Abstract
Cisplatin is a chemotherapeutic drug used to treat a variety of cancers. Both intrinsic and acquired resistance to cisplatin, as well as toxicity, limit its effectiveness. Molecular mechanisms that underlie cisplatin resistance are poorly understood. Here we demonstrate that deletion of the yeast CTR1 gene, which encodes a high-affinity copper transporter, results in increased cisplatin resistance and reduced intracellular accumulation of cisplatin. Copper, which causes degradation and internalization of Ctr1 protein (Ctr1p), enhances survival of wild-type yeast cells exposed to cisplatin and reduces cellular accumulation of the drug. Cisplatin also causes degradation and delocalization of Ctr1p and interferes with copper uptake in wild-type yeast cells. Mouse cell lines lacking one or both mouse Ctr1 (mCtr1) alleles exhibit increased cisplatin resistance and decreased cisplatin accumulation in parallel with mCtr1 gene dosage. We propose that cisplatin uptake is mediated by the copper transporter Ctr1p in yeast and mammals. The link between Ctr1p and cisplatin transport may explain some cases of cisplatin resistance in humans and suggests ways of modulating sensitivity and toxicity to this important anticancer drug.
Cisplatin [cis-diamminedichloroplatinum(II)] chemotherapy is curative for the majority of patients with advanced testicular cancer, which was almost uniformly fatal before its use, and is also effective for ovarian, bladder, cervical, head and neck, and small-cell lung cancers (1). Many patients with these cancers, however, eventually relapse and become refractory to this drug (2). In contrast to the cancers treatable with cisplatin, common cancers such as colorectal are intrinsically resistant to this drug. Increasing dosage to overcome resistance can cause serious nephrotoxicity and ototoxicity. Understanding the mechanism of intrinsic and acquired resistance to cisplatin is thus critical for developing more effective treatment for cancer.
The mechanism by which cisplatin enters cells is unknown. The drug becomes activated once it enters the cytoplasm, where the chloride atoms on cisplatin are displaced by water molecules. This aquated product is a potent electrophile that can react with any nucleophile, including the sulfhydryl groups on proteins and nitrogen donor atoms on nucleic acids. Work from many laboratories has implicated DNA as a critical target for cisplatin cytotoxicity, the most revealing evidence being the hypersensitivity to cisplatin of both prokaryotic and eukaryotic cells deficient in DNA repair (3–5). The most prevalent cisplatin-induced DNA adduct is an intrastrand crosslink in which the platinum is covalently bound to the N7 positions of the imidazole ring of two adjacent guanines (6). The intrastrand crosslinks are repaired by the nucleotide excision repair pathway (7).
Studies in mammalian cells have identified several genes that affect sensitivity of cells to cisplatin (8). Overexpression of a multidrug resistance protein MRP2 or a copper-transporting P-type ATPase ATP7B results in increased resistance and decreased cellular accumulation of cisplatin, suggesting that MRP2 and ATP7B may facilitate cisplatin efflux (9, 10). Genes that alter intracellular levels of thiols, which are highly nucleophilic and can react with intracellular cisplatin, have been demonstrated to affect levels of resistance to cisplatin by sequestering intracellular cisplatin (11). Mouse cell lines deleted for MSH2, a mismatch repair protein, exhibit increased cisplatin resistance (12). It is hypothesized that mismatch repair proteins detect the cisplatin-induced damage and initiate a series of events that result in cell cycle arrest and death.
In the budding yeast Saccharomyces cerevisiae, biochemical and genetic screens have identified several genes that govern cisplatin resistance (8, 13–15). The mechanisms by which most of these genes affect cisplatin resistance are not known. It has been suggested that the Ixr1 protein, which binds cisplatinated DNA (16) and whose absence leads to increased cisplatin resistance, shields cisplatin-DNA adducts from repair (5).
Here we describe the identification of the CTR1 gene as a major determinant governing cisplatin resistance and accumulation in yeast and mice.
Materials and Methods
Strains.
Isogenic strains carrying deletions in MAC1, CTR1, FRE1, FRE7, CTA1, CTT1, ATX1, CCC2, FET3, LYS7, SOD1, COX17, SCO1, CTR2, FET4, and RAD2 were generated in YSI1 (W303 background; MATaade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 GAL+ psi+) by the protocol of Sakumoto et al. (17). A PCR fragment was used to replace the entire ORF with the Candida glabrata HIS3 gene. A ctr1Δ rad2Δ strain (YSI27) was the product of a cross between YSI22 (MATactr1Δ) and YSI17 [MATα rad2Δ; obtained from a cross of wild-type MATα (YSI2) and YSI16 [MATarad2)]. Strains CTR1-HA (YSI37) and CTR1-GFP (YSI38) were generated in YSI1 by the protocol of Longtine et al. (18). A PCR fragment was used to add a hemagglutinin (HA) epitope sequence or the GFP gene, followed by the HIS3 gene, at the C terminus of the genomic CTR1 gene. Both CTR1-HA and CTR1-GFP complemented the cisplatin-resistant phenotype of the ctr1Δ mutant.
Selection for Cisplatin-Resistant Mutants.
The mTn-lacZ/LEU2-mutagenized library was a generous gift from M. Snyder (Yale University) (19). Strain YSI1 was mutagenized with the transposon library and resistant mutants were selected on SC-Leu plates containing 0.2 mM cisplatin and 2.7 mg/ml NaCl at 30°C.
Cisplatin Survival Curves.
Logarithmic-phase yeast cells were exposed for 2 h to different concentrations of cisplatin (Bristol Laboratories) in rich medium [yeast extract/peptone/dextrose (YPD)] and incubated on YPD plates at 30°C for 2 days to allow formation of colonies. Data are expressed as percentages of colonies formed compared with control cultures not exposed to cisplatin. For determining survival of mouse cells, cisplatin was added 24 h after plating of cells (80% confluence) and incubated for 2 h. Cells were detached with 10 mM EDTA in PBS and stained with trypan blue to detect dead cells; unstained cells were counted using a hemocytometer. Data are expressed as percentages of unstained cells compared with control cultures not exposed to cisplatin.
Preparation of Cells for Platinum Measurement.
After cisplatin treatment, cells were washed twice with PBS and lysed with 1% Triton X-100/0.1% SDS. After a 3-min centrifugation at 15,000 rpm at 4°C in a Microfuge (Beckman), the supernatant was used to determine protein concentration by the bicinchoninic acid (BCA) protein assay (Pierce) and platinum content by atomic absorption spectrophotometry. The platinum reading was normalized to protein concentration.
Atomic Absorption Spectrophotometry.
Platinum was measured by using a Perkin–Elmer atomic absorption spectrometer (AAnalyst 100 and 3300) with an HGA-800 and HGA-400 graphite furnace system. A volume of 20 μl was introduced into the graphite furnace and the peak area was read during a 5-s atomization step at 2,500°C. The amount of platinum in the samples was determined from a calibration curve prepared by using cisplatin solutions.
Tissue Culture.
Mouse cell lines were cultured in DMEM (GIBCO) with 20% FBS, 110 mg/liter pyruvate, 50 mg/liter uridine, 1 mM nonessential amino acids, 1× antibiotic/antimycotic solution (GIBCO), and 55 μM 2-mercaptoethanol.
64Cu Uptake Assay
Logarithmic-phase cells were treated with 64Cu (5 μM as CuSO4) in the presence of 100 μM or 1 mM cisplatin, or 10 μM unlabeled CuSO4 for 2 h. 64Cu uptake was quenched by adding ice-cold 50 mM EDTA/0.1 M Tris succinate, pH 6.0. Cells were then washed on filters twice with ice-cold 10 mM EDTA/0.1 M Tris succinate, pH 6.0, solution. The radioactivity on the filters was measured with a γ-counter, and the values were divided by cell OD600.
Results
Yeast Mutants with Increased Cisplatin Resistance.
To identify genes involved in cisplatin resistance, we mutagenized wild-type yeast cells with a transposon library and selected for mutants able to grow in the presence of a toxic dose of cisplatin. The mutant that showed the highest degree of resistance was defective in the MAC1 gene. Deletion of the complete MAC1 ORF resulted in a 2.5-fold increase in cisplatin resistance relative to wild-type cells; the number of cells that survived a 2-h treatment with 1 mM cisplatin was 1,000 times greater in the mac1Δ mutant than in wild-type cells (Fig. 1A). MAC1 encodes a protein that activates transcription of catalase genes and genes involved in uptake of copper and iron (20–23). To address whether increased cisplatin resistance of the mac1Δ mutant is due to the inability to transcribe a Mac1p target gene, we deleted each of its known target genes, namely, CTR1, FRE1, FRE7, CTA1, and CTT1, and measured the sensitivity of the mutants to cisplatin. Of the five mutants, only the ctr1Δ strain showed a level of resistance similar to the mac1Δ mutant (Fig. 1A). These observations suggest that CTR1 is the major target for Mac1p with respect to cisplatin resistance.
Fig 1.
Cisplatin resistance of yeast mutants. Logarithmic-phase cells were exposed for 2 (A–C) or 3 (D) h to different concentrations of cisplatin and incubated on YPD plates at 30°C for 2 days to allow formation of colonies. Data are expressed as percentages of colonies formed compared with control cultures not exposed to cisplatin. (A) Cisplatin resistance of mutants defective in Mac1p target genes. Strains are wild-type (YSI1), mac1Δ (YSI9), ctr1Δ (YSI22), fre1Δ (YSI23), fre7Δ (YSI24), cta1Δ (YSI25), and ctt1Δ (YSI26). (B) Cisplatin resistance of mutants defective in intracellular copper trafficking and utilization. Strains are wild-type (YSI1), ctr1Δ (YSI22), atx1Δ (YSI28), ccc2Δ (YSI29), fet3Δ (YSI30), lys7Δ (YSI31), sod1Δ (YSI32), cox17Δ (YSI33), and sco1Δ (YSI34). (C) Cisplatin sensitivity of low-affinity copper transporter mutants, ctr2Δ (YSI35) and fet4Δ (YSI36). (D) Cisplatin sensitivity of high-affinity copper transporter mutants in the BR10 strain background (22). Strains were DTY1 (CTR1 CTR3), SKY52 (ctr1Δ CTR3), SKY44 (CTR1 ctr3Δ), and SKY46 (ctr1Δ ctr3Δ).
CTR1 encodes a high-affinity copper transporter (24). Cells deleted for CTR1 are resistant to growth inhibition caused by high levels of copper in the medium, fail to grow on low-copper medium, and are defective in enzymes such as superoxide dismutase and in cellular activities such as iron uptake and respiration that require copper (24, 25). To determine whether the cisplatin resistance of ctr1Δ mutants results from an effect on intracellular proteins that mediate intracellular copper trafficking and utilization, we measured cisplatin resistance of atx1Δ, ccc2Δ, fet3Δ, lys7Δ, sod1Δ, cox17Δ, and sco1Δ mutants (26). Significant increases in cisplatin resistance as compared with the ctr1Δ mutant in quantitative assays were not observed (Fig. 1B). Thus, defects in proteins involved in intracellular copper distribution or utilization apparently do not play a major role in cisplatin resistance in yeast.
We also tested for involvement of other yeast copper transporters in cisplatin sensitivity: another high-affinity transporter, Ctr3p, and two low-affinity transporters, Ctr2p and Fet4p (27–29). Deletion of CTR2 or FET4 did not affect cisplatin resistance (Fig. 1C). Deletion of CTR3 resulted in a slight increase in cisplatin resistance (Fig. 1D). Although the yeast Ctr1 and Ctr3 proteins have redundant roles in copper uptake, they have distinct structural features that could be responsible for a difference in their function in cisplatin resistance. These observations indicate that, of the yeast copper transporters, Ctr1p plays the most prominent role in governing cisplatin resistance.
Cisplatin Accumulation in Yeast ctr1Δ Mutants.
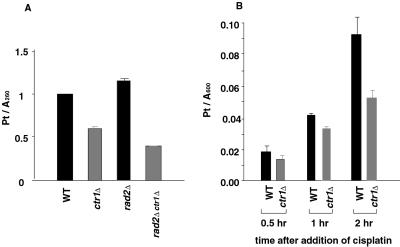
To understand the basis for cisplatin resistance in ctr1Δ strains, we first compared the level of cisplatin-DNA adducts in wild-type and ctr1Δ mutant cells, because the formation of DNA adducts is the major cause of cisplatin toxicity (30). As shown in Fig. 2A, the amount of cisplatin bound to DNA during a 2-h incubation with 1 mM cisplatin in the mutant cells was approximately 59% of that in wild-type cells, in which platinum was calculated to be bound to DNA at a ratio of 1 molecule per 14 base pairs. To determine whether the reduced level of cisplatin-DNA adducts in the mutant is due to enhanced excision repair, we examined the level of adduct formed in a ctr1Δ strain incapable of excision repair because of deletion of RAD2 (31). We observed that the level of adduct was also reduced in the ctr1Δ rad2Δ strain, to 35% of the level formed by the isogenic CTR1 rad2Δ strain (Fig. 2A), indicating that enhanced excision repair is not responsible for the increased cisplatin resistance exhibited by ctr1Δ mutants. To determine whether cellular accumulation of cisplatin is affected in the ctr1Δ mutant, we measured the level of platinum in wild-type and mutant cells treated with 1 mM cisplatin for 2 h. Fig. 2B shows that the level of cisplatin accumulation in the ctr1Δ mutant was only 56% of that exhibited by wild-type cells. This decrease in cellular accumulation of cisplatin is likely to reflect decreased drug uptake and not increased efflux, because we did not see a significant increase in drug clearance in the ctr1Δ mutant relative to wild-type after cisplatin was removed from the medium and cells were further incubated for 6 h (S.I. and I.H., unpublished observations). Therefore, decreased drug uptake is likely to be responsible for the lower cisplatin adduct level observed in the ctr1Δ mutant and consequent increased survival.
Fig 2.
Cisplatin adduct formation and accumulation in yeast ctr1Δ mutants. (A) Accumulation of cisplatin-DNA adducts in wild-type (WT) and isogenic ctr1Δ mutant strains. Strains were YSI1 (wild-type), YSI22 (ctr1Δ), YSI16 (rad2Δ), and YSI27 (rad2Δ ctr1Δ). Genomic DNA was purified from cells treated with 1 mM cisplatin for 2 h in YPD. Platinum was measured using an atomic absorption spectrometer. Numbers represent absorption by atomized platinum divided by A260 of each DNA sample. (B) Cellular platinum accumulation in wild-type (YSI1) and ctr1Δ mutant (YSI22) strains. The amount of platinum in whole cells was measured using the atomic absorption spectrophotometer after incubating cells with 1 mM cisplatin for the indicated times. Numbers were obtained by dividing the reading from the spectrophotometer by the OD600 of the culture at each time point.
A Link between Cisplatin and Copper Transport in Wild-Type Yeast Cells.
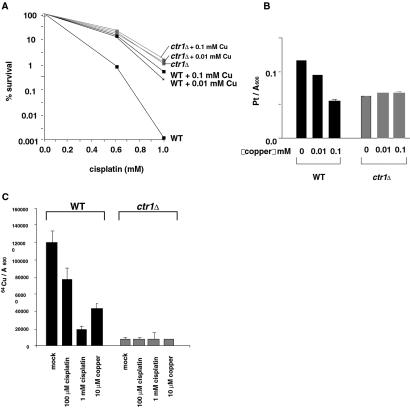
We observed striking effects of copper on cisplatin resistance in wild-type cells. When 0.1 mM copper was added to the medium, survival of wild-type cells was increased 2-fold (Fig. 3A); cellular accumulation of cisplatin was decreased 2-fold (Fig. 3B). We have also found that cisplatin reduced uptake of 64Cu into wild-type cells; when treated for 2 h with 1 mM cisplatin, the level of 64Cu uptake was 16% of that of mock-treated cells (Fig. 3C). Such a decrease was not observed in the ctr1Δ mutant. These observations demonstrate that copper and cisplatin have the ability to reduce each other's uptake and that this reduction is dependent on CTR1.
Fig 3.
A Ctr1p-mediated link between cisplatin and copper transport in yeast. (A) Effect of copper on survival of wild-type (WT) (YSI1) and the ctr1Δ mutant (YSI22) exposed to cisplatin. Cells were treated for 2 h with various concentrations of cisplatin in the presence of 0.01 mM, 0.1 mM, or no CuSO4. Data were analyzed as in Fig. 1. (B) Effect of copper on cellular accumulation of platinum in wild-type and ctr1Δ mutants. Cells were exposed to 1 mM cisplatin in the presence of 0.01 mM, 0.1 mM, or no CuSO4. Data were analyzed as in Fig. 2B. (C) Effect of cisplatin on copper uptake in wild-type and ctr1Δ mutants. Cells were treated with 64Cu (5 μM as CuSO4) in the presence of 100 μM or 1 mM cisplatin, or 10 μM unlabeled CuSO4 for 2 h. Cellular 64Cu level was measured with a γ-counter, and the values were divided by the cell OD600.
Prior studies have shown that the level of Ctr1p is regulated by copper concentration; high copper triggers internalization and degradation of Ctr1p (25, 32). We found that cisplatin also caused the level of Ctr1p to decrease; incubation of wild-type cells with 0.1 mM cisplatin for 2 h in the presence of a protein synthesis inhibitor led to a 3-fold decrease in Ctr1p (Fig. 4A). Furthermore, when cells that produce a Ctr1p-GFP fusion protein were treated with 1 mM cisplatin, a reduced level of the fusion protein was detected at the plasma membrane, and clustering of Ctr1p-GFP appearing as a punctate signal was observed (Fig. 4B). These observations provide further support for a link between Ctr1p and cisplatin transport.
Fig 4.
Effect of cisplatin on yeast Ctr1 protein. (A) Degradation of yeast Ctr1p upon cisplatin treatment. The Ctr1 protein was tagged at its C terminus with an HA epitope by modification of the genomic CTR1 locus (YSI37). Cycloheximide (100 μg/ml) was added to the CTR1-HA cells 30 min before cisplatin treatment to block new protein synthesis. Cells were then exposed to 0.1 mM or no cisplatin, and samples were taken at the indicated times. The level of Ctr1 protein was determined by Western blotting using anti-HA antibodies. The loading of proteins in each lane appeared equal by Coomassie blue staining. (B) Localization of yeast Ctr1p after cisplatin or copper treatment. The Ctr1 protein was tagged at its C terminus with GFP by modification of the genomic CTR1 locus (YSI38). Cells were incubated with 100 μg/ml cycloheximide for 30 min to block new protein synthesis, treated with 1 mM cisplatin for 4 h, and analyzed by fluorescence microscopy.
Analysis of Mouse Ctr1 Knockout Cell Lines.
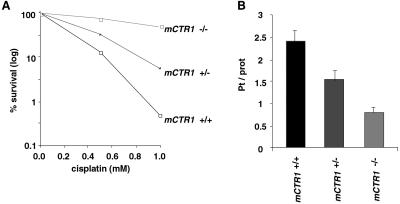
Mammals contain functional homologues of the yeast CTR1 gene, hCtr1 in humans and mCtr1 in mice, which complement a yeast ctr1 mutant for intracellular copper deficiency (33, 34). To test for a potential role of mammalian Ctr1 in cisplatin resistance, we prepared mouse cell lines from two independent embryos that were wild-type, heterozygous, or homozygous for a knockout allele of mCtr1 (35) and tested their cisplatin sensitivity. As shown in Fig. 5A, homozygous mutant cells were 8-fold more resistant than wild-type cells to cisplatin, and heterozygous cells were 4-fold more resistant than wild-type cells to cisplatin. Similar results for survival were obtained using an independent pair of wild-type and homozygous mutant cell lines. The levels of cisplatin accumulation were also reduced accordingly (Fig. 5B): heterozygous cells exhibited a 35% reduction in cisplatin accumulation compared with wild-type cells; homozygous mutant cells exhibited a 70% decrease. A similar graded reduction in copper uptake was also observed with the wild-type, heterozygous, and homozygous mutant cell lines (J.L. and D.J.T., unpublished observations). These observations suggest that mCtr1 protein functions as a cisplatin transporter in mouse cells and that its human homologue, hCtr1, which is 92% identical to mCtr1, may mediate cisplatin uptake as well.
Fig 5.
Cisplatin resistance and accumulation by mouse embryonic cell lines lacking mCtr1. Cells were incubated for 2 h with cisplatin. For determining survival (A), cells were stained with trypan blue to detect dead cells; unstained cells were counted using a hemocytometer. Data are expressed as percentages of unstained cells compared with control cultures not exposed to cisplatin. For measuring cisplatin accumulation (B), cells were lysed and, after a centrifugation, the supernatant was used to determine platinum content. The platinum reading was normalized to protein concentration.
Discussion
Our genetic observations on yeast and mouse demonstrate that the high-affinity copper transporter Ctr1 plays a major role in cisplatin uptake. The effects of a ctr1 deletion on cisplatin resistance in yeast seem to be specific to this mutation and not secondary to starvation for intracellular copper, because mutants defective in other copper transporters and/or copper utilization did not display an increase in cisplatin resistance similar to that in the ctr1Δ mutant. The ability of copper and cisplatin to decrease each other's uptake in wild-type yeast cells but not in the ctr1Δ mutant and the ability of cisplatin to down-regulate and delocalize Ctr1 protein support a direct involvement of Ctr1p in cisplatin transport.
The mechanism of cisplatin uptake has been unclear. Inability to saturate the rate of cisplatin uptake has been taken to indicate a simple diffusion mechanism, whereas the presence of a variety of agents that do not alter plasma membrane permeability but affect cisplatin uptake has suggested that uptake may be mediated by plasma membrane proteins (36). It has been proposed that ≈50% of the initial rate of uptake is due to passive diffusion and that the remaining 50% is due to facilitated diffusion through an as-yet-unidentified gated channel (36). Our work has identified Ctr1 to be a major protein governing cisplatin uptake in yeast and mice that could be responsible for facilitated uptake of cisplatin. The residual cisplatin uptake observed in yeast and mouse ctr1Δ mutants (Figs. 2B and 5B) could be due to diffusion across the plasma membrane or to additional transport proteins. We do not yet understand how Ctr1p mediates uptake of cisplatin. Ctr1p has three putative transmembrane domains and forms at least a dimer in yeast and mice (25, 37). Hence, Ctr1p might serve as a channel. Another possibility is that cisplatin uptake is coupled to endocytosis and/or degradation of Ctr1p. Although we cannot exclude the possibility that Ctr1p plays a much less direct role in cisplatin uptake, our genetic studies have ruled out involvement of several effectors downstream of Ctr1 and of other copper transporters in cisplatin uptake.
Our work offers additional perspectives on the molecular mechanism of intrinsic and acquired resistance to cisplatin, which represents a major impediment to successful treatment of cancer, and may facilitate development of more effective therapy. Differences in the effectiveness of cisplatin treatment among different cancers or individuals may reflect the activity of Ctr1p. Our findings on Ctr1p as a major determinant of cisplatin uptake may make it possible to enhance the efficacy of the drug by, for example, selectively increasing the activity of Ctr1p in tumors that are intrinsically resistant or that have acquired resistance to cisplatin. Similarly, it may be possible to reduce cisplatin-induced nephrotoxicity and ototoxicity by down-regulating Ctr1p in normal cells.
Acknowledgments
We thank Marc Shuman for cisplatin and for leading our initial foray into cisplatin resistance in yeast, Michael Snyder for the mTn-lacZ/LEU2-mutagenized library, Yien Kuo and Jane Gitschier for use of the atomic absorption spectrophotometer in initial experiments, and members of our laboratories for helpful discussions. We especially thank Erin O'Shea and Liz Blackburn for critical reading of the manuscript. This study was supported by a Stewart Trust Cancer Research Award and a Sandler Basic Science Award (to I.H.), and by National Institutes of Health Grants GM62555 and GM41840 (to D.J.T.) and GM61390 (to I.H.). S.I. was a Howard Hughes Medical Institute Predoctoral Fellow. J.L. was supported by an American Heart Association Postdoctoral Fellowship.
Abbreviations
HA, hemagglutinin
See commentary on page 13963.
References
- 1.Loehrer P. J. & Einhorn, L. H. (1984) Ann. Intern. Med. 100, 704-713. [DOI] [PubMed] [Google Scholar]
- 2.Giaccone G. (2000) Drugs 59, Suppl. 4, 9-17. [DOI] [PubMed] [Google Scholar]
- 3.Beck D. J. & Brubaker, R. R. (1973) J. Bacteriol. 116, 1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraval H. N. A., Rawlings, C. J. & Roberts, J. J. (1978) Mutat. Res. 51, 121-132. [DOI] [PubMed] [Google Scholar]
- 5.McA'Nulty M. M. & Lippard, S. J. (1996) Mutat. Res. 362, 75-86. [DOI] [PubMed] [Google Scholar]
- 6.Fichtinger-Schepman A. M. J., van der Veer, J. L., den Hartog, J. H. J., Lohman, P. H. M. & Reedijk, J. (1985) Biochemistry 24, 707-713. [DOI] [PubMed] [Google Scholar]
- 7.Beck D. J., Popoff, S., Sancar, A. & Rupp, W. D. (1985) Nucleic Acids Res. 13, 7394-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishida S. & Herskowitz, I. (2003) in Yeast as a Tool in Cancer Research, eds. Heitman, J. & Nitiss, J. L. (Kluwer, Boston), in press.
- 9.Kawabe T, Chen, Z., S., Wada, M., Uchiumi, T., Ono, M., Akiyama, S. & Kuwano, M. (1999) FEBS Lett. 456, 327-331. [DOI] [PubMed] [Google Scholar]
- 10.Komatsu M., Sumizawa, T., Mutoh, M., Chen, Z. S., Terada, K., Furukawa, T., Yang, X. L., Gao, H., Miura, N., Sugiyama, T. & Akiyama, S. (2000) Cancer Res. 60, 1312-1316. [PubMed] [Google Scholar]
- 11.Kurokawa H., Ishida, T., Nishio, K., Arioka, H., Sata, M., Fukumoto, H., Miura, M. & Saijo, N. (1995) Biochem. Biophys. Res. Commun. 216, 258-264. [DOI] [PubMed] [Google Scholar]
- 12.Fink D., Nebel, S., Aebi, S., Nehme, A. & Howell, S. B. (1997) Int. J. Oncol. 11, 539-542. [DOI] [PubMed] [Google Scholar]
- 13.Burger H., Capello, A., Schenk, P., W., Stoter, G., Brouwer, J. & Nooter, K. (2000) Biochem. Biophys. Res. Commun. 269, 767-774. [DOI] [PubMed] [Google Scholar]
- 14.Furuchi T., Ishikawa, H., Miura, N., Ishizuka, M., Kajiya, K., Kuge, S. & Naganuma, A. (2001) Mol. Pharmacol. 59, 470-474. [DOI] [PubMed] [Google Scholar]
- 15.Schenk P. W., Boersma, A. W., Brandsma, J. A., den Dulk, H., Burger, H., Stoter, G., Brouwer, J. & Nooter, K. (2001) Cancer Res. 61, 6982-6986. [PubMed] [Google Scholar]
- 16.Brown S. J., Kellet, P. J. & Lippard, S. J. (1993) Science 261, 603-605. [DOI] [PubMed] [Google Scholar]
- 17.Sakumoto N., Mukai, Y., Uchida, K., Kouchi, T., Kuwajima, J., Nakagawa, Y., Sugioka, S., Yamamoto, E., Furuyama, T., Mizubuchi, H., et al. (1999) Yeast 15, 1669-1679. [DOI] [PubMed] [Google Scholar]
- 18.Longtine M. S., McKenzie, A., Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. & Pringle, J. R. (1998) Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- 19.Burns N., Grimwade, B., Ross-Macdonald, P. B., Choi, E. Y., Finberg, K., Roeder, G. S. & Snyder, M. (1994) Genes Dev. 8, 1087-1105. [DOI] [PubMed] [Google Scholar]
- 20.Jungmann J., Reins, H. A., Lee, J., Romeo, A., Hassett, R., Kosman, D. & Jentsch, S. (1993) EMBO J. 12, 5051-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi-Iwai Y., Serpe, M., Haile, D., Yang, W., Kosman, D. J., Klausner, R. D. & Dancis, A. (1997) J. Biol. Chem. 272, 17711-17718. [DOI] [PubMed] [Google Scholar]
- 22.Labbe S., Zhu, Z. & Thiele, D. J. (1997) J. Biol. Chem. 272, 15951-15958. [DOI] [PubMed] [Google Scholar]
- 23.Martins L. J., Jensen, L. T., Simons, J. R., Keller, G. L. & Winge, D. R. (1998) J. Biol. Chem. 273, 23716-23721. [DOI] [PubMed] [Google Scholar]
- 24.Dancis A., Yuan, D. S., Haile, D., Askwith, C., Eide, D., Moehle, C., Kaplan, J. & Klausner, R. D. (1994) Cell 76, 393-402. [DOI] [PubMed] [Google Scholar]
- 25.Dancis A., Haile, D., Yuan, D. S. & Klausner, R. D. (1994) J. Biol. Chem. 269, 25660-25667. [PubMed] [Google Scholar]
- 26.Valentine J. S. & Gralla, E. B. (1997) Science 278, 817-818. [DOI] [PubMed] [Google Scholar]
- 27.Knight S. A., Labbe, S., Kwon, L. F., Kosman, D. J. & Thiele, D. J. (1996) Genes Dev. 10, 1917-1929. [DOI] [PubMed] [Google Scholar]
- 28.Kampfenkel K., Kushnir, S., Babiychuk, E., Inze, D. & Van Montagu, M. (1995) J. Biol. Chem. 270, 28479-28486. [DOI] [PubMed] [Google Scholar]
- 29.Hassett R., Dix, D. R., Eide, D. J. & Kosman, D. J. (2000) Biochem. J. 351, 477-484. [PMC free article] [PubMed] [Google Scholar]
- 30.Zamble D. & Lippard, S. J. (1995) Trends Biol. Sci. 20, 435-439. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z., Wu, X. & Friedberg, E. C. (1993) Proc. Natl. Acad. Sci. USA 90, 4907-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ooi C. E., Rabinovich, E., Dancis, A., Bonifacino, J. S. & Klausner, R. D. (1996) EMBO J. 15, 3515-3523. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou B. & Gitschier, J. (1997) Proc. Natl. Acad. Sci. USA 94, 7481-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J., Prohaska, J. R., Dagenais, S. L., Glover, T. W. & Thiele, D. J. (2000) Gene 254, 87-96. [DOI] [PubMed] [Google Scholar]
- 35.Lee J., Prohaska, J. R. & Thiele, D. J. (2001) Proc. Natl. Acad. Sci. USA 98, 6842-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gately D. P. & Howell, S. B. (1993) Br. J. Cancer 67, 1171-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J., Pena, M. M. O., Nose, Y. & Thiele, D. J. (2002) J. Biol. Chem. 277, 4380-4387. [DOI] [PubMed] [Google Scholar]