Abstract
Regulation of the proteasome system, which is responsible for the generation of most MHC class I-bound peptides, occurs through the interaction of the 20S proteasome with several regulatory proteins. One of these is PI31, which acts in vitro as an inhibitor of proteasome activity. Here, we demonstrate that, rather than inhibiting proteasome function, PI31 acts as a selective modulator of the proteasome-mediated steps in MHC class I antigen processing. Overexpression of PI31 in mouse embryonic cells has no impact on proteasome-mediated proteolysis. Instead, PI31, which localizes at the nuclear envelope/endoplasmic reticulum membrane, selectively interferes with the maturation of immunoproteasome precursor complexes. Consequently, overexpression of PI31 abrogates MHC class I presentation of an immunoproteasome-dependent cytotoxic T lymphocyte epitope and reduces the surface MHC class I levels on IFN-γ-treated mouse embryonic cells. Thus, PI31 represents a cellular regulator of proteasome formation and of proteasome-mediated antigen processing.
The proteasome system plays a central role in the degradation of intracellular proteins. Proteasomes remove abnormal proteins from the cytosol, play a role in transcription factor activation, in cell-cycle progression and, to an important extent, in the proteolytic processes involved in MHC class I antigen processing (1, 2). Proteasomes are composed of a 20S catalytic core that is attached to regulatory complexes, the 19S complex and the proteasome activator PA28 (3, 4). The 20S complex consists of four heptameric rings. The outer rings are occupied by α-subunits, which serve as interaction sites for the docking of regulatory particles. Enzymatic activity is mediated by three subunits that are located in the two inner proteasomal β-rings, β1/δ, β2/MC14, and β5/MB1. These subunits enter the proteasome complex as proforms preceded by a prosequence, which is removed during maturation of the precursor complex into a functional 20S proteasome (5, 6).
Whereas most of the functions exerted by proteasomes require proteolytic activity but no particular cleavage specificity, the liberation of MHC class I ligands from full-length antigens or shorter antigenic sequences demands a highly specific cleavage site usage (7). To accomplish this, cells are equipped with three IFN-γ-inducible facultative catalytic proteasome subunits, β1-induced (i)/LMP2, β2i/MECL-1, and β5i/LMP7, which are preferentially incorporated into newly assembled 20S complexes and thereby replace the three constitutive catalytic subunits (8). Both constitutive proteasomes as well as β1i/LMP2, β2i/MECL-1, and β5i/LMP7-containing proteasomes, so-called immunoproteasomes, are capable of antigenic peptide production. However, because of an only partial overlap in cleavage specificities and frequencies of cleavage site usage, it seems that the set of peptides produced by immunoproteasomes differs quantitatively and qualitatively from the one produced by constitutive proteasomes (9, 10). These processing differences can lead to an enhanced or reduced cytotoxic T lymphocyte (CTL) recognition of infected or tumor cells (11–16).
Binding of the 19S regulatory particle confers protein-degrading activity onto the 20S complex, which is essential for proteasomal functioning. PA28 attached to the other side of the 20S can modulate proteolytic cleavages. Although PA28 does not change the cleavage specificity of 20S, it influences both the quantity and the quality of the peptide repertoire displayed by MHC class I molecules on the cell surface (13, 17).
Recently, the function of a previously described (18) inhibitor of proteasome activity, PI31, was further examined (19, 20). These in vitro studies showed that both purified and recombinant PI31 diminished the enzymatic activity of purified 20S proteasomes. Recombinant PI31 was found to compete with the proteasome regulators PA28 (19) and 19S/PA700 (20) for binding to 20S, indicating that PI31 binds the 20S α-rings and may function by hindering substrate access to the 20S catalytic channel. Because these studies raised the possibility that PI31 might modulate cellular proteasome activity, we investigated the effects of PI31 on proteasome functioning in intact cells.
Methods
Cell Lines and Transfectant Cells.
Cells were cultured as described (11). To establish (m)PI31 transfectant cells, the pSG5-(m)PI3 expression construct (19) was introduced into MEC or B8 cells by using calcium phosphate precipitation. Cells were placed into limiting dilution, and transfectant clones were selected with puromycin (Ad5E1 MEC, 2 μg/ml) or zeocin (MEC217, MECPA28, B8, 100 μg/ml).
CFSE Labeling.
Cells were resuspended in 1 ml of PBS containing 5 μM carboxy-fluorescein diacetate-succinimidyl ester (CFSE; Molecular Probes) and labeled for 60 s at room temperature. Labeled cells were washed with medium and analyzed immediately or placed at 37°C and analyzed at the time intervals specified in the figure legends. Fluorescence was measured with a FACScalibur and analyzed with a CELLQUEST application program (Becton Dickinson).
GFP Degradation.
Ad5E1 MECs and PI31-transfectant clones were transfected with a Ub-R-GFP construct (21) and LLnL (50 μM final concentration) was added after 24 h. Three and one-half hours later, cells were trypsinized and then incubated in medium containing cyclohexamid (40 μg/ml) and anisomycin (30 μg/ml) at 37°C. Samples were taken at different time points and analyzed by flow cytometry in PBA buffer (PBS/0.5% BSA/0.02% NaN3) containing LLnL, cycloheximide, and anisomycin.
IκBα Degradation.
Ad5E1 MECs and PI31-transfectant clones were resuspended in medium at a concentration of 2.5 × 106 cells per ml. Samples of 200 μl were incubated without or with 20 ng/ml recombinant (rec) mouse tumor necrosis factor-α (mTNFα) for 30 min at 37°C. Then, 1 ml of ice cold PBS was added, and the cells were pelleted and frozen in liquid nitrogen. IkBα degradation was examined by Western blot analysis.
Western Blot Analysis.
Immunoblot analyses were performed as described (22). For detection of ubiquitinylated proteins, samples of 4 × 105 cells were incubated at the temperatures specified in the figure legends and then lysed in lysis buffer with 5 mM NEM. Extracted proteins were separated on 9% polyacrylamide gels and transferred to nitrocellulose. Membranes were probed with FK1 mouse mAb, specific for polyubiquitin-linked proteins (Affiniti, U.K.). To analyze subunit composition of proteasome precursor and 20S complexes, pellets of 4 × 108 cells were lysed in 1 ml of lysis buffer and applied to 10–40% glycerol gradients (13). Gradient fractions of 600 μl were collected and subjected to immunoblot analysis with polyclonal rabbit antisera against α2/MC3, β1i/LMP2, β5i/LMP7, β2i/MECL-1, PA28α, PA28β (11, 22), IκBα (Santa Cruz Biotechnology), or PI31. PI31-specific antisera were raised by immunization of rabbits with purified recombinant GST-PI31 fusion protein following standard procedures.
Metabolic Labeling Experiments.
Metabolic labeling of 20S complexes and 20S immunoprecipitation were performed as described (11).
To analyze viral protein stability, flasks with 2 × 106 parental Ad5E1 MEC or -PI31 clone 5 cells were washed with PBS, placed in methionine-free medium for 30 min, and then pulse-labeled with 1 mCi (1 Ci = 37 GBq) [35S]Met. Labeled cells were washed twice with PBS and then chased for 15 min (t:0) plus 1, 2, 3, or 9 h before lysis in 1 ml of 0.5% Nonidet P-40/0.2% sodium-deoxycholic acid/20 mM tri-ethanolamine·HCl, pH 8.0/700 mM NaCl/with protease inhibitors and 100 μg/ml DNase). Centrifuged detergent lysates were precleared overnight and immunoprecipitated with protein A-Sepharose and 200 μl of the mouse monoclonal antibodies M73, specific for Ad5E1A, or A1C6, specific for Ad5E1B.
To analyze H2-Kd trafficking, IFN-γ-treated cells were labeled with 200 μCi [35S]Met for 30 min and then immediately lysed or chased for 90 or 180 min before lysis. H2-Kd HC were immunoprecipitated with anti-exon 8 antiserum and analyzed by SDS/PAGE.
Immunofluorescence.
B8 cells were grown on glass slides and immunofluorescence analysis was performed as described (23) with a Leica TCS-NT Confocal laser scanning microscope. PI31 was detected with affinity-purified rabbit anti-PI31 antiserum and PDI with mouse anti-PDI antibody, clone 1D3 (Stressgene Biotechnologies, Victoria, BC, Canada). Secondary antibodies used were fluorescein-conjugated goat anti-rabbit IgG (Dianova, Hamburg, Germany) and Cy3-conjugated goat anti-mouse IgG (The Jackson Laboratory).
T Cell Assays.
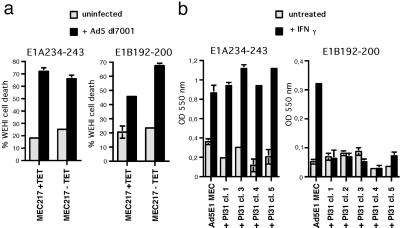
TNFα-production assays were performed as described (11). In short, uninfected and infected MEC217 cells cultured without or with tetracyclin (TET) were cocultured with 2 × 103 Ad5E1A-specific CTL (24) or 5 × 103 Ad5E1B-specific CTL (25) for 9 h or 22 h, respectively, in medium containing 30 units/ml rec (m)IL-2. TNFα-secretion was quantified as described (11), and percentages of specific WEHI cell death were calculated.
LacZ-inducible Ad5-specific T-cell hybridomas (26) were subcloned before usage. To test E1 peptide presentation, 5 × 104 hybridoma cells and 1 × 105 target cells were cocultured, and induction of LacZ expression was assayed as described (26).
Flow Cytometry.
Samples of 2 × 105 Ad5E1 MECs and of PI31-transfectant clones 1–5 were incubated with saturating amounts of the mouse monoclonal antibodies AF6–88.5 and 28.14.8S, specific for H2-Kb and -Db, respectively, with rat monoclonal antibody anti-mouse CD119/IFN-γ receptor α-chain (PharMingen) or without antibody for 45 min on ice. Cells were washed with PBA and then incubated with FITC-labeled goat-anti mouse IgG FAB2-fragments (Dianova) or FITC anti-rat IgG2a (PharMingen) for 45 min, washed, and analyzed with a FACScalibur.
Results
PI31 Does Not Inhibit Cellular Proteasome Activity.
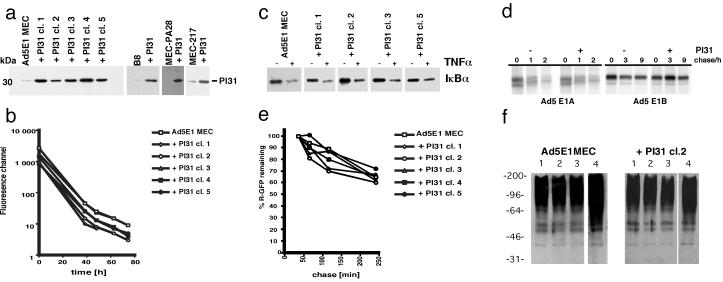
In vitro studies have shown that both purified and recombinant PI31 compete with the proteasome regulators PA28 and PA700 for binding to the 20S complex, and that PI31 inhibits 20S enzymatic activity (18–20). To explore the effects of this cell-derived proteasome inhibitor in intact cells, we transfected the mouse embryonic cell line Ad5E1 MEC (24), which expresses the adenovirus type 5 early 1 (E1) region and exhibits a low endogenous PI31 expression, with the coding sequences of mouse PI31. Transfectant cells were identified by immunoblot analysis, and five random clones with significantly increased PI31 levels were chosen for further analyses (Fig. 1a). Overall, PI31 transfectant clones grew remarkably well; the doubling time of the selected five clones (24 h) was identical to that of untransfected Ad5E1 MECs (data not shown). To investigate their growth behavior more precisely, cells were labeled with CFSE and diminution of fluorescence was followed by flow cytometry (Fig. 1b). No differences in decay of fluorescence signal were detected between parental Ad5E1 MECs and PI31-transfectants, indicating identical growth rates. Because proteasome activity is essential for cell-cycle progression, these findings imply that PI31 has no major impact on the general proteasome function in Ad5E1 cells.
Fig 1.
(a) Ad5E1 MEC, MEC217, MEC-PA28, and B8 cells were transfected with pSG5-mPI31 and cloned by limiting dilution. Parental cells and clones derived from single cells were extracted, the lysates were separated by SDS/PAGE, and PI31 was detected by Western blotting with PI31-specific antiserum. (b) Ad5E1 MECs and derived PI31-transfectant clones 1–5 were labeled with CFSE. Cells were cultured for the specified time periods, after which fluorescence was measured by flow cytometry. (c) Ad5E1 parental and PI31-transfectant cells were treated with 20 ng/ml TNFα for 30 min or left untreated and then extracted. IκBα was detected by Western blot analysis. (d) Ad5E1 MECs and PI31 transfectant clone 5 were pulse-labeled with [35S]Met and then chased for the indicated time periods. Ad5E1A and Ad5E1B proteins were immunoprecipitated from the detergent lysates, separated by SDS/PAGE, and visualized by autoradiography. Of note, the signals detected in Ad5 E1A lanes after 1 and 2 h of chase are of higher molecular weight than the E1A proteins (t:0) and have to be regarded as unspecific. (e) Ad5E1 MECs and derived PI31-transfectant clones 1–5 were transfected with a Ub-R-GFP expression construct; the N-end-rule substrate R-GFP was accumulated in the presence of LLnL. Degradation of R-GFP was measured by flow cytometry after removal of LLnL. (f) Ad5 E1 MECs and PI31 transfectant clone 2 cells were incubated for 30 min at 37°C (lane 1), 30 min at 42°C (lane 2), 30 min at 42°C and then 30 min at 37°C (lane 3), or for 30 min at 37°C in the presence of 250 μM LLnL (lane 4) and then extracted. Polyubiquitin-linked proteins were detected by Western blot analysis with FK1 antibody.
To examine whether the overexpression of PI31 affects other proteasome-mediated processes, Ad5E1 MECs were treated with TNFα. Degradation of IκBα, which is induced by activation of the NFκB-signal transduction pathway and has been shown to be exerted by 26S proteasomes (27), was analyzed by Western blotting and scan densitometry. Similar decreases in IκBα levels were observed for Ad5E1 MECs and PI31 transfectants (Fig. 1c), arguing against an impairment of 26S proteasome function despite high PI31 expression.
Because most nuclear and cytoplasmic proteins are degraded by 26S proteasomes, we investigated further the degradation of the Ad5 E1A and E1B proteins in the presence of PI31. Metabolic labeling and chase analysis revealed comparable degradation rates of the viral proteins between parental and PI31-transfected Ad5E1 MECs (Fig. 1d). Whereas the two E1A proteins completely diminished within the first 2 h of chase, E1B appeared relatively stable over time, in concordance with the earlier reported long half-life of this protein (28).
As an alternative approach to monitor PI31 effects on proteasomal proteolysis, we examined degradation of the ubiquitin (Ub)-R-GFP fusion protein, a relatively short-lived N-end-rule substrate (21), in PI31 transfectant cells. Cells were transiently transfected with a Ub-R-GFP cDNA construct, and R-GFP was accumulated by treatment of the cells with the reversible proteasome inhibitor LLnL (Fig. 1e). Upon removal of LLnL from the culture medium, GFP signals declined over time, with no discernible differences between parental and PI31 transfectant cells (Fig. 1e), indicating that protein degradation proceeds normally in the presence of PI31. In addition, no major impact of PI31 on the breakdown of other cellular proteins like c-jun or other viral proteins was detected (data not shown).
It is possible that proteasome redundancy masks potential inhibitory effects of PI31. To test whether stress conditions that lead to increased proteolysis uncover so-far-undetected effects of PI31 on constitutive proteasomes, Ad5 E1 parental and PI31 transfectant cells were subjected to heat shock. Accumulation of polyubiquitin-linked proteins was examined by Western analysis (Fig. 1f). These experiments showed that incubation of the cells for 30 min at 42°C (Fig. 1f), or up to 3 h at 44 or 45°C (not shown), did not significantly increase the amounts of polyubiquitin-linked proteins in PI31-transfectant cells. Contents of ubiquitinylated proteins remained unchanged during subsequent culture of the cells at 37°C. Thus, PI31-transfectants show no apparent defects in degradation of ubiquitinylated proteins. In contrast, cells treated with the synthetic proteasome inhibitor LLnL did accumulate multiubiquitin conjugates (Fig. 1f).
Taken together, these data demonstrate that in contrast to previously obtained in vitro evidence, PI31 does not function as a general inhibitor of proteasome activity in vivo.
Cellular PI31 Interferes with Immunoproteasome Maturation.
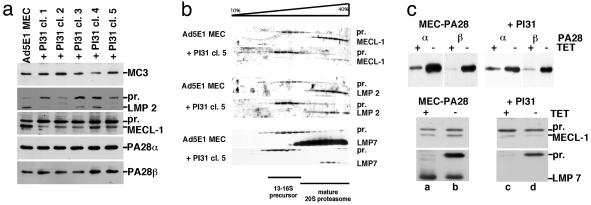
To examine whether PI31 influences the amount of intracellular proteasomes of PA28 or proteasome subunit composition, Ad5E1 parental and transfectant cells were treated with IFN-γ to induce immunosubunit expression, and total cell lysates were subjected to Western analysis. As shown in Fig. 2a, equal amounts of the proteasomal subunit α2/MC3 were detected in parental and PI31-transfectant cells, which indicates that possible effects of PI31 were not compensated for by an increase in proteasome quantity. Moreover, PI31 did not exert any discernible effects on the quantity of the PA28α and PA28β subunits. In contrast, a marked increase in the ratios between precursor and mature, N-terminally processed β1i/LMP2 and β2i/MECL-1 was observed in PI31 transfectants (Fig. 2a), indicating a reduced or delayed formation of 20S immunoproteasomes in the PI31-transfectant cells. To explore this finding in more detail, precursor and mature 20S complexes of IFN-γ-induced cells were separated by glycerol gradient centrifugation and then analyzed for the presence of immunosubunits. As shown for a representative clone in Fig. 2b, an accumulation of β1i/LMP2-, β2i/MECL-1-, and β5i/LMP7-containing proteasome precursor complexes was detected in PI31 transfectants, indicating that the immunosubunit proforms enter the 16S proteasome precursor complexes (6) but are inefficiently processed. Although we did not detect any differences in β1i/LMP2 and β2i/MECL-1 mRNA levels between Ad5E1 MECs and PI31-transfectant clones (data not shown), we infer that PI31 directly interferes with the maturation of immunoproteasomes.
Fig 2.
Ad5E1 MECs and -PI31 transfectants were treated for 3 days with 60 units/ml rec (m)IFN-γ. (a) Total cell lysates were subjected to Western blot analysis with antisera specific for α2/MC3, β1i/LMP2, β2i/MECL-1, PA28α, and PA28β. (b) Lysates were centrifuged on a glycerol gradient (10–40%), and collected fractions (600 μl) were probed by Western blot analysis with antisera specific for the immunosubunits. Mature and precursor (pr) forms are indicated. Of note, the strong signals obtained for β5i/LMP7 in Ad5E1 MEC fractions are explained by the high affinity of the β5i/LMP7-specific antiserum rather than by protein quantity. (c) MEC-PA28 and -PI31 transfectant cells cultured for 3 days with or without TET in the absence (Upper) or presence (Lower) of IFN-γ were extracted and probed with antisera specific for PA28α and PA28β or β2i/MECL-1 and β5i/LMP7.
As PA28 was suggested to enhance immunoproteasome formation (29), one could speculate that the negative effects of PI31 on immunoproteasome assembly result from competitive inhibition of PA28 binding. However, analysis of a PI31-transfected MEC clone with TET-regulated PA28 expression (Figs. 1a and 2c Upper) showed that PI31 impairs immunosubunit incorporation (Fig. 2c Lower) even in the presence of large quantities of PA28 (lanes 2 and 4). From these data and the results of Murata et al. (30) showing that the absence of PA28 does not affect immunoproteasome assembly, we infer that PI31 is unlikely to exert its effects by competing with PA28 for 20S binding.
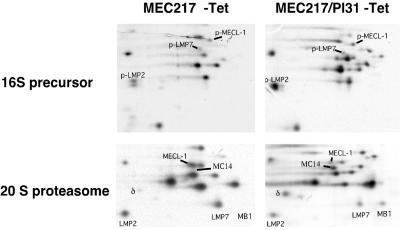
To confirm our finding that PI31 diminishes immunoproteasome maturation, we transfected MEC217 cells (11) that up-regulate immunosubunit expression in response to removal of TET from the culture medium with PI31. Parental MEC217 and a PI31 transfectant clone (Fig. 1a), cultured without TET, were metabolically labeled. Cell lysates were fractionated on glycerol gradients, and proteasomes were immunoprecipitated. Analysis of the 16S precursor fractions (Fig. 3 Upper) revealed that both MEC217 and PI31-transfectant cells incorporated the immunosubunit proforms in their proteasome precursor complexes. Proforms of β1/δ, β2/MC14, and β5/MB1 were not detected, indicating that at most a minor fraction of precursor complexes contained the constitutive catalytic subunits. However, whereas the immunoproteasome precursors matured into 20S immunoproteasomes in MEC217 cells (Fig. 3 Lower), only approximate ratios of 1:1 between constitutive subunits and immunosubunits were reached in 20S proteasomes of MEC217-PI31 cells. Thus, PI31 allows the formation of 16S immunoproteasome precursor complexes but severely impairs their maturation to functional 20S complexes, as is evident from the preservation of constitutive β1/δ, β2/MC14, and β5/MB1 in 20S proteasomes of MEC217-PI31.
Fig 3.
MEC217 and MEC217-PI31 cells were cultured in the absence of TET for 1 day to induce immunosubunit expression and then labeled for 4 h with [35S]Met and extracted. Detergent lysates were centrifuged on glycerol gradients (10–40%) and collected as 600-μl fractions. Proteasome complexes were immunoprecipitated from the fractions, separated by NEPHGE-SDS/PAGE two-dimensional gel electrophoresis, and visualized by autoradiography; the subunits were assigned according to Elenich et al. (35).
PI31 Localizes at the Nuclear Envelope/Endoplasmic Reticulum (ER) Membrane.
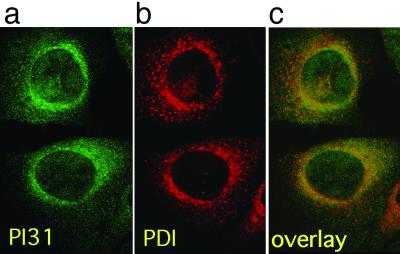
To investigate whether PI31 is scattered throughout cells or confined to specific subcellular compartments, we analyzed the intracellular localization of PI31 by immunofluorescence microscopy. Cells grown on micro-slides were stained with anti-PI31 antiserum and FITC-labeled secondary antibody. Confocal microscopic analysis (Fig. 4) showed an unequal distribution of PI31 throughout cells, with a predominant localization at the cytosolic site of the nuclear envelope/ER membrane. Colocalization studies with the ER markers PDI (Fig. 4), calnexin, and gp96 (not shown) confirmed this presumed localization. Interestingly, immunoproteasomes also have been described to reside at the nuclear envelope/ER membrane (31), making this the most likely location for immunoproteasomes to assemble and mature. Therefore, our results suggest that the intracellular localization of PI31 is in agreement with the observed effects of this molecule on immunoproteasome precursor maturation.
Fig 4.
(a) PI31 was visualized in B8 mouse fibroblasts by immunofluorescence staining with affinity-purified rabbit anti-PI31 antiserum and FITC-labeled secondary antibody (green). (b) The nuclear envelope/ER membrane marker PDI was visualized with monoclonal mouse anti-PDI antibody and Cy3-labeled secondary antibody (red). (c) a and b superimposed; colocalization appears in yellow.
PI31 Diminishes Processing of an Immunoproteasome-Dependent CTL Epitope.
The Ad5E1 proteins contain two antigenic peptides, E1A 234–243 (24) and E1B 192–200 (25), that are presented by H2-Db MHC-I molecules and are important targets for Ad5-specific murine CTL. We recently showed (11) that infected cells processed E1B 192–200 efficiently only when the IFN-γ-inducible proteasome immunosubunits were expressed. In contrast, the liberation of E1A 234–243 requires proteasome activity (not shown) but occurs independently of the presence of immunoproteasomes. As shown in Fig. 5a, TET-regulated expression of the immunosubunits in Ad5-infected MEC217 cells does not influence the presentation of E1A 234–243 to specific CTL, whereas immunosubunits enhance the presentation of the E1B 192–200 epitope, as reported (11).
Fig 5.
(a) MEC217 cells, cultured for 2 days with TET to repress or without TET to induce immunosubunit expression, were infected with Ad5 dl7001 or left uninfected and tested for recognition by E1A 234–243- and E1B 192–200-specific CTL in a TNFα-production assay. Percentages of specific WEHI cell death, a measure for TNFα-secretion by activated CTL, are depicted on the y axis. Mean values and SDs of triplicate wells are shown. (b) Ad5E1 MECs and Ad5E1 MEC-PI31 clones, untreated or treated with 60 units/ml IFN-γ for 3 days, were tested for recognition by E1A- and E1B-specific T cell hybridomas. Lac Z activity was quantified by measuring the conversion of chlorophenol red β-galactoside in hybridoma lysates at OD at 550 nm (y axis).
To investigate whether PI31-inhibition of IFN-γ-induced immunoproteasome formation has functional consequences, we analyzed antigen processing of the two Ad5 CTL epitopes in parental and transfectant Ad5E1 MECs. IFN-γ-treated and untreated cells were cultured overnight with E1A 234–243 or E1B 192–200-specific T cell hybridomas (26). Whereas untreated MECs were not recognized above background levels (because of low MHC class I expression; see Fig. 6a), all IFN-γ-treated cells exhibited a similar ability to stimulate E1A-specific hybridoma cells (Fig. 5b). In contrast, E1B 192–200 was recognized on IFN-γ-treated parental Ad5E1 MECs (Fig. 5b), but none of the PI31 transfectant clones was able to stimulate E1B-specific hybridoma cells (Fig. 5b). Thus, PI31 does not affect E1A 234–243 presentation, but interferes with antigen processing of the immunoproteasome-dependent E1B CTL epitope.
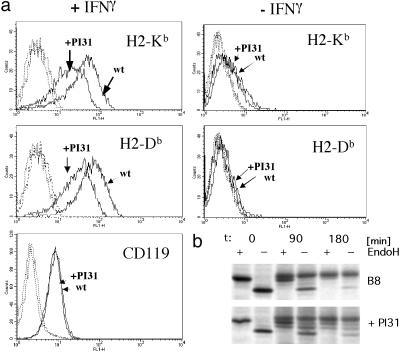
Fig 6.
(a) Parental Ad5E1 MEC and Ad5E1 MEC-PI31 clone 3 cells were cultured with or without IFN-γ for 3 days. Cells were stained with H2-Kb-, H2-Db-, and CD119-specific antibodies and then with FITC-conjugated secondary antibody. Fluorescence was measured with a FACScalibur (Becton Dickinson). Histograms obtained for parental cells (wt) and MEC-PI31 transfectant clone 3 (+PI31) are depicted. Dashed lines, background staining obtained with secondary antibody only; solid lines, untreated cells (Right) or IFN-γ-treated cells (Left). (b) H2-Kd HC of IFN-γ-treated B8 and B8-PI31 transfectant cells were immunoprecipitated with anti-exon 8 antiserum at indicated time points after pulse-labeling and treated with or without EndoH.
One explanation of the above results could be that PI31 also acts as a transcription or translation inhibitor. However, Northern analysis revealed no differences in mRNA levels of E1B and H2-Db heavy chain (HC) between IFN-γ-treated Ad5E1 MECs and derived transfectants (not shown). Also, levels of E1B protein synthesis are similar in parental and transfectant cells (Fig. 1d), and PI31-transfected cells up-regulated the TAP2 protein after IFN-γ-stimulation, indicating that the IFN-γ-signaling pathway is functional in the presence of excessive amounts of PI31 (not shown). Taken together, our results imply that diminished presentation of the immunoproteasome-dependent E1B 192–200 on PI31-Ad5E1 MECs is most likely the result of reduced generation of this antigenic peptide in the absence of 20S immunoproteasomes.
PI31 Decreases IFN-γ-Induced MHC Class I Up-Regulation.
To examine whether PI31 influences the generation of only a few or a great array of CTL epitopes, we analyzed the cell-surface expression of H2-Kb and H2-Db MHC class I molecules on the Ad5E1 MEC clones. Unstimulated MECs transcribe the MHC class I HCs at low levels, resulting in a barely detectable MHC class I cell-surface expression (for parental Ad5E1 MEC and a representative transfectant clone, see Fig. 6a). Both parental and PI31-transfected Ad5E1 MECs markedly up-regulated MHC class I surface expression after IFN-γ-stimulation (Fig. 6a and Table 1); however, the absolute levels of H2-Kb and H2-Db reached on PI31-transfected MECs were substantially lower than those on parental cells. In contrast, similar amounts of IFN-γ receptor α chain (CD119) were detected on transfectant and parental cells, indicating that PI31 does not inhibit cell-surface transport of glycoproteins in general. IFN-γ-induced up-regulation of cell-surface molecules was further analyzed on PI31-transfected B8 mouse fibroblast cells (Fig. 1a), which confirmed that enhanced PI31 expression significantly diminishes up-regulation of MHC class I molecules after IFN-γ stimulation without affecting CD119 cell-surface levels (not shown). Although metabolic labeling failed to reveal differences in MHC class I HC synthesis between parental and transfectant cells, chase analyses showed a delayed trafficking of H2-Kd in B8-PI31 cells and of H2-Kb complexes in Ad5E1 MEC-PI31 cells, as evidenced from the accumulation of HC with partially processed oligosaccharide chains after 90 and 180 min chase (Fig. 6b and data not shown). Therefore, we infer that defective immunoproteasome maturation in PI31 transfectant cells leads to insufficient production of class I ligands which impairs the IFN-γ-induced up-regulation of MHC class I cell-surface expression.
Table 1.
MHC class I and CD119 cell-surface expression on PI31-transfectant cells
| Cells | H2-Kb | H2-Db | CD119 |
|---|---|---|---|
| Ad5E1 MEC | 13,1† | 19,5 | 3,3 |
| + PI31 clone 1 | 6,3 | 10,7 | 3,5 |
| + PI31 clone 2 | 6,2 | 9,2 | ND |
| + PI31 clone 3 | 7,0 | 10,4 | 3,3 |
| + PI31 clone 4 | 4,8 | 9,5 | 2,8 |
| + PI31 clone 5 | 8,2 | 14,0 | 3,1 |
ND, not determined.
Ad5E1 MEC and PI31 transfectant clones were treated with 60 units/ml (m)IFN-γ for 3 days and analyzed for cell-surface expression of MHC class I H2-Kb and -Db and CD119 by immunofluorescence staining.
† Fluorescence indexes: mean fluorescence channel-stained cells/mean fluorescence channel of cells incubated with second antibody only.
Discussion
Our experiments show that, in contrast to the observed inhibitory effects of PI31 on purified 20S complexes in vitro, PI31 does not inhibit the general 20S/26S proteasome activity in intact cells. Instead, we found that PI31 specifically interferes with 20S immunoproteasome formation and, in parallel, with MHC class I-presentation of immunoproteasome-dependent CTL epitopes. The formation of 20S constitutive proteasomes seems unaffected. Remarkably, the inhibitory effects of PI31 occur at the level of proteasome precursor-complex processing and become apparent only upon analysis of the mature 20S proteasome population. Thus, unprocessed proforms of β1i/LMP2, β5i/LMP7, and β2i/MECL-1 enter the proteasome precursor in PI31-expressing cells, but only a small percentage of immunosubunit-containing precursor complexes reaches the mature stage (Figs. 2b and 3). Nevertheless, the total cellular proteasome contents are not detectably altered under conditions of PI31 overexpression, implying that PI31 selectively hinders immunoproteasome maturation.
Prosequence removal, leading to proteasome activation, takes place in the 16S precursor complex and is a multistep process (5) in which the N-terminal sequences are removed by autocatalytic cleavages (6). Our finding that immunoproteasome precursor complexes are assembled but fail to mature properly in PI31-transfectant cells implies that PI31 specifically interferes with the processing of immunosubunit prosequences. Therefore, our present data suggest that, rather than inhibiting 20/26S proteasome activity as such, intracellular PI31 interferes with the autocatalytic steps that result in the formation of active 20S immunoproteasomes.
In parallel to impaired immunoproteasome formation, IFN-γ-treated PI312+ cells exhibit a severely reduced MHC class I presentation of the immunoproteasome-dependent E1B CTL epitope (Fig. 5b). In contrast, MHC class I antigen processing of the immunoproteasome-independent E1A 234–342 epitope, which is presented by the same MHC class I molecule, is not impaired in these cells (Fig. 5b), indicating the intact functioning of the non-proteasome-related components of the antigen processing pathway, and that the amount of class I molecules available for peptide binding is not limiting. Instead, the reduced up-regulation of both H2-Db and H2-Kb cell-surface expression after IFN-γ stimulation (Fig. 6 and Table 1) suggests that generation of a large number of CTL epitopes is inhibited in PI31-transfected cells, resulting in insufficient amounts of antigenic peptides to load newly synthesized MHC class I molecules. This observation was unexpected, because we found previously that immunosubunit introduction into MECs does not alter MHC class I cell-surface expression (unpublished observations). On the other hand, both down-regulation of β1i/LMP2 expression in lymphoma T cells and the disruption of β5i/LMP7 expression in mice were shown to diminish the MHC class I cell-surface levels (32, 33). Taken together, and in agreement with other reports, these findings imply that, in cells with high MHC class I expression levels, MHC class I assembly is limited by peptide availability. Moreover, because MHC class I cell-surface expression is down-regulated upon inhibition of immunoproteasome formation, we infer that immunosubunit incorporation increases the yield of antigenic peptides. In agreement with our findings, a previous study (34) also suggested that MHC class I loading in IFN-γ-induced cells requires full immunoproteasome activity.
As both PI31 and immunoproteasomes (or their precursors) localize at the ER membrane (31), we speculate that PI31 may serve to control immunoproteasome formation and may thereby maintain an intracellular balance between constitutive and immunoproteasomes. Whereas proteasome components that play a central role in cellular metabolism are highly conserved between yeast and mammals, these components of the proteasome system that are involved in MHC class I antigen processing are found in mammals only. Interestingly, like PA28 and the immunosubunits, PI31 also has no homologue in yeast, which may be indicative of a specific function of this molecule in the antigen-processing pathway. Interestingly, many cell types, for example, fibroblasts, display a low constitutive expression of the proteasome immunosubunits. Therefore, we suggest that PI31 plays a regulatory role by preventing the formation of immunoproteasomes in the absence of infection.
Acknowledgments
We thank R. Toes, U. Schaible, J. Howard, N. Dantuma, M. Masucci, and J. Yewdell for the gift of reagents and helpful discussion. This work was supported by the Deutsche Forschungs Gemeinschaft Grants KL421 10-2 and SFB421-A1.
Abbreviations
CTL, cytotoxic T lymphocytes
TET, tetracyclin
ER, endoplasmic reticulum
TNFα, tumor necrosis factor α
HC, heavy chain
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rock K. L., York, I. A., Saric, T. & Goldberg, A. L. (2002) Adv. Immunol. 80, 1-70. [DOI] [PubMed] [Google Scholar]
- 2.Shastri N., Schwab, S. & Serwold, T. (2002) Annu. Rev. Immunol. 20, 463-493. [DOI] [PubMed] [Google Scholar]
- 3.Tanahashi N., Kawahara, H., Murakami, Y. & Tanaka, K. (1999) Mol. Biol. Rep. 26, 3-9. [DOI] [PubMed] [Google Scholar]
- 4.Kloetzel P. M. (2001) Nat. Rev. Mol. Cell. Biol. 2, 179-187. [DOI] [PubMed] [Google Scholar]
- 5.Nandi D., Woodward, E., Ginsburg, D. B. & Monaco, J. J. (1997) EMBO J. 16, 5363-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidtke G., Kraft, R., Kostka, S., Henklein, P., Frommel, C., Lowe, J., Huber, R., Kloetzel, P. M. & Schmidt, M. (1996) EMBO J. 15, 6887-6898. [PMC free article] [PubMed] [Google Scholar]
- 7.Sijts A., Zaiss, D. & Kloetzel, P. (2001) Curr. Mol. Med. 1, 665-676. [DOI] [PubMed] [Google Scholar]
- 8.Fruh K., Gossen, M., Wang, K., Bujard, H., Peterson, P. A. & Yang, Y. (1994) EMBO J. 13, 3236-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toes R. E., Nussbaum, A. K., Degermann, S., Schirle, M., Emmerich, N. P., Kraft, M., Laplace, C., Zwinderman, A., Dick, T. P., Muller, J., et al. (2001) J. Exp. Med. 194, 1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cascio P., Hilton, C., Kisselev, A. F., Rock, K. L. & Goldberg, A. L. (2001) EMBO J. 20, 2357-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sijts A. J. A. M., Standera, S., Toes, R. E., Ruppert, T., Beekmann, N. J., van Veelen, P. A., Ossendorp, F. A., Melief, C. J. & Kloetzel, P.-M. (2000) J. Immunol. 164, 4500-4506. [DOI] [PubMed] [Google Scholar]
- 12.Sijts A. J. A. M., Ruppert, T., Rehermann, B., Schmidt, M., Koszinowski, U. & Kloetzel, P.-M. (2000) J. Exp. Med. 191, 503-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Hall T., Sijts, A. J. A. M., Camps, M., Melief, C., Kloetzel, P.-M. & Ossendorp, F. (2000) J. Exp. Med. 192, 483-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W., Norbury, C. C., Cho, Y., Yewdell, J. W. & Bennink, J. R. (2001) J. Exp. Med. 193, 1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwarz K., Van Den Broek, M., Kostka, S., Kraft, R., Soza, A., Schmidtke, G., Kloetzel, P.-M. & Groettrup, M. (2000) J. Immunol. 165, 768-778. [DOI] [PubMed] [Google Scholar]
- 16.Morel S., Levy, F., Burlet-Schiltz, O., Brasseur, F., Probst-Kepper, M., Peitrequin, A. L., Monsarrat, B., Van Velthoven, R., Cerottini, J. C., Boon, T., et al. (2000) Immunity 12, 107-117. [DOI] [PubMed] [Google Scholar]
- 17.Groettrup M., Soza, A., Eggers, M., Kuehn, L., Dick, T. P., Schild, H., Rammensee, H. G., Koszinowski, U. H. & Kloetzel, P. M. (1996) Nature 381, 166-168. [DOI] [PubMed] [Google Scholar]
- 18.Chu-Ping M., Slaughter, C. A. & DeMartino, G. N. (1992) Biochim. Biophys. Acta 1119, 303-311. [DOI] [PubMed] [Google Scholar]
- 19.Zaiss D. M., Standera, S., Holzhutter, H., Kloetzel, P. & Sijts, A. J. (1999) FEBS Lett. 457, 333-338. [DOI] [PubMed] [Google Scholar]
- 20.McCutchen-Maloney S. L., Matsuda, K., Shimbara, N., Bins, D. D., Tanaka, K., Slaughter, C. A. & DeMartino, G. (2000) J. Biol. Chem. 275, 18557-18565. [DOI] [PubMed] [Google Scholar]
- 21.Dantuma N. P., Lindsten, K., Glas, R., Jellne, M. & Masucci, M. G. (2000) Nat. Biotechnol. 18, 538-543. [DOI] [PubMed] [Google Scholar]
- 22.Groettrup M., Standera, S., Stohwasser, R. & Kloetzel, P. M. (1997) Proc. Natl. Acad. Sci. USA 94, 8970-8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knuehl C., Seelig, A., Brecht, B., Henklein, P. & Kloetzel, P.-M. (1996) Exp. Cell Res. 225, 67-74. [DOI] [PubMed] [Google Scholar]
- 24.Kast W. M., Offringa, R., Peters, P. J., Voordouw, A. C., Meloen, R. H., van der Eb, A. J. & Melief, C. J. (1989) Cell 59, 603-614. [DOI] [PubMed] [Google Scholar]
- 25.Toes R. E., Offringa, R., Blom, R. J., Brandt, R. M., van der Eb, A. J., Melief, C. J. & Kast, W. M. (1995) J. Immunol. 154, 3396-3405. [PubMed] [Google Scholar]
- 26.Schoenberger S. P., Jonges, L. E., Mooijaart, R. J., Hartgers, F., Toes, R. E., Kast, W. M., Melief, C. J. & Offringa, R. (1998) Cancer Res. 58, 3094-3100. [PubMed] [Google Scholar]
- 27.Ciechanover A., Gonen, H., Bercovich, B., Cohen, S., Fajerman, I., Israel, A., Mercurio, F., Kahana, C., Schwartz, A. L., Iwai, K. & Orian, A. (2001) Biochimie 83, 341-349. [DOI] [PubMed] [Google Scholar]
- 28.Branton P. E. & Rowe, D. T. (1985) J. Virol. 56, 633-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preckel T., Fung-Leung, W. P., Cai, Z., Vitiello, A., Salter-Cid, L., Winqvist, O., Wolfe, T. G., Von Herrath, M., Angulo, A., Ghazal, P., et al. (1999) Science 286, 2162-2165. [DOI] [PubMed] [Google Scholar]
- 30.Murata S., Udono, H., Tanahashi, N., Hamada, N., Watanabe, K., Adachi, K., Yamano, T., Yui, K., Kobayashi, N., Kasahara, M., et al. (2001) EMBO J. 20, 5898-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks P., Fuertes, G., Murray, R. Z., Bose, S., Knecht, E., Rechsteiner, M. C., Hendil, K. B., Tanaka, K., Dyson, J. & Rivett, J. (2000) Biochem. J. 346 Pt 1, 155-161. [PMC free article] [PubMed] [Google Scholar]
- 32.Sibille C., Gould, K. G., Willard-Gallo, K., Thomson, S., Rivett, A. J., Powis, S., Butcher, G. W. & De Baetselier, P. (1995) Curr. Biol. 5, 923-930. [DOI] [PubMed] [Google Scholar]
- 33.Fehling H. J., Swat, W., Laplace, C., Kuhn, R., Rajewsky, K., Muller, U. & von Boehmer, H. (1994) Science 265, 1234-1237. [DOI] [PubMed] [Google Scholar]
- 34.Benham A. M. & Neefjes, J. J. (1997) J. Immunol. 159, 5896-5904. [PubMed] [Google Scholar]
- 35.Elenich L. A., Nandi, D., Kent, A. E., McCluskey, T. S., Cruz, M., Iyer, M. N., Woodward, E. C., Conn, C. W., Ochoa, A. L., Ginsburg, D. B. & Monaco, J. J. (1999) Immunogenetics 49, 835-842. [DOI] [PubMed] [Google Scholar]