Abstract
Heterochromatin protein 1 (HP1), first discovered in Drosophila melanogaster, is a highly conserved chromosomal protein implicated in both heterochromatin formation and gene silencing. We report here characterization of an HP1-interacting protein, heterochromatin protein 2 (HP2), which codistributes with HP1 in the pericentric heterochromatin. HP2 is a large protein with two major isoforms of approximately 356 and 176 kDa. The smaller isoform is produced from an alternative splicing pattern in which two exons are skipped. Both isoforms contain the domain that interacts with HP1; the larger isoform contains two AT-hook motifs. Mutations recovered in HP2 act as dominant suppressors of position effect variegation, confirming a role in heterochromatin spreading and gene silencing.
Sequencing and analysis of eukaryotic genomes has confirmed that the bulk of the DNA does not code for proteins. Significant portions, including centromeric and telomeric regions, are packaged as constitutive heterochromatin. Heterochromatic domains generally are associated with a condensed appearance, late replication in S phase, and a low level of meiotic recombination. Such regions are gene-poor (although not devoid of genes), being made up primarily of repetitious sequences. Packaging of a euchromatic gene in a heterochromatic structure results in gene silencing, an epigenetic regulation that appears to be general, affecting most promoters. A well studied example is position effect variegation (PEV) in Drosophila melanogaster. PEV occurs when a gene normally found in a euchromatic domain is placed close to or within a heterochromatic domain by virtue of a chromosome rearrangement or transposition event. The gene is silenced in some of the cells in which it normally is expressed, resulting in a variegating phenotype (1). A silenced reporter gene shows an altered chromatin structure, including loss of accessibility in promoter regions and adoption of a repetitive nucleosome array (2, 3). In addition to its role in chromatin condensation and epigenetic regulation, heterochromatin formation plays an essential role in sister chromatid cohesion at centromeres (4, 5).
Many studies, particularly in yeast and Drosophila, have shown that a multiprotein complex is required to establish a heterochromatic domain. Proteins involved often have been identified by screening for dominant mutations that affect the level of silencing of a reporter gene, suppressing the variegating phenotype [Su(var) mutations] (1, 6, 7). Cytological approaches also have been used. One of the best-characterized structural proteins, heterochromatin protein 1 (HP1), was discovered in a screen by using mAbs to analyze the distribution of proteins on polytene chromosomes (8). Loss-of-function mutations in the gene encoding HP1, Su(var)2–5, result in loss of silencing at a reporter gene, whereas extra copies of the gene result in increased silencing (9, 10). HP1 is located principally in the pericentric heterochromatin and in a banded pattern along the small fourth chromosome, with lesser immunofluorescent staining of some euchromatic sites and the telomeres (11). The dosage response and specific localization argue that HP1 is directly involved in producing the silent phenotype of variegating loci. Loss of HP1 results in increased accessibility in the regulatory region of a variegating reporter, indicating that HP1 makes a direct contribution to the chromatin structure required for silencing (12).
HP1 is highly conserved, with homologues from yeast (Schizosaccharomyces pombe) to humans, and is associated consistently with the pericentric heterochromatin (13). HP1 (212 aa in Drosophila) contains a chromo domain, which binds specifically to histone H3 methylated at lysine 9 (H3-mLys9) (14, 15), and a chromo shadow domain, which forms a homodimer (16). The latter domain interacts with a number of chromosomal proteins (13), including SU(VAR)3–9, shown to possess histone H3 methyltransferase activity (17–19). This finding suggests that HP1 plays a key role in maintaining the heterochromatic state, interacting both with the modified histone and the enzyme capable of providing that modification (20). Mapping studies, using both cytological and biochemical approaches, have shown HP1 and H3-mLys9 to be important markers of heterochromatin. However, important questions remain concerning how the complex is targeted to sites of heterochromatin formation and how large-scale condensation is achieved. Hence, there is a need to identify additional proteins that interact with HP1.
We report here identification of a Drosophila HP1-interacting protein, heterochromatin protein 2 (HP2). HP2 interacts with HP1 in a yeast two-hybrid assay; it coprecipitates with HP1 from a Drosophila embryo extract, and it colocalizes with HP1 on Drosophila polytene chromosomes. Sequence analysis and characterization of the gene reveal two isoforms. Both proteins are large, 176 and 356 kDa, and, for the most part, are devoid of recognizable sequence motifs. However, the larger protein contains two AT-hook domains. Mutations in HP2 act as dominant suppressors of variegation, demonstrating that HP2 is required for spreading of heterochromatin and establishment and/or maintenance of the silent state.
Materials and Methods
Two-Hybrid Screen.
The yeast strain EGY48 was used in a screen of a D. melanogaster cDNA library of 0- to 21-h embryo mRNA cloned into the pJG4–5 activation domain plasmid (21). The HP1 “bait” construct was made by cloning amino acids 2–206 of HP1 into plasmid pEG202 containing DNA coding for the LexA-binding site. The final construct was confirmed by sequencing. The LexA-HP1 fusion protein enters the nucleus and binds to the ColE1 sites upstream of the reporter genes but does not activate the reporter genes (data not shown).
Antibodies, Immunoprecipitation, and Chromosome Staining.
Antiserum Ab 2–90 was produced against the protein encoded by the 2–90 cDNA recovered in the above screen. This cDNA was cloned into the pET28a protein expression plasmid, and the 6-HIS-tagged recombinant 2–90 protein was expressed in Escherichia coli. The protein was purified by using Ni-NTA spin columns (Qiagen, Chatsworth, CA) and size-separated on polyacrylamide gels, and the band corresponding to the 2–90 protein was cut from the gel and used as an immunogen in rabbits. Antibodies from this serum were affinity-purified by using the purified 2–90 protein. Antiserum Ab P-6 was made in guinea pigs against a peptide (amino acids 1336–1353 of the larger isoform) covalently crosslinked to keyhole limpet hemocyanin. These antibodies were purified by protein G affinity chromatography (Pierce) and used directly.
For immunoprecipitation assays, cDNAs isolated from the two-hybrid library were subcloned in the expression vector pET28. Protein produced from each plasmid with a coupled transcription/translation system (Promega) in the presence of S35-Met was mixed with soluble proteins from a Drosophila embryo extract (22), and anti-HP1 immunoprecipitation was carried out with Protein A-Sepharose 4 Fast Flow (Amersham Pharmacia; manufacturer's protocol). Bound proteins were eluted in 4% SDS and analyzed on 7% acrylamide gels.
Immunofluorescent staining of polytene chromosomes from third instar larvae of D. melanogaster was carried out as described (23). Secondary antibodies (Molecular Probes) were labeled with Alexa Fluor 594 (red) and Alexa Fluor 488 (green). Third instar larvae carrying the β-galactosidase (β-gal)–HP1–Polycomb fusion protein were treated at 37°C for 30 min and allowed to recover at room temperature for 24 h before collection and staining.
Genetic Screen and DNA Sequencing.
A lethal P element insertion, l(2)07214, just distal to the HP2 gene, was used in a male recombination scheme to generate adjacent deletions and duplications (24). Adult males carrying a second chromosome with cn l(2)07214 heterozygous with a bw chromosome, and a source of transposase, were crossed to females carrying a balancer chromosome marked with cn and bw. Progeny showing recombination between cn and bw were recovered, and the recombinant second chromosomes were placed in stock. Recombinant chromosomes were analyzed by PCR to assess whether a deletion or duplication had been produced, and inverse PCR was used to map the novel end of the P element in each case (25). Three lines were recovered. The B11 chromosome [designated Df(2R)B11] has a deletion of 42,934 bp, removing all of the coding sequence for HP2 as well as a few other genes (see below). A second, larger deletion (B7), removing approximately 152 kb, was not analyzed further. Analysis of line 214B revealed a duplication of 41,500 bp, with the duplication closely matching the region deleted in Df(2R)B11.
To recover point mutations in the HP2 gene, isogenic cn bw flies were fed a 1% sucrose solution containing 0.025 M ethyl methanesulfonate for 24 h and then mated to virgins carrying a Cy cn bw balancer chromosome. Individual male progeny with a newly mutagenized chromosome over the Cy cn bw chromosome were mated to 2–3 Df(2R)B11/Cy cn bw virgins. In a cross carrying a new mutation lethal over the B11 deletion, only curly winged flies will survive. The mutations identified were placed in stock over the Cy cn bw chromosome and retested with Df(2R)B11 for the lethal phenotype.
For sequencing, each putative mutation was placed over a Cy-GFP balancer chromosome and crossed to a line carrying the Df(2R)B11 deletion over the Cy-GFP balancer chromosome; first instar larvae with the mutated chromosome over the deletion (i.e., not expressing GFP) were collected. DNA from these individuals was sequenced by using various primer pairs to generate PCR fragments covering the entire HP2 gene. The sequences were compared with each other as well as with the genomic sequence found in the P1 clone DS00857. Putative mutations were confirmed by sequencing separate PCR products from at least three individuals.
Results
Identification of an HP1-Interacting Protein.
To identify proteins that interact with HP1, we undertook a yeast two-hybrid screen of a D. melanogaster early embryo cDNA library by using a LexA fusion protein containing the D. melanogaster HP1 coding sequence (amino acids 2–206) as bait (21). Forty-five putative interacting clones were isolated in the initial screen; secondary screening indicated five independent clones producing proteins that interacted with the HP1 fusion protein but not with control proteins in the yeast two-hybrid assay. To test further the interaction of these proteins with HP1, each of the five cDNAs was cloned in-frame in pET28a, and radiolabeled protein produced from these constructs was used in an in vitro coprecipitation assay. Of the five candidates, two showed significant interaction with HP1, as assayed by recovery of labeled protein of appropriate size in the immunoprecipitate (analyzed by gel electrophoresis; data not shown). The cDNA for the weaker interactor was sequenced and found to code for HOAP, a protein previously shown to interact with HP1 (26). The sequence of the stronger interactor, clone 2–90, indicated a unique protein that we have named HP2.
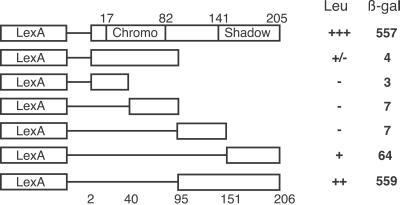
To characterize the detected interaction more fully, we used the yeast two-hybrid system to map the domain in HP1 responsible for interaction with the 2–90 product, testing LexA fusion proteins containing HP1 subdomains (27) as bait. The ability of these fusion proteins to interact with 2–90 and induce expression of the reporter genes was compared with that of full-length HP1. The results (Fig. 1) show that the binding activity is localized to the C-terminal half of HP1 (amino acids 95–206), containing part of the “hinge” and all of the shadow domain (amino acids 110–170). Partial activity is observed with the C-terminal quarter of HP1, which contains the amino acids responsible for HP1 self-dimerization and interaction with some other chromosomal proteins (16). Thus, the shadow domain and/or regions downstream are required for full interaction; the chromo domain does not appear to be involved.
Fig 1.
Interacting domain of HP1. Subdomains of the HP1 coding region fused to the LexA DNA-binding domain were tested for interaction with the polypeptide expressed by clone 2–90. The rate of growth of yeast carrying each construct on medium lacking leucine is indicated (Leu column) [no growth (−); strong growth (+++)], as is the specific activity of the induced β-gal reporter gene (β-gal column). Numbers above the lines indicate chromo and shadow domain boundaries, whereas those below the lines indicate the boundaries of subdomains used in the fusion constructs, with the exception of construct 4, which starts at amino acid 41, and construct 5, which starts at amino acid 93.
Analysis of the Gene for HP2.
Preliminary data generated by Northern blots of RNA isolated from embryos (0–16 h) and adult females hybridized with clone 2–90 cDNA suggested that the gene for HP2 encodes two transcripts of ca. 5 kb and 10 kb (data not shown), much larger than the 2-kb cDNA insert found in clone 2–90. To isolate the entire gene, we used the 2–90 cDNA insert to probe a macroarray of D. melanogaster genomic P1 clones; we found two (overlapping) P1 clones that hybridized to 2–90 cDNA. These clones, DS00857 and DS00968, previously had been mapped to a contig containing tra2 (28). Restriction maps of the smaller (DS00857) suggested that the HP2 gene was located near the P element insertion site l(2)07214. The sequence we determined for this region (unsubmitted) exactly matches the sequence submitted by Celera (GenBank accession no. AE003814).
Computer analysis by genescan (29) identified a likely gene 10,589 bp in length (CG12864). The predicted gene has nine exons coding for a transcript of 9,953 bases, with the end of the last exon matching the end of the 2–90 cDNA (Fig. 2). The size of the predicted gene agrees with the estimated size of the larger HP2 transcript of approximately 10 kb. genescan did not predict the smaller transcript, which is not surprising given the difficulty of predicting alternative splicing events (30). Sequencing revealed that a complex rearrangement had occurred within the 2–90 cDNA before isolation. The 2–90 cDNA is in the following order, defining base 1 as the first base of transcription predicted by genescan: 7490–7652, 6873–7096, 7136–7471, 8645–9953. Although the rearrangement is complex, there remains a single ORF of 2,028 bp (676 aa) that matches the reading frame for the native HP2 transcript.
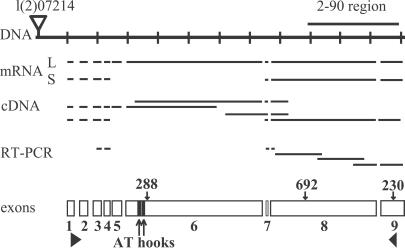
Fig 2.
The structure of the gene encoding HP2. A portion of genomic P1 clone DS00857 is shown (DNA), with the location of the 2–90 clone distal to the insertion site for P element l(2)07214. Various cDNAs and RT-PCR products sequenced to confirm the intron–exon boundaries are indicated. mRNA shows the predicted structures of the two transcripts produced by this locus (L, long; S, short). The nine exons are diagrammed below; solid triangles mark the start and stop of translation, and two solid rectangles mark the AT hooks. The locations of the genetic lesions in Su(var)2-HP2230, Su(var)2-HP2288, and Su(var)2-HP2692 also are indicated.
To confirm the predicted structure of the HP2 gene, we completely sequenced several cDNA clones mapped to this region (31) as well as several RT-PCR products that we generated. The sequences of these clones and products matched the predicted intron–exon boundaries for the regions covered (Fig. 2), with the exception that intron 6 was not removed in one cDNA clone. This cDNA may be derived from an incompletely spliced transcript or may represent a rare product containing exons 1–6 (a stop codon is present in intron 6). The 5′ end of one of the cDNA clones matches the start of transcription predicted by genescan. Using RT-PCR primers in the third and eighth exons, we were able to amplify a product of 384 bases; sequencing showed an alternative splice junction in which the end of the fourth exon is spliced to the seventh exon, eliminating the fifth and sixth exons. The predicted size of a transcript derived from this splicing pattern (containing exons 1–4 and 7–9) is 5,081 bases, the size of the smaller transcript observed.
The expression of two protein products was confirmed by Western blots of nuclear protein extracts from Drosophila embryos. A rabbit antiserum derived from the expressed 2–90 protein (Ab 2–90) detects two polypeptides of ≈175 kDa and ≈350 kDa, whereas a guinea pig antiserum (Ab P-6) derived from a synthetic sixth exon peptide (amino acids 1336–1353) detects only the larger polypeptide (data not shown). This finding confirms that both the smaller and larger transcripts are translated into protein (HP2-S and HP2-L, respectively) and verifies that HP2-S lacks exon 6.
HP2 Is Codistributed with HP1.
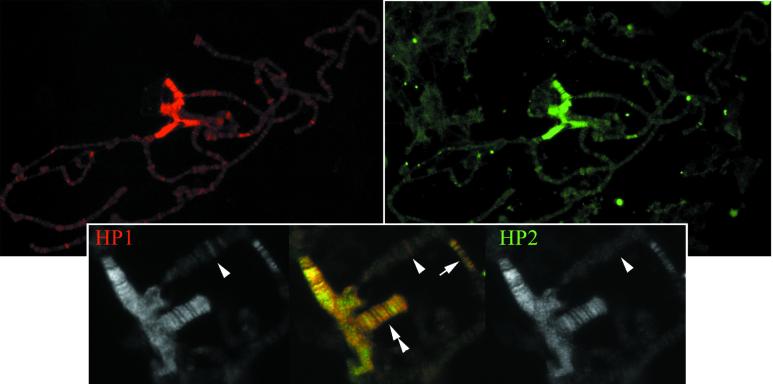
Both Ab 2–90 and Ab P-6 were used to label WT third instar polytene chromosomes with the same results; as shown (Fig. 3), the distribution patterns of HP1 and HP2 are strikingly similar. Both show predominant staining in the pericentric heterochromatin, including a banded pattern on the small fourth chromosome, with staining of minor peripheral sites in the euchromatic arms. Precise colocalization is observed along the fourth chromosome and in region 31, the most prominent of the euchromatic sites. Several other minor euchromatic sites show a higher immunofluorescent signal for HP1 than for HP2.
Fig 3.
Immunolocalization of HP1 and HP2. (Upper) Polytene chromosomes stained with anti-HP1 (red, Left) and anti-HP2-L (Ab P-6; green, Right). (Lower) Close-up view of the chromocenter. Single antibody signals are shown in black and white; the color merge of HP1 signal (red) and HP2-L signal (green) is shown in the center. Note the staining of the chromocenter, in a banded pattern along the fourth chromosome (double arrowhead) and of 5–6 bands in 31B (arrow). The arrowhead identifies a euchromatic band positive for HP1 but not HP2.
To confirm in vivo the interaction between HP1 and HP2, we used a Drosophila strain in which HP1 is recruited to ectopic sites in the euchromatin after heat shock, asking whether HP2 also would be recruited to these sites. This line contains a β-gal-HP1-Polycomb (β-gal-HP1-Pc) hybrid protein, a fusion of the β-gal protein upstream of a full-length HP1 protein in which the HP1 chromo domain (amino acids 21–67) has been replaced by the Pc chromo domain (amino acids 24–69) (32). Expression of this transgene is regulated by an Hsp70 heat shock promoter. After heat shock, the β-gal-HP1-Pc protein localizes to both normal Pc and normal HP1 sites on polytene chromosomes. In addition, WT HP1 is redistributed to the euchromatic Pc sites, indicating that the chimeric protein has the ability to recruit HP1 (or complexes containing HP1) (32). Before heat shock, HP1 and HP2 show an essentially WT staining pattern in this line; after heat shock, both the hybrid β-gal-HP1-Pc protein and HP2 can be seen at a variety of new euchromatic sites (Fig. 4). The recruitment of HP2 to these new sites upon expression of the β-gal-HP1-Pc chimera indicates that HP2 is either interacting directly with HP1 or is part of a complex containing HP1 recruited by the chimeric protein.
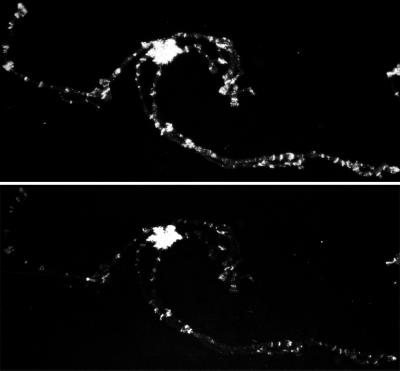
Fig 4.
β-gal (Upper) and HP2 (Lower) immunofluorescent staining of polytene chromosomes from heat shock-treated flies containing a transgene expressing a chimeric β-gal-HP1-Polycomb fusion protein. The staining pattern in the absence of heat shock shows only minor amounts of euchromatic staining by antibodies against HP2 (similar to Fig. 3), whereas the staining pattern after heat shock shows extensive relocalization of HP2 (Lower) to euchromatic regions marked by the presence of the fusion protein (Upper).
HP2-L Contains Two AT Hooks.
Conceptual translation of the two transcripts predicts the larger isoform to be 3,257 aa (355,974 Da) and the smaller isoform to be 1,633 aa (176,103 Da). Both proteins are serine-rich (HP2-L, 13.9%; HP2-S, 14.3%), and both contain a large number of charged amino acids (DEKR) (HP2-L, 30%; HP2-S, 25%). Statistical analysis shows that both proteins have serine contents above the 95th percentile for all proteins (33). However, the proteins differ in their net charge at neutral pH; the predicted pI of HP2-L is 5.9, whereas that of HP2-S is 9.1. Because of the high compositional bias and, therefore, low complexity, these proteins are recalcitrant to typical computer analysis. At stringent levels of filtering, the seg computer program (34) indicates only nine small regions of high complexity for HP2-L, none of which showed significant homology to any member of the nonredundant protein database.
Domain search algorithms did, however, reveal a pair of AT hooks, centered around amino acids 490 and 520; these motifs are within the sixth exon and, therefore, are exclusively in HP2-L. AT hooks are small, 9-aa motifs with basic residues surrounding an invariant glycine–arginine–proline central core sequence; three types of AT hooks, differing in the pattern of conserved flanking sequences, have been identified (35, 36). The first AT hook in HP2 appears to be a type I (LPKRGRPRGQKQR; 10 of 13 aa match to the consensus), whereas the second appears to be a type II (KRKRGRPRK; 9 of 9 aa match to the consensus).
Mutations in HP2 Result in Suppression of PEV.
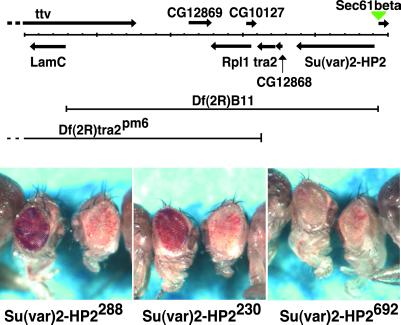
The B11 deletion, Df(2R)B11, acts as a dominant suppressor of PEV (data not shown). To determine whether mutations in the gene that codes for HP2 also result in suppression of PEV, we used chemical mutagenesis to isolate new mutations lethal over Df(2R)B11 (details of this screen to be published elsewhere). From 11,827 chromosomes screened, we identified, placed in stock, and confirmed 69 lines that had mutagenized chromosomes that were completely lethal over the Df(2R)B11 chromosome. From these, 17 lines were identified in which the lethal mutation mapped to the right of tra2 [by complementation to Df(2R)tra2pm6] (Fig. 5) in a complementation group distinct from Sec61beta [as defined by l(2)07214]. Members of this group most likely have mutations in the gene for HP2.
Fig 5.
Genetic analysis of Su(var)2-HP2. (Upper) The chromosomal region around the gene for HP2 is shown. Known and predicted genes are indicated by solid arrows (locations of putative primary transcripts). The location of the P element l(2)07214 is indicated by the green triangle. The boundaries of the two deletions used are shown. In addition to Su(var)2-HP2, Df(2R)B11 deletes tra2 and RpI1, three putative genes, CG12868, CG12869, and CG10127, and the last five exons of ttv, and ends 94 bases upstream of the start of transcription of LamC [map based on Adams et al. (37) as interpreted by Flybase (http://flybase.bio.indiana.edu/) (38)]. (Lower) Eye phenotypes of the three Su(var)2-HP2 alleles in males carrying a wm4h X chromosome. In each picture, the left fly carries the indicated mutation, whereas the right fly is a sibling carrying a WT copy of Su(var)2-HP2. Both Su(var)2-HP2230 and Su(var)2-HP2288 suppress variegated wm4h expression, whereas Su(var)2-HP2692 does not.
Three of these lines, 230, 288, and 692, have been sequenced and tested for the ability to dominantly suppress variegation of wm4h (Fig. 5). The DNA sequences from these three lines revealed 52 polymorphisms (common differences between the three mutant lines and the published genomic sequence, GenBank accession no. AE003814); 18 of the polymorphisms were silent, 33 changed a single amino acid, and one changed three adjacent amino acids. We also found a single mutation in the gene for HP2 specific to each line, confirmed by sequencing PCR amplicons from at least two heterozygous adults (Fig. 2). Line 288, with the strongest Su(var) activity, has a C-to-T change about 120 bases downstream of the second AT hook, changing amino acid 588 from threonine to isoleucine. Because this mutation is in the sixth exon, only HP2-L is affected. Line 230, also showing Su(var) activity, has an A-to-T change 110 bases (37 aa) upstream from the stop codon, changing amino acid 3220 from asparagine to isoleucine. Mutations 230 and 692 both are in the last exon, affecting both HP2-L and HP2-S. Line 692 contains an insertion of a single A just after amino acid codon 2370, resulting in removal of the last 887 aa. These are replaced by 28 aa, sequence TNQGITDFIK ASCTRWKEPA TCFKKESG. Interestingly, the mutagenized chromosome that carries the 692 allele does not suppress variegation of wm4h. This lack of suppression may be caused by genetic background effects from other mutations on the same chromosome; alternatively, it suggests a vital function(s) of HP2 separate from heterochromatin-induced gene silencing. Given that two separate mutations in the gene appear to suppress PEV of wm4h, we have named the gene Su(var)2-HP2 to indicate both its activity and location on the second chromosome.
Discussion
We have identified and characterized HP2, a large chromosomal protein that interacts with HP1 both in a yeast two-hybrid assay (Fig. 1) and by coimmunoprecipitation. The two proteins colocalize on polytene chromosomes (Fig. 3); HP2 is recruited to ectopic HP1 sites in vivo (Fig. 4). HP1 both recognizes H3-mK9, a marker of heterochromatin, and interacts with the modifying methyltransferase, SU(VAR)3–9. These interactions suggest a model for the spread of this packaging form along the chromatin fiber, a property of heterochromatin inferred from the observation of PEV (14, 15, 20). Our finding that mutations in HP2 can lead to suppression of PEV indicates a similar requirement for its participation in heterochromatin formation and spreading, with associated gene silencing. HP1 also has been shown to play a role in silencing at some euchromatic sites, including genes within region 31 (39). HP2 is observed in that region of the genome as well (Fig. 3), although, overall, HP2 association with euchromatic sites appears to be less than that found for HP1.
The gene for HP2 produces two transcripts, generated by inclusion or omission of exons 5 and 6; two proteins are detected with the predicted sizes of 176 and 356 kDa. The larger protein contains two AT hooks, protein motifs thought to contribute to heterochromatin assembly and stability by binding to AT-rich satellite DNA through the minor groove (40–44). One of the point mutations in HP2 leading to suppression of PEV is located close to the AT hooks (Figs. 2 and 5). Although there are no other recognizable motifs, HP2 does have a recognizable and unusual amino acid composition. Both isoforms are very rich in serine, as well as the four charged amino acids. blast searches of the nonredundant protein database with HP2-L find no proteins with significant similarity (E < 0.1) when compositional bias-based statistics are used. blast searches without compositional bias compensation find a wide variety of proteins rich in serine and the charged amino acids, including another HP1-binding protein, mouse ATRX (45).
ATRX, a transcription regulator, is localized to pericentric heterochromatin and the short arms of acrocentric chromosomes; mutations in the gene result in changes in DNA methylation patterns (46, 47). The segment of the protein (amino acids 325-1176) that interacts with a mouse HP1 homologue does not possess any recognizable structure, but also is rich in serine and the charged amino acids (53.7%). This suggests that HP1 may interact with proteins such as HP2 and ATRX on the basis of a compositional motif and suggests further that HP2 might have multiple interaction sites for HP1. Although HP1 may well act as a dimer (16), interactions of HP1 with a very large protein such as HP2 could be important in condensation of large chromatin domains. The identification of a novel protein of this type, and demonstration that mutations in the protein result in suppression of PEV, indicates the importance of identifying and characterizing additional partners of HP1 to further our exploration of the mechanisms involved in heterochromatin condensation and gene silencing.
Acknowledgments
We thank David Landsman (National Library of Medicine and National Institutes of Health) for confirming assignment of the AT-hook motif, William Mattox (M.D. Anderson Cancer Center, University of Texas, Houston) for providing the Drosophila strain with tra2pm6, and Lori Wallrath, Joel Eissenberg, Dale Dorsett, and members of the Elgin lab for critical review. This work was supported by National Institutes of Health Grant HD23844 (to S.C.R.E.). B.A.T., J.A.B., and L.F. were supported in part by Washington University/Howard Hughes Medical Institute Summer Undergraduate Research Fellowships provided under a grant from the Howard Hughes Medical Institute to Washington University. G.E.S. has been supported by National Institutes of Health Training Grant T32 GM07067.
Abbreviations
PEV, position effect variegation
HP1 and HP2, heterochromatin proteins 1 and 2
β-gal, β-galactosidase
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Weiler K. S. & Wakimoto, B. T. (1995) Annu. Rev. Genet. 29, 577-605. [DOI] [PubMed] [Google Scholar]
- 2.Wallrath L. L. & Elgin, S. C. R. (1995) Genes Dev. 9, 1263-1277. [DOI] [PubMed] [Google Scholar]
- 3.Sun F. L., Cuaycong, M. H. & Elgin, S. C. R. (2001) Mol. Cell. Biol. 21, 2867-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard P., Maure, J. F., Partridge, J. F., Genier, S., Javerzat, J. P. & Allshire, R. C. (2001) Science 294, 2539-2542. [DOI] [PubMed] [Google Scholar]
- 5.Nonaka N., Kitajima, T., Yokobayashi, S., Xiao, G., Yamamoto, M., Grewal, S. L. & Watanabe, Y. (2002) Nat. Cell Biol. 4, 89-93. [DOI] [PubMed] [Google Scholar]
- 6.Sinclair D. A., Lloyd, V. K. & Grigliatti, T. A. (1989) Mol. Gen. Genet. 216, 328-333. [DOI] [PubMed] [Google Scholar]
- 7.Wustmann G., Szidonya, J., Taubert, H. & Reuter, G. (1989) Mol. Gen. Genet. 217, 520-527. [DOI] [PubMed] [Google Scholar]
- 8.James T. C. & Elgin, S. C. R. (1986) Mol. Cell. Biol. 6, 3862-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eissenberg J. C., James, T. C., Foster-Hartnett, D. M., Hartnett, T., Ngan, V. & Elgin, S. C. R. (1990) Proc. Natl. Acad. Sci. USA 87, 9923-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eissenberg J. C., Morris, G. D., Reuter, G. & Hartnett, T. (1992) Genetics 131, 345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.James T. C., Eissenberg, J. C., Craig, C., Dietrich, V., Hobson, A. & Elgin, S. C. R. (1989) Eur. J. Cell Biol. 50, 170-180. [PubMed] [Google Scholar]
- 12.Cryderman D. E., Cuaycong, M. H., Elgin, S. C. R. & Wallrath, L. L. (1998) Chromosoma 107, 277-285. [DOI] [PubMed] [Google Scholar]
- 13.Eissenberg J. C. & Elgin, S. C. R. (2000) Curr. Opin. Genet. Dev. 10, 204-210. [DOI] [PubMed] [Google Scholar]
- 14.Bannister A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C. & Kouzarides, T. (2001) Nature 410, 120-124. [DOI] [PubMed] [Google Scholar]
- 15.Lachner M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. (2001) Nature 410, 116-120. [DOI] [PubMed] [Google Scholar]
- 16.Brasher S. V., Smith, B. O., Fogh, R. H., Nietlispach, D., Thiru, A., Nielsen, P. R., Broadhurst, R. W., Ball, L. J., Murzina, M. V. & Laue, E. D. (2000) EMBO J. 19, 1587-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aagaard L., Laible, G., Selenko, P., Schmid, M., Dorn, R., Schotta, G., Kuhfittig, S., Wolf, A., Lebersorger, A., Singh, P. B., et al. (1999) EMBO J. 18, 1923-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rea S., Eisenhaber, F., O'Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C. P., Allis, C. D., et al. (2000) Nature 406, 593-599. [DOI] [PubMed] [Google Scholar]
- 19.Schotta G., Ebert, A., Krauss, V., Fischer, A., Hoffman, J., Rea, S., Jenuwein, T., Dorn, R. & Reuter, G. (2002) EMBO J. 21, 1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richards E. J. & Elgin, S. C. R. (2002) Cell 108, 489-500. [DOI] [PubMed] [Google Scholar]
- 21.Gyuris J., Golemis, E., Chertkov, H. & Brent, R. (1993) Cell 75, 791-803. [DOI] [PubMed] [Google Scholar]
- 22.Becker P. B., Tsukiyama, T. & Wu, C. (1994) Methods Cell Biol. 44, 207-223. [DOI] [PubMed] [Google Scholar]
- 23.Clark R. F., Wagner, C. R., Craig, C. A. & Elgin, S. C. (1991) Methods Cell Biol. 35, 203-227. [DOI] [PubMed] [Google Scholar]
- 24.Preston C. R., Sved, J. A. & Engels, W. R. (1996) Genetics 144, 1623-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spradling A. C., Stern, D., Beaton, A., Rhem, E. J., Laverty, T., Mozden, N., Misra, S. & Rubin, G. M. (1999) Genetics 153, 135-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shareef M. M., King, C., Damaj, M., Badau, R., Huang, D. E. & Kellum, R. (2001) Mol. Biol. Cell 12, 1671-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powers J. & Eissenberg, J. C. (1993) J. Cell Biol. 120, 291-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimmerly W., Stultz, K., Lewis, S., Lewis, K., Lustre, V., Romero, R., Benke, J., Sun, D., Shirley, G., Martin, C., et al. (1996) Genome Res. 6, 414-430. [DOI] [PubMed] [Google Scholar]
- 29.Burge C. & Karlin, S. (1997) J. Mol. Biol. 268, 78-94. [DOI] [PubMed] [Google Scholar]
- 30.Black D. L. (2000) Cell 103, 367-370. [DOI] [PubMed] [Google Scholar]
- 31.Rubin G. M., Hong, L., Brokstein, P., Evans-Holm, M., Frise, E., Stapleton, M. & Harvey, D. A. (2000) Science 287, 2222-2224. [DOI] [PubMed] [Google Scholar]
- 32.Platero J. S., Hartnett, T. & Eissenberg, J. C. (1995) EMBO J. 14, 3977-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brendel V., Bucher, P., Nourbakhsh, I., Blaisdell, B. E. & Karlin, S. (1992) Proc. Natl. Acad. Sci. USA 89, 2002-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wootton J. C. & Federhen, S. (1996) Methods Enzymol. 266, 554-571. [DOI] [PubMed] [Google Scholar]
- 35.Aravind L. & Landsman, D. (1998) Nucleic Acids Res. 26, 4413-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huth J. R., Bewley, C. A., Nissen, M. S., Evans, J. N., Reeves, R., Gronenborn, A. M. & Clore, G. M. (1997) Nat. Struct. Biol. 4, 657-665. [DOI] [PubMed] [Google Scholar]
- 37.Adams M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185-2195. [DOI] [PubMed] [Google Scholar]
- 38.The Flybase Consortium (1999) Nucleic Acids Res. 27, 85-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang K. K., Eissenberg, J. C. & Worman, H. J. (2001) Proc. Natl. Acad. Sci. USA 98, 11423-11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strick R. & Laemmli, U. K. (1995) Cell 83, 1137-1148. [DOI] [PubMed] [Google Scholar]
- 41.Girard F., Bello, B., Laemmli, U. K. & Gehring, W. J. (1998) EMBO J. 17, 2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janssen S., Durussel, T. & Laemmli, U. K. (2000) Mol. Cell 6, 999-1011. [DOI] [PubMed] [Google Scholar]
- 43.Janssen S., Cuvier, O., Muller, M. & Laemmli, U. K. (2000) Mol. Cell 6, 1013-1024. [DOI] [PubMed] [Google Scholar]
- 44.Aulner N., Monod, C., Mandicourt, G., Jullien, D., Cuvier, O., Sall, A., Janssen, S., Laemmli, U. K. & Käs, E. (2002) Mol. Cell. Biol. 22, 1218-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Douarin B., Nielsen, A. L., Garnier, J. M., Ichinose, H., Jeanmougin, F., Losson, R. & Chambon, P. (1996) EMBO J. 15, 6701-6715. [PMC free article] [PubMed] [Google Scholar]
- 46.McDowell T. L, Gibbons, R. J., Sutherland, H., O'Rourke, D. M., Bickmore, W. A., Pombo, A., Turley, H., Gatter, K., Picketts, D. J., Buckle, V. J., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 13983-13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibbons R. J., McDowell, T. L., Raman, S., O'Rourke, D. M., Garrick, D., Ayyub, H. & Higgs, D. R. (2000) Nat. Genet. 24, 368-371. [DOI] [PubMed] [Google Scholar]