative size of the two dorsal forewing eyespots of Bicyclus anynana. (a) Directions of artificial selection imposed on the anterior (A) and posterior (P) eyespots. (b) Distribution of phenotypes in females from the original unselected stock population. All directions of selection were derived from this group, which showed a significant positive phenotypic correlation between the size of the two target eyespots.
Selection Criteria.
Artificial selection targeted the ratios between eyespot diameter and a linear measurement of wing size. To impose selection on both eyespots simultaneously, we have used an additive combination of the rank values for A/wing (RA) and P/wing (RP); RA+RP for the + direction (A+P+) and RA−RP for the − directions (A+P− and A−P+). In each line, individuals with the most extreme values in the desired direction were selected.
To achieve an increase in size of one eyespot while the other maintains the same size (the = directions: A+P = and A=P+), we applied directional selection on one eyespot combined with stabilizing selection on the other. The criterion was to select a group of individuals with extreme values for one eyespot (“eyespot +”), whereas the mean value for the other (“eyespot =”) was as close to the population mean as possible. Choice of parents was done in an iterative manner; at each step, the mean value for eyespot = was calculated for the group of individuals with extreme values for eyespot +. If this mean was larger than that of the parental population (because of the positive correlation between eyespots; Fig. 1b), the individual with the largest value for eyespot = was removed from the selected pool, and the individual with the next highest value for eyespot + was added from the remainder population. This process was repeated until the average value for eyespot = in the selected group was closest to the population mean.
Statistical Analysis.
Statistical tests used the MINITAB statistical package and followed ref. 31. To assess the response to artificial selection across selection directions, we performed ANOVAs on G10 eyespot/wing phenotypes by using the mean values for each of the two (or three for UCs) replicate lines in each direction. ANOVAs were followed by Dunnett's comparisons between each selection direction and the UC values, separately for the two eyespots. Pearson correlation coefficients between A/wing and P/wing values were calculated for the base population (G0) and for each line at G10. Their analysis followed the method for multiple comparisons in ref. 31.
To test for differences across the +, =, and − lines, we compared not only the final phenotypes but also the response to selection relative to the cumulative selection differential (hereafter referred to as rate of response) within each of the two groups of lines, i.e., those selected for a larger anterior eyespot (A+P+, A+P=, and A+P−) and those selected for a larger posterior eyespot (A+P+, A=P+, and A−P+). Differences in phenotype across selection directions were tested with ANOVAs on G10 eyespot/wing values by using the mean values for the two replicate lines in each direction. ANOVAs were followed by Tukey pairwise comparisons. For each direction of selection, least squares regression lines were fitted to the average points for eyespot/wing size (taken as the difference to UC values) on cumulative selection differential. Analysis of covariance (ANCOVA) was used to compare the slopes of the regression lines with direction as a fixed factor and cumulative selection differential as a covariate (31). The ANCOVA, testing for the significance of the interaction between these two factors on eyespot/wing size, was followed by Tukey pairwise comparisons of slopes between directions.
Results
Response to Selection.
The phenotypic variation for dorsal eyespot size in the base population was characterized by a significant positive correlation between the values of eyespot diameter/wing size for the anterior and posterior eyespots (Fig. 1b; rPearson = 0.52 ± 0.02, P < 0.0005). Responses to selection were progressive in all directions and gradually led to butterflies with highly distinct phenotypes (Fig. 2). After 10 generations, there were statistically significant differences in eyespot/wing phenotypes across directions; both for the anterior eyespot [F(5,7) = 79.36, P < 0.0005] and the posterior eyespot [F(5,7) = 82.62, P < 0.0005]. Dunnett's test comparing mean values between each of the directional selection groups and the UCs showed that, for both eyespots at a family error rate of 0.05, all lines had eyespot/wing values significantly different from the UC mean except for the eyespots under stabilizing selection (i.e., posterior eyespot in direction A+P= and anterior eyespot in A=P+).
Fig 2.
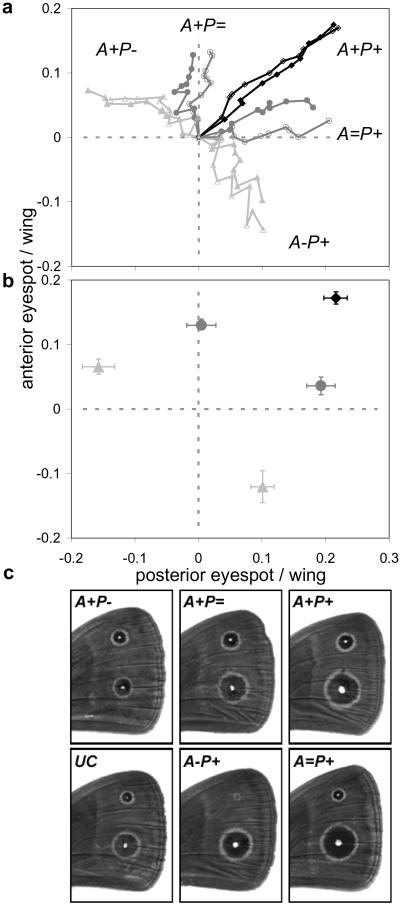
Response to 10 generations of artificial selection on eyespot size. (a) Eyespot/wing size relative to UC values are given for each generation of selection. Each point represents the mean (±SE) for the two target eyespots simultaneously in each generation for each of the two replicate lines. Lines join points for consecutive generations, all starting from the same original population (intersection of dashed lines). (b) Eyespot/wing size at G10 relative to UC values (dashed lines) are given for the different directions of selection. Each point represents the mean (±SE) for the anterior (A) and the posterior (P) eyespots simultaneously for each direction (+ direction in black, = in medium gray, and − in light gray). (c) Illustrative photos of G10 phenotypes in each direction (shown is the distal-most part of the dorsal surface of the right forewing of females).
The phenotypic correlations between the two target traits in the initial population (G0) and all final (G10) selection lines are given in Table 1. Statistical analysis reveals significant differences in phenotypic correlation among lines (original population and all G10 selection lines; χ2 = 42.48, DF = 13, P < 0.0005). This heterogeneity becomes not significant if both A−P+ replicate lines (χ2 = 8.07, DF = 11, P = 0.71), or only replicate one (χ2 = 18.77, DF = 12, P = 0.09), are excluded.
Table 1.
Correlation between eyespot sizes before and after selection
| Line | N | rPearson |
|---|---|---|
| Stock | 2254 | 0.518 |
| A+P− 1 | 174 | 0.531 |
| A+P− 2 | 162 | 0.512 |
| A+P= 1 | 70 | 0.435 |
| A+P= 2 | 70 | 0.539 |
| A+P+ 1 | 172 | 0.585 |
| A+P+ 2 | 183 | 0.520 |
| A=P+ 1 | 70 | 0.425 |
| A=P+ 2 | 70 | 0.429 |
| A−P+ 1 | 184 | 0.175 |
| A−P+ 2 | 186 | 0.306 |
| UC 1 | 147 | 0.598 |
| UC 2 | 167 | 0.423 |
| UC 3 | 147 | 0.501 |
Pearson correlation coefficients are given between eyespot/wing values for the anterior and posterior eyespots in females from the base Stock population (G0) and all selection lines (replicates 1–3) from the last generation (G10). Sample sizes are also given. All correlation coefficients are significantly different from zero with P < 0.0005, except that for line A−P+ 1 with P = 0.017.
Comparing +, =, and − Directions.
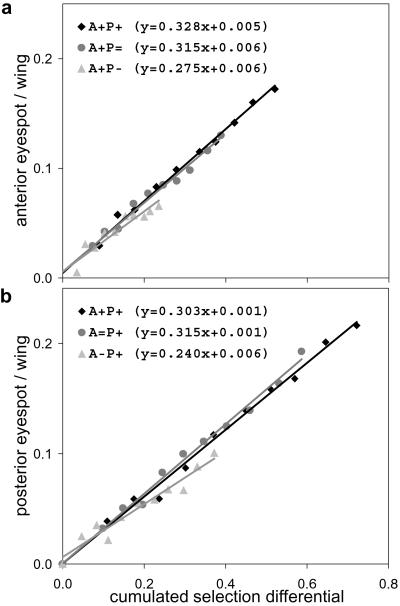
To test how much one eyespot's response to selection depended on the direction of selection imposed on the other eyespot, we made comparisons among the three types of lines selected for larger anterior eyespot (A+P+, A+P=, and A+P−), and among those selected for larger posterior eyespot (A+P+, A=P+, and A−P+). Analysis was made both in terms of G10 phenotypes and of rate of response to selection. After 10 generations of selection, the different directions in each group showed distinct eyespot size phenotypes (Fig. 2; Table 2). The response to selection relative to the cumulative selection differential (Fig. 3) was lower in the − directions than in the + and = directions. By using Tukey's pairwise comparisons below a 0.05 error level, the rate of response to selection for a larger posterior eyespot (P+) was significantly lower in direction A−P+ relative to the two other directions, which did not differ significantly (Fig. 3b; ANCOVA F(2,27) = 7.36, P = 0.003). Although there was a similar trend (i.e., lower rate for −) for the lines selected for a larger anterior eyespot (A+), there were no significant differences in response between the +, =, and − directions (Fig. 3a; ANCOVA F(2,27) = 2.36, P = 0.114). Tukey comparisons revealed no differences between pairs of A+ directions.
Table 2.
Comparison of eyespot diameter/wing size across directions of selection at G10
| Direction (other eyespot)
|
Larger anterior (A+) | Larger posterior (P+) | ||
|---|---|---|---|---|
| A/wing | P/wing | A/wing | P/wing | |
| + | 0.458 ± 0.002 | 0.847 ± 0.004 | 0.458 ± 0.002 | 0.847 ± 0.004 |
| = | 0.416 ± 0.002 | 0.636 ± 0.013 | 0.321 ± 0.010 | 0.824 ± 0.012 |
| − | 0.351 ± 0.007 | 0.474 ± 0.018 | 0.165 ± 0.023 | 0.732 ± 0.001 |
| ANOVA F(2, 3) | 129.67 | 210.61 | 100.18 | 66.12 |
| P value | 0.001 | 0.001 | 0.002 | 0.003 |
Mean phenotypes across the two replicate lines (±SE) are given for the females of the different selection directions. ANOVA F values are given for the comparison of phenotypes among the three groups selected for a larger anterior eyespot (A+P+, A+P=, and A+P−) and among the three groups selected for a larger posterior eyespot (A+P+, A=P+, and A−P+).
Values that are not significantly different under Tukey's pairwise comparisons at a 5% error rate; all other pairs within each column are significantly different.
Fig 3.
Rates of response to selection for increased size of one eyespot under different selection conditions for the other eyespot. (a) Response for a larger anterior eyespot. (b) Response for a larger posterior eyespot. Least squares linear regression lines were fitted for response to selection (measured as eyespot/wing size relative to unselected controls) on cumulated selection differential (see Materials and Methods); equations are displayed (a: R2 = 91–99%; b: R2 = 95–99%; lowest levels for the − directions for both eyespots). All regression coefficients are significantly different from zero, with P < 0.0005.
Discussion
Limitations on independent evolutionary change arising from the genetic and developmental coupling between traits have often been postulated to explain patterns of existing morphological variation but seldom tested directly. Several lines of evidence have suggested that the two eyespots on the dorsal forewing of B. anynana butterflies are coupled in this manner and, thus, are constrained to evolve in concert (23, 27). Genetic correlations between these eyespots are supported by their concerted responses to artificial selection targeting a single eyespot and by the effects of most single mutations on both eyespots (23). In addition, developmental studies of eyespot formation revealed some of the mechanistic basis for this coupling; all eyespots are formed by the same cellular mechanism and coexpress several developmental genes in preadult wing primordia (reviewed in refs. 20 and 21). Here, we have used artificial selection to explore the evolutionary potential available in an outbred laboratory stock of B. anynana for independent changes of individual eyespot size. To what extent is change resulting from selection on one eyespot limited by the type of selection imposed on the other?
Response to Selection on Eyespot Size.
The pattern of variation in dorsal eyespot size in our base B. anynana population is characterized by a positive phenotypic correlation between eyespots (Fig. 1b) and by high levels of additive genetic variance (Figs. 2 and 3). High heritabilities are not uncommon for morphological traits (6) and occur in B. anynana not only for eyespot size (19, 29, 32) but also for other features of eyespot morphology (22). To test how the response to selection on one eyespot was influenced by selection on the other, we have compared the response for increased size of one eyespot in three situations that differed with respect to the selection simultaneously imposed on the other eyespot: (i) concerted selection for increased size (+ direction); (ii) independent stabilizing selection on size (= directions); and (iii) antagonistic selection for decreased size (− directions).
Response to 10 generations of artificial selection yielded highly differentiated phenotypes across directions (Fig. 2, Table 2). The fact that more extreme phenotypes were produced in the + direction (Fig. 2) does not per se reflect the operation of constraints, because of the higher phenotypic variation along this axis (Fig. 1b). When the amount of response to selection relative to the cumulative selection differential was compared across directions, there were no consistent indication of an easier response along the + direction (Fig. 3). There was a lower rate of response in − directions (significant only for the P+ group) but no difference between + and =. In another experiment, we have shown that antagonizing selection (i.e., in different directions for the two target eyespots) can sometimes yield higher rates of response than synergistic selection (29).
These results are not consistent with the prediction that changes would be easier in the + direction, nor with the idea that the developmental and genetic integration between different eyespots significantly constrains wing pattern evolution in Bicyclus butterflies. This flexibility matches well with the diversity of eyespot size patterns found across Bicyclus species (29, 33) and must have facilitated the evolution of the spectacular diversity in butterfly wing patterns (17, 21) and the frequent evolution of remarkably accurate mimicry (34, 35).
Time Frame for the Influence of Constraints.
Our results strongly suggest that it is unlikely that the developmental coupling between eyespots can constrain responses to prolonged periods of natural selection on eyespot size in any direction. The speed of response to selection that is possible in the different directions might, nonetheless, influence the success of adaptive evolution to rapidly changing environments (12, 36). For the same number of generations of selection, our + direction has produced butterflies with more extreme phenotypes (Fig. 2), because there is more phenotypic variation along this axis. For a given proportion of the population being selected, it will clearly take longer to achieve a particular amount of phenotypic change in some directions than in others. Consequently, if there is only a limited time window for selection to be successful, the among-trait variation could influence the chance of successful adaptive evolution. This type of effect has been quantified in a recent study that showed that the rates of evolutionary change in directions antagonistic to among-trait correlations in a prairie plant were predicted to be slower than the expected rate of environmental change caused by global warming (37).
For situations in which time is less limiting, it will always be difficult to make a firm prediction that among-trait correlations will constrain evolutionary change. The patterns of genetic variances and covariances of traits can only be used to predict short-term evolution, because it is clear from different data sets that they can change with environment (38) or time (39), or because of selection (40), mutation (41), inbreeding (42), and/or drift (43).
Compartments on Butterfly Wings.
Our results show that differences in response among directions of selection are minor and demonstrate high potential for independent evolution of eyespot sizes. The developmental and evolutionary independence of different wing regions has been demonstrated in other insects (e.g., ref. 44). In butterflies, such flexibility is probably related to the compartmentalization of each of these iterated homologous pattern elements within wing regions bounded by veins (45). This individualization might involve the lack of physical communication between such compartments and/or compartment-specific genetic compositions that regulate the expression of the eyespot-forming genes (20, 46). There is, as yet, little experimental evidence for such ideas, but there is evidence for compartment-specific gene effects. Those effects have been reported for genes involved in the presence or absence of eyespots in B. anynana; the Spotty mutant has extra eyespots on only two of the compartments that are characteristically without eyespots (25), and the 3–4 mutant loses two eyespots without affecting others (20). Furthermore, the magnitude of the allelic effects for at least one gene known to be involved in eyespot size variation in this species is also eyespot-specific: alleles mapped to Distal-less had larger effects on the posterior eyespot than on the anterior (47). Accumulating data from gene-mapping studies have shown that even though many quantitative trait loci affecting correlated traits do map to the same location, there are often character-specific magnitudes of gene effects (e.g., refs. 47–49).
The Genetic Basis of Correlations Among Eyespots.
Genetic correlations among traits can result from linkage between the loci that affect them and/or pleiotropy of the alleles at those loci (50). The step-like “progress” in Fig. 2 for the uncoupling directions (particularly clear for A−P+) suggests that linkage might be an important factor determining the positive correlation between our two target traits. Break up of linkage between eyespot-regulating alleles could then explain at least part of the more erratic response in the − relative to the + directions; recombination can make genetic variation available for response in directions antagonistic to the initial among-eyespot correlations. This type of effect might occur together with changes in epistatic interactions as allelic frequencies change during selection to cause pulses of response (see ref. 51). Positive pleiotropy is also very likely to contribute to eyespot coupling. However, even though some genes are likely to have similar effects on both eyespots, the magnitude of these effects probably tends to be eyespot-specific (47). Furthermore, genes with negative or no pleiotropic effects also might be available. In terms of quantitative genetics, our +, =, and − lines will favor genes with different types of pleiotropic effects. Although + directions favor positive pleiotropy and − directions favor negative pleiotropy, = directions select against all pleiotropy. It has been proposed that a combination of positive directional selection among groups of traits with stabilizing selection within other groups (as in our = selective regime) is the probable mechanism underlying the origin of modularity (52).
The observation of genetic correlations between traits and the knowledge of shared developmental pathways clearly give only a partial picture of the genetic architecture of modular traits. The individual units of such traits can be somewhat divergent because of the presence of nonpleiotropic modifiers of their morphology. These modifiers (compare ref. 53) can be responsible for independent variability of individual eyespots and could account for the moderate phenotypic correlations and the dissociable responses we observed. Future gene-mapping experiments will unravel details of the distribution of gene effects for our target traits (compare ref. 54).
Nested Hierarchical Modularity.
Different bodies of data led to the suggestion that the whole of the eyespot pattern in B. anynana behaved as a single module or character (27). Eyespots share a common underlying developmental mechanism (21) and are characterized by positive phenotypic and genetic correlations (23). We argue that the organization of the eyespot pattern can be better visualized as a nested hierarchical module (i.e., modules within modules; ref. 5), wherein different levels of integration can be identified: (i) the whole eyespot pattern forms one level, because there is more interdependence among individual eyespots than between these and any other pattern elements; (ii) those eyespots on the same wing surface appear to be more strongly coupled than those on different wing surfaces, possibly in association with different ecological pressures (23, 55); (iii) within a wing surface, eyespots in close vicinity tend to be more coupled than distant ones (e.g., the mutations Spotty and 3–4 each affect only two adjacent eyespots; see also ref. 5); (iv) in addition, our results demonstrate high potential for independent variation of eyespot size, and thus, some individuality at the single-eyespot level (see also refs. 29 and 46). The hierarchical organization is thought to make the independent modification of iterated traits possible and seems to be a general property of modular traits (5). Moreover, this organization is not fixed in time but is itself flexible; the strength of the genetic integration within each level as well as the overall depth of the hierarchical complexity may change as evolution progresses.
Evolution of Individuality.
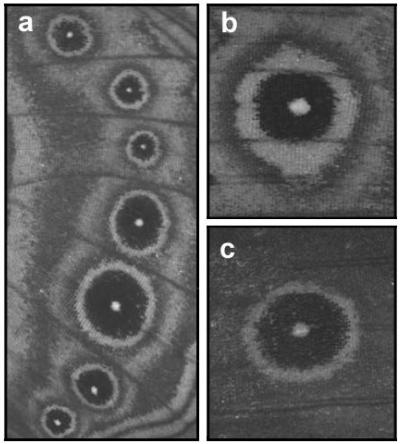
The absence of constraints on the evolutionary divergence of dorsal eyespot size that we have documented could result from a previous history of natural selection for genes conferring individuality (i.e., evolutionary and developmental independence) to different eyespots. It is likely to be functionally advantageous for subsets of eyespots to differ in size; for example, in the context of visual communication with different predators or with multiple types of interactions (e.g., predation and sexual selection acting simultaneously). Specific examples could include predators attacking either from above or below and, thus, targeting different eyespots, and predators that attack from different distances and, thus, may favor a range of eyespot sizes. Studies on other organisms have shown that different features of animal color patterns sometimes do have distinct functional values (56, 57). The ancestral state in butterfly eyespot patterns may have been that of full integration, i.e., the whole pattern as a single module with little further complexity or differentiation among repeats (compare nymphalid groundplan; ref. 17). In time, the complexity and depth of organization have probably increased with the evolution of differences in eyespot morphology among wings, wing surfaces, and, finally, subsets of eyespots in response to different functional demands. Through varying selective pressures on subsets of eyespots, evolution will have favored genes of localized effects across wing surfaces, thus enabling diversification of eyespot morphologies within individuals (Fig. 4). Indeed, some butterfly species show extreme divergence of individual eyespots not only with respect to size but also to other features, such as color composition and shape (see Precis coenia and Smyrna blomfildia in refs. 17 and 26). Examples of serial repeats acquiring different morphologies associated with specialized functions include the divergent morphology of subsets of teeth in mammals and leg morphology in insects.
Fig 4.
Individuality of Bicyclus anynana eyespots. (a) Ventral surface of hindwing showing the sequence of eyespots of different sizes along the anterior-posterior axis (the distal wing margin is located at the right, and the anterior is located at the top). (b and c) Comparison of morphology of ventral (b) and dorsal (c) eyespots, which also have different physiological and ecological properties (55, 60). Photos show the anterior forewing eyespot from the same individual.
Conclusion
Much literature on modularity has focused on how the functional coupling between traits can lead to their genetic coupling (8, 9, 52). Morphological integration is expected to evolve for traits that collectively serve a common functional role (9, 52). Our results suggest that the issue addressed here is the reverse: the evolution of the genetic decoupling of traits that arose presumably as iterated developmentally homologous elements. The general problem of how serially homologous structures acquire individuality through evolutionary time is a fascinating topic for evo-devo research (5). Perhaps the most popular example is the differentiation of body segments in arthropods (58, 59), but those issues can be more readily addressed for the experimentally manipulatable eyespots of butterflies.
Acknowledgments
We thank V. French, M. Matos, J. A. J. Metz, H. Teotónio, G. P. Wagner, and B. Zwaan for discussions on experimental design; J. Ng for the computer program for the = selection; N. Wurzer and colleagues for maize; M. Britijn for help with figures; and M. Matos, M. J. West-Eberhard and two anonymous referees for comments on the text. This work was supported by the Portuguese Foundation for Science and Technology and the Gulbenkian Ph.D. Program (P.B.) and the Human Frontier Science Program (P.M.B.).
Abbreviations
UC, unselected control
ANCOVA, analysis of covariance
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wagner G. P. & Altenberg, L. (1996) Evolution (Lawrence, Kans.) 50, 967-976. [DOI] [PubMed] [Google Scholar]
- 2.Raff R., (1996) The Shape of Life (Univ. of Chicago Press, Chicago).
- 3.Bolker J. A. (2000) Am. Zool. 40, 770-776. [Google Scholar]
- 4.Wagner G. P., (2001) The Character Concept in Evolutionary Biology (Academic, San Diego).
- 5.West-Eberhard M. J., (2003) Developmental Plasticity and Evolution (Oxford Univ. Press, New York).
- 6.Roff D., (1999) Evolutionary Quantitative Genetics (Chapman & Hall, New York).
- 7.Lande R. (1979) Evolution (Lawrence, Kans.) 33, 402-416. [DOI] [PubMed] [Google Scholar]
- 8.Cheverud J. (1984) J. Theor. Biol. 110, 155-171. [DOI] [PubMed] [Google Scholar]
- 9.Cheverud J. M. (1996) Am. Zool. 36, 44-50. [Google Scholar]
- 10.Paulsen S. M. (1994) Dev. Genet. 15, 79-91. [Google Scholar]
- 11.Donohue K., Pyle, E. H., Messiqua, D., Heschel, M. S. & Schmitt, J. (2000) Evolution (Lawrence, Kans.) 54, 1969-1981. [DOI] [PubMed] [Google Scholar]
- 12.Wagner G. P. (1988) J. Evol. Biol. 1, 45-66. [Google Scholar]
- 13.Wolf J. B., Frankino, W. A., Agrawal, A. F., Brodie, E. D. & Moore, A. J. (2001) Evolution (Lawrence, Kans.) 55, 232-245. [DOI] [PubMed] [Google Scholar]
- 14.Maynard-Smith J., Burian, R., Kaufman, S., Alberch, P., Campbell, J., Goodwin, B., Lande, R., Raup, D. & Wolpert, L. (1985) Q. Rev. Biol. 60, 265-287. [Google Scholar]
- 15.Raff R. A. (2000) Nat. Rev. Genet. 1, 74-79. [DOI] [PubMed] [Google Scholar]
- 16.Arthur W. (2002) Nature 415, 757-764. [DOI] [PubMed] [Google Scholar]
- 17.Nijhout H. F., (1991) The Development and Evolution of Butterfly Wing Patterns (Smithsonian Institute, Washington, DC).
- 18.Brakefield P. (1984) in The Biology of Butterflies, eds. Vane-Wright, R. & Ackery, P. (Academic, London).
- 19.Monteiro A. F., Brakefield, P. M. & French, V. (1994) Evolution (Lawrence, Kans.) 48, 1147-1157. [DOI] [PubMed] [Google Scholar]
- 20.McMillan W. O., Monteiro, A. & Kapan, D. D. (2002) Trends Ecol. Evol. 17, 125-133. [Google Scholar]
- 21.Beldade P. & Brakefield, P. M. (2002) Nat. Rev. Genet. 3, 442-452. [DOI] [PubMed] [Google Scholar]
- 22.Monteiro A., Brakefield, P. M. & French, V. (1997) Evolution (Lawrence, Kans.) 51, 1207-1216. [DOI] [PubMed] [Google Scholar]
- 23.Brakefield P. M. (1998) Heredity 80, 265-272. [Google Scholar]
- 24.French V. & Brakefield, P. M. (1995) Dev. Biol. 168, 112-123. [DOI] [PubMed] [Google Scholar]
- 25.Brakefield P. M., Gates, J., Keys, D., Kesbeke, F., Wijngaarden, P. J., Monteiro, A., French, V. & Carroll, S. B. (1996) Nature 384, 236-242. [DOI] [PubMed] [Google Scholar]
- 26.Brunetti C. R., Selegue, J. E., Monteiro, A., French, V., Brakefield, P. M. & Carroll, S. B. (2001) Curr. Biol. 11, 1578-1585. [DOI] [PubMed] [Google Scholar]
- 27.Brakefield P. M. (2001) J. Exp. Zool. 291, 93-104. [DOI] [PubMed] [Google Scholar]
- 28.Monteiro A., Brakefield, P. M. & French, V. (1997) Genetics 146, 287-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beldade P., Koops, K. & Brakefield, P. M. (2002) Nature 416, 844-847. [DOI] [PubMed] [Google Scholar]
- 30.Saccheri I. J., Brakefield, P. M. & Nichols, R. A. (1996) Evolution (Lawrence, Kans.) 50, 2000-2013. [DOI] [PubMed] [Google Scholar]
- 31.Zar J., (1999) Biostatistical Analysis (Prentice–Hall, Englewood Cliffs, NJ).
- 32.Holloway G. J., Brakefield, P. M. & Kofman, S. (1993) Heredity 70, 179-186. [Google Scholar]
- 33.Condamin M., (1973) Monographie du Genre Bicyclus (Lepidoptera, Satyridae) (Inst. Fond. Afr. Noire, Dakar).
- 34.Nijhout H. F. (1994) BioScience 44, 148-157. [Google Scholar]
- 35.Joron M. & Mallet, J. L. B. (1998) Trends Ecol. Evol. 13, 461-466. [DOI] [PubMed] [Google Scholar]
- 36.Houle D. (2001) in The Character Concept in Evolutionary Biology, ed. Wagner, G. P. (Academic, San Diego), pp. 109–140.
- 37.Etterson J. R. & Shaw, R. G. (2001) Science 294, 151-154. [DOI] [PubMed] [Google Scholar]
- 38.Kause A. & Morin, J. P. (2001) Genet. Res. 78, 31-40. [DOI] [PubMed] [Google Scholar]
- 39.Steppan S. J., Phillips, P. C. & Houle, D. (2002) Trends Ecol. Evol. 17, 320-327. [Google Scholar]
- 40.Wilkinson G. S., Fowler, K. & Partridge, L. (1990) Evolution (Lawrence, Kans.) 44, 1990-2003. [DOI] [PubMed] [Google Scholar]
- 41.Camara M. D. & Pigliucci, M. (1999) Evolution (Lawrence, Kans.) 53, 1692-1703. [DOI] [PubMed] [Google Scholar]
- 42.Phillips P. C., Whitlock, M. C. & Fowler, K. (2001) Genetics 158, 1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roff D. (2000) Heredity 84, 135-142. [DOI] [PubMed] [Google Scholar]
- 44.Weber K. E. (1992) Genetics 130, 345-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nijhout H. F., (1994) Development (Cambridge, U.K.), Suppl., 225–233.
- 46.Nijhout H. F. (2001) J. Exp. Zool. 291, 213-225. [DOI] [PubMed] [Google Scholar]
- 47.Beldade P., Brakefield, P. M. & Long, A. D. (2002) Nature 415, 315-318. [DOI] [PubMed] [Google Scholar]
- 48.Doebley J. & Stec, A. (1991) Genetics 129, 285-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juenger T., Purugganan, M. & Mackay, T. F. C. (2000) Genetics 156, 1379-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falconer D. S. & Mackay, T. F. C., (1996) Introduction to Quantitative Genetics (Addison–Wesley, Essex).
- 51.Nijhout H. F. & Paulsen, S. M. (1997) Am. Nat. 149, 394-405. [Google Scholar]
- 52.Wagner G. P. (1996) Am. Zool. 36, 36-43. [Google Scholar]
- 53.Turner J. R. G. (1977) Evol. Biol. 10, 163-205. [Google Scholar]
- 54.Mezey J. G., Cheverud, J. M. & Wagner, G. P. (2000) Genetics 156, 305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brakefield P. M., Kesbeke, F. & Koch, P. B. (1998) Am. Nat. 152, 853-860. [DOI] [PubMed] [Google Scholar]
- 56.Kuwamura T., Karino, K. & Nakashima, Y. (2000) J. Ethol. 18, 17-23. [Google Scholar]
- 57.Badyaev A. V., Hill, G. E., Dunn, P. O. & Glen, J. C. (2001) Am. Nat. 158, 221-235. [DOI] [PubMed] [Google Scholar]
- 58.Gibert P., Moreteau, B. & David, J. R. (2000) Evol. Dev. 2, 249-260. [DOI] [PubMed] [Google Scholar]
- 59.Williams T. A. & Nagy, L. M. (2001) J. Exp. Zool. 291, 241-257. [DOI] [PubMed] [Google Scholar]
- 60.Brakefield P. M. & French, V. (1999) BioEssays 21, 391-401. [Google Scholar]