Abstract
The proteasome inhibitor PS-341 inhibits IκB degradation, prevents NF-κB activation, and induces apoptosis in several types of cancer cells, including chemoresistant multiple myeloma (MM) cells. PS-341 has marked clinical activity even in the setting of relapsed refractory MM. However, PS-341-induced apoptotic cascade(s) are not yet fully defined. By using gene expression profiling, we characterized the molecular sequelae of PS-341 treatment in MM cells and further focused on molecular pathways responsible for the anticancer actions of this promising agent. The transcriptional profile of PS-341-treated cells involved down-regulation of growth/survival signaling pathways, and up-regulation of molecules implicated in proapoptotic cascades (which are both consistent with the proapoptotic effect of proteasome inhibition), as well as up-regulation of heat-shock proteins and ubiquitin/proteasome pathway members (which can correspond to stress responses against proteasome inhibition). Further studies on these pathways showed that PS-341 decreases the levels of several antiapoptotic proteins and triggers a dual apoptotic pathway of mitochondrial cytochrome c release and caspase-9 activation, as well as activation of Jun kinase and a Fas/caspase-8-dependent apoptotic pathway [which is inhibited by a dominant negative (decoy) Fas construct]. Stimulation with IGF-1, as well as overexpression of Bcl-2 or constitutively active Akt in MM cells also modestly attenuates PS-341-induced cell death, whereas inhibitors of the BH3 domain of Bcl-2 family members or the heat-shock protein 90 enhance tumor cell sensitivity to proteasome inhibition. These data provide both insight into the molecular mechanisms of antitumor activity of PS-341 and the rationale for future clinical trials of PS-341, in combination with conventional and novel therapies, to improve patient outcome in MM.
In eukaryotes, a highly conserved multienzyme system covalently links ubiquitin to intracellular proteins targeted for degradation. The resulting ubiquitin-protein conjugates are degraded by the 26S proteasome, a large ATP-dependent protease (1–5). Proteasome inhibitors constitute a class of antitumor agents with preclinical evidence of activity against hematologic malignancies and solid tumors (6–11). Specifically PS-341, a boronic acid dipeptide with selective activity as a proteasome inhibitor, has activity against multiple myeloma (MM) cells in vitro (11); and inhibits tumor growth in a murine plasmacytoma model (12). In a multicenter Phase II clinical trial in MM patients with very ominous prognosis due to rapidly progressing relapsed refractory disease, PS-341 has demonstrated remarkable antitumor activity, including objective responses (even complete ones) in ≈55% of patients and disease stabilization in another ≈25% of patients (13, ∗∗). To date, however, the precise molecular targets mediating the anti-MM activity of PS-341 are not fully defined.
Proteasome inhibition abrogates degradation and induces cytoplasmic accumulation of IκB, which blocks the nuclear translocation and transcriptional activity of NF-κB. This effect may account in part for the anti-MM effects of PS-341: NF-κB, a potential therapeutic target in MM, regulates cell adhesion molecule expression and IL-6 production in the bone marrow milieu (11); and its constitutive activity enhances MM cell survival and resistance to cytotoxic agents, by transcription of inhibitors of apoptosis such as Bcl-2, A1, cIAP-2, and XIAP (14); conversely, certain anti-MM therapies, e.g., dexamethasone, thalidomide, and its immunomodulatory analogs (IMiDs), inhibit NF-κB activity (11, 15–19). Comparison of the effects of PS-341 vs. PS-1145, a specific IκB kinase inhibitor, on MM cells, suggests that NF-κB inhibition may not be the sole mediator of PS-341 anti-MM activity (20). Further delineation of the molecular targets correlating with response and resistance to PS-341 may both delineate the mechanism(s) of its antitumor activity and allow for the development of more specific, less toxic, targeted therapies.
Transcript profiling and population genomics in Saccharomyces cerevisiae identified the transcription factor Rpn4p as a mediator of response to PS-341 (21). More importantly, that study, performed by the same group that developed PS-341, demonstrated that only a limited number of genes is involved in the PS-341-induced sequelae in that model (21). Because the genome of S. cerevisiae is fully sequenced and well explored genetically, it is unlikely that any significant PS-341-induced interactions in that model were missed, highlighting a striking selectivity in the actions of this proteasome inhibitor and supporting its role as a clinically applicable agent. Because of differences in cellular physiology between S. cerevisiae and human neoplastic cells (e.g., human MM cells undergo apoptosis after treatment with PS-341 at concentrations 10,000- to 100,000-fold lower than those used in ref. 21), we focused in this study on the molecular mechanisms of the antitumor cell actions of PS-341 that are most relevant to its use in our patients with MM, which is currently considered the prototypic disease model of antitumor activity of PS-341. Specifically, we characterized by oligonucleotide microarrays the gene expression profile of proteasome inhibitor-treated MM cells and defined molecular pathways with a putative role in the proapoptotic effects of proteasome inhibition. Further mechanistic studies on these pathways showed that PS-341 decreases the levels of several antiapoptotic proteins and induces a dual apoptotic pathway of mitochondrial cytochrome c release and caspase-9 activation, as well as activation of Jun kinase (JNK) and a Fas/caspase-8-dependent apoptotic pathway [which is inhibited by a JNK inhibitor and a dominant negative (decoy) Fas construct, respectively]. Overexpression of Bcl-2 or constitutively active Akt in MM cells also modestly attenuates PS-341-induced cell death, whereas inhibitors of the BH3 domain of Bcl-2 or of the chaperoning activity of the heat-shock protein 90 (hsp90) enhance tumor cell sensitivity to proteasome inhibition. These studies therefore both delineate molecular targets of proteasome inhibitors and provide the framework for development of more effective proteasome inhibitor-based therapies, which will aim at potentiating the proapoptotic molecular sequelae induced by PS-341 and/or neutralizing protective mechanisms against its action.
Materials and Methods
Tissue Culture and Materials.
The human MM cell line MM.1S was kindly provided by S. Rosen (Northwestern University, Chicago) and cultured as described (14). PS-341 was provided by Millennium Pharmaceuticals (Cambridge, MA). The hsp90 inhibitor NSC683664 was obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, National Cancer Institute (Bethesda).
Affymetrix Chip Hybridization.
cRNA (20 μg) was fragmented by incubating in a buffer containing 200 mM Tris-acetate (pH 8.1), 500 mM KOAc, and 150 mM MgOAc at 94°C for 35 min. The hybridization mixture containing 15 μg adjusted fragmented cRNA mixed with Eukaryotic Hybridization controls (contains control cRNA and oligonucleotide B2) was hybridized with a preequilibrated human U95Av2 Affymetrix chip at 45°C for 16 h. After the hybridization cocktails were removed, the chips were washed in a fluidic station with low-stringency buffer (6× standard saline phosphate with EDTA, 0.01% Tween 20, and 0.005% antifoam) for 10 cycles (two mixes per cycle) and high-stringency buffer (100 mM N-morpholinoethanesulfonic acid, 0.1 M NaCl, and 0.01% Tween 20) for four cycles (15 mixes per cycle) and stained with streptavidin phycoerythrin. This process was followed by incubation with normal goat IgG and biotinylated mouse antistreptavidin antibody and restaining with streptavidin phycoerythrin. The chips were scanned in an HP ChipScanner (Affymetrix, Santa Clara, CA) to detect hybridization signals.
Caspase Cleavage.
The involvement of caspases in PS-341-induced apoptosis was studied by evaluating the levels of procaspases-8 and -9, and the emergence of their cleaved active forms, by immunoblotting of lysates of cells treated with PS-341 (100 nM) for 2–16 h.
Transfection of Fas-Fc, Bcl-2, and Constitutively Active Akt Constructs.
To evaluate the role of the Fas apoptotic pathway in PS-341-induced apoptosis, a soluble Fas-Fc chimeric molecule, which acts as a decoy receptor and inhibits Fas activation (23), was expressed in MM.1S cells by transient transfection by using Lipofectamine 2000 (Life Technologies, Grand Island, NY), according to the instructions of the manufacturer. Forty-eight hours later, cells were treated with PS-341 or vehicle as indicated. Cell survival was assessed by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, as described (24).
To evaluate the role of Akt kinase and Bcl-2 in PS-341-induced apoptosis, MM.1S cells were stably transfected with vectors for a myristoylated (constitutively active) Akt construct or a Bcl-2 construct (Upstate Biotechnologies), respectively, or the empty (neo) vector by using Lipofectamine 2000 according to manufacturer's instructions. Forty-eight hours later, cells were incubated in growth medium containing G418 (500 μg/ml, Life Technologies) to select pools of stable clones.
Statistical Analysis.
Statistical significance was examined by a two-way ANOVA, followed by Duncan's post hoc test. In all analyses, P < 0.05 was considered statistically significant.
Further Details.
For further details, see supporting Materials and Methods, which is published on the PNAS web site, www.pnas.org.
Results
Transcriptional Profile of PS-341-Treated MM.1S Cells.
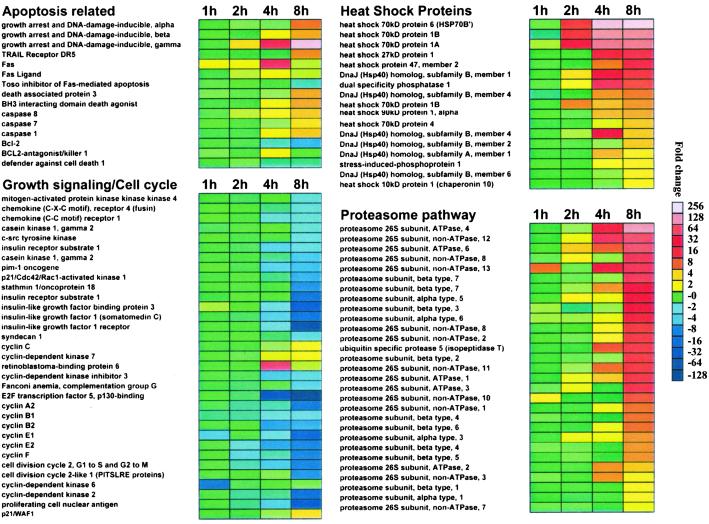
To define molecular pathways regulating PS-341-induced apoptosis, we characterized, by oligonucleotide microarray analysis, the gene expression profiles of MM cells treated with PS-341 (100 nM for 0–8 h) vs. control cells by using the human U95Av2 Affymetrix genechip. MM cells do not exhibit morphological features of apoptosis until after 12 h of treatment with PS-341; cell viability was thus maintained throughout the experiment. PS-341 did not suppress transcriptional activity of tumor cells indiscriminately, but rather induced distinct patterns of coordinated changes in a range of transcripts, including down-regulation of growth and antiapoptotic transcripts; induction of members of apoptotic cascades; up-regulation of components of the proteasome/ubiquitin pathway; and an especially potent up-regulation of hsp transcripts. The selectivity of the actions of PS-341 on mammalian cells is consistent with the results of Fleming et al. in S. cerevisiae (21), where PS-341-induced sequelae involved distinct patterns of molecular events, and all of the identified genes were encoding well characterized substrates or regulators of the 26S proteasome. Understandably, such a specific response pattern highlights the functional impact of PS-341 on select intracellular molecular pathways, and further supports the use of this proteasome inhibitor as a tool for further mechanistic studies, as well as a clinically applicable therapeutic agent. A summary of PS-341-induced changes in transcripts regulating these pathways is presented in Fig. 1.
Fig 1.
Transcriptional profile of PS-341-treated MM-1S cells by oligonucleotide-microarray analysis. Representative functional clusters of transcriptional changes induced by PS-341 (100 nM, 1–8 h) include molecules implicated in caspase-mediated apoptosis, growth factor signaling and cell cycle control, heat shock proteins, and proteasome subunits. Color saturation is proportional to magnitude of the difference from the respective control.
PS-341 Down-Regulates Expression of Growth-Signaling Pathway Components.
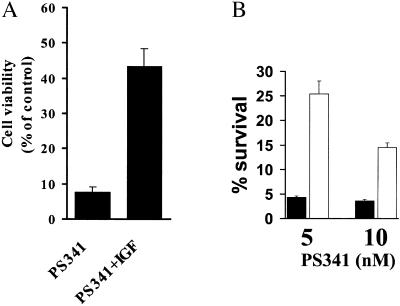
PS-341 potently down-regulates transcription of key molecules implicated in growth and survival cascades, e.g., 4- to 16-fold reductions in expression of insulin-like growth factor 1 (IGF-1), its receptor (IGF-1R), and molecules regulating IGF-1 signaling, including insulin receptor substrate-1 (IRS-1) and IGFBP-3 (Fig. 1), indicating that PS-341 targets the IGF-1-signaling pathway, which promotes MM cell growth, survival, and resistance to drugs including dexamethasone (25), Fas crosslinking, and TRAIL/Apo2L (26). These data are also consistent with the in vitro synergy of PS-341 and TRAIL/Apo2L (27), and the additive anti-MM effect of PS-341 and dexamethasone, either in vitro (11) or in vivo.** We extended these findings by further investigating the effect of exogenous IGF-1 on PS-341-induced apoptosis: IGF-1 (200 ng/ml) decreased the PS-341-sensitivity of MM cells (Fig. 2A). Because Akt is a key mediator of antiapoptotic effects of IGF-1 (26), we stably transfected MM.1S with constitutively active Akt and found that the transfectants were less sensitive to PS-341 than control vector cells (Fig. 2B). This finding suggested that the antitumor activity of PS-341 can be influenced by paracrine factors present in the MM microenvironment, such as IGF-1; and that the clinical efficacy of PS-341 may be enhanced when used in combination with agents inhibiting these factors and/or their signaling pathways.
Fig 2.
Protective effect of the IGF/Akt pathway against apoptosis induced by proteasome inhibition. (A) IGF-1 (200 ng/ml) lowers the sensitivity to PS-341 (10 nM). Cells were serum-starved overnight and then incubated with or without PS-341 in serum-free medium for additional 18 h. (B) PS-341 (5 and 10 nM)-induced cell death (quantified by MTT, mean ± SD) in MM.1S cells transfected with a vector expressing constitutively active Akt (open bars), or control vector (filled bars). After overnight serum starvation, cells were incubated with or without PS-341 in serum-free medium for additional 18 h.
PS-341 Induces Dual Apoptotic Signaling.
Our oligonucleotide microarray studies also revealed that PS-341 modulates the expression of genes related to death cascades, e.g., PS-341 induces pro-caspase-8, pro-caspase-1, pro-caspase-7 (mch3), caspase-4, caspase-9 (mch6), and pro-caspase-5 mRNA; increases the death-inducing receptors DR5 and Fas and the death ligand FasL; decreases the antiapoptotic proteins Bcl-2 and BIRC3, as well as Toso and FLIP, which are negative regulators of Fas-mediated apoptosis (28); and increases GADD34 (29–31) and GADD45, which mediate growth arrest and apoptosis triggered by UV radiation or chemotherapy (32) (Fig. 1). PS-341 also induces an increase in p21/WAF1; suppression of transcripts for XBP-1, a key transcription factor for plasma cell differentiation; and down-regulates syndecan-1 (CD138), which is expressed on the surface of viable MM cells, stimulates the growth and survival of MM cells, and is down-regulated during induction of apoptosis (33).
These results prompted us to focus further on apoptotic signaling induced by proteasome inhibition, and, specifically, the cascades of caspase activation. Having previously characterized the caspase cascades triggered by conventional therapies, e.g., dexamethasone (34), and novel agents, e.g., thalidomide (Thal) and its immunomodulatory derivatives (IMiDs) (19), Fas (35), and Apo2L/TRAIL (24), the goal of this study was to delineate how PS-341 overcomes drug resistance, and provide the basis for enhancing its activity by combination with other therapies. PS-341 rapidly decreased protein expression of Bcl-2, A1, cIAP-2, and (at higher concentration) FLIP and XIAP (Fig. 7A, which is published as supporting information on the PNAS web site), but did not change the protein levels of cIAP-1 and Bcl-XL. These changes are reminiscent of those observed in MM cells after specific inhibition of NF-κB (14). PS-341 inhibits tumor necrosis factor-α-induced NF-κB activation (36) and we now show that it also down-regulates constitutive NF-κB activity (Fig. 7B) in a dose-dependent manner that parallels the down-regulation of the aforementioned apoptosis inhibitors. Because the observed in PS-341-induced decreases in antiapoptotic Bcl-2 and A1 proteins suggest mitochondrial involvement in the induction of apoptosis, we next confirmed that PS-341 induces cytoplasmic release of mitochondrial cytochrome c (Fig. 7C).
Further investigating the role of caspases in PS-341-induced apoptosis, we confirmed by immunoblotting, that PS-341 induces an early cleavage of caspase-9, and later cleavage of caspase-8 (Fig. 5A, which is published as supporting information on the PNAS web site). The pan-caspase inhibitor ZVAD-FMK completely abolishes PS-341-induced apoptosis, whereas the caspase-8 inhibitor IETD-FMK and the caspase-9 inhibitor LEHD-FMK each mediate partial protection (Fig. 5B). These studies confirm dual apoptotic signaling triggered by PS-341.
Our finding of mitochondrial involvement, evidenced by down-regulation of Bcl-2 and A1 expression associated with cytochrome c release, prompted us to investigate further whether overexpression of Bcl-2 conferred protection against PS-341. We therefore stably transfected MM.1S cells with Bcl-2 expressing or control vectors (Fig. 8A, which is published as supporting information on the PNAS web site). The effect of PS-341 on survival was modestly attenuated by Bcl-2 overexpression in transfectants (Fig. 8B). Conversely, the BH3I-2′ inhibitor, a cell-permeable peptide which specifically prevents BH3 domain-mediated interaction between proapoptotic and antiapoptotic members of the Bcl-2 family (37) and by itself has no anti-MM activity, sensitizes MM.1S cells to PS-341 (Fig. 8C). Collectively, these data support the role of the Bcl-2/mitochondria/cytochrome c/caspase-9 axis as one of the mediators of PS-341-induced apoptosis.
Because our oligonucleotide microarray profiling and immunoblotting data both show involvement of the death receptor Fas and its downstream effector caspase 8, we next examined the role of Fas-mediated signaling in PS-341-induced apoptosis. Pretreatment with subtoxic concentrations of PS-341 (5 nM for 4 h) sensitizes MM.1S cells to subtoxic concentrations of the Fas crosslinking antibody CH11 (Fig. 9A, which is published as supporting information on the PNAS web site). Immunoblotting also reveals that PS-341 induces up-regulation of FasL on MM.1S cells (Fig. 9B). Because proteasome inhibitors stabilize c-myc (38), which regulates FasL expression (39, 40), we next confirmed that c-myc protein levels are also increased in PS-341-treated MM.1S cells (Fig. 9B). To confirm functional involvement of the Fas-signaling pathway in PS-341 induced MM cell apoptosis, we transiently transfected MM.1S cells with an expression vector containing a cDNA encoding the extracellular domain of human Fas ligated to the constant region of the Fc fragment of human IgG1 (Fas-IgG1), which acts as a decoy inhibitor of Fas activation (23). Transient expression of Fas-Fc reduces PS-341-sensitivity in MM.1S cells, compared with cells transfected with the control vector (Fig. 9C). These data support the notion that the Fas/FasL system is an additional mediator of PS-341-induced apoptosis.
To characterize the mechanism of up-regulation of Fas, we evaluated the role of c-Jun and AP-1, because AP-1 has been reported to regulate Fas expression (41). PS-341 treatment (8–12 h) of MM.1S cells stabilizes JNK (data not shown) and increases c-Jun phosphorylation and DNA-binding activity of the transcription factor AP-1 (Fig. 6A, which is published as supporting information on the PNAS web site), correlating with increased protein levels of Fas (Fig. 6B). The JNK inhibitor SP600125 partially blocks PS341-induced Fas up-regulation (Fig. 6B) and apoptosis (Fig. 6C). These data are in agreement with Meriin et al. (42), who described a crucial role for JNK, a proteasome substrate, in apoptosis induced by the proteasome inhibitor MG132 in U937 cells, because it was suppressed by a dominant-negative SEK1 mutant and a JNK antisense oligonucleotide. Taken together, these data suggest that JNK is activated during and contributes to PS-341-induced apoptosis by stimulating the Fas pathway.
PS-341 Increases Transcription of Components of the Ubiquitin/Proteasome Pathway in MM.1S Cells.
Our oligonucleotide microarray analyses also reveal that PS-341 stimulates transcription of molecules in the ubiquitin/proteasome pathway (Fig. 1). Most notably, PS-341 up-regulates expression of ubiquitin, as well as a wide range of proteasome subunits. These include: 26S proteasome subunits p44.5 and p55, which are required for cell survival (43); proteasome subunits HsN3, HsC7-I and HsC10-II; p112, the largest regulatory subunit of the 26S proteasome (44); p40.5 subunit, which protects against heat stress (45); p31 regulatory subunit, a homolog of the S. cerevisiae protein Nin1p which is required for G1/S and G2/M cell cycle transition (46); HC5, HC8, and p58 subunits; proteasome-associated pad1 homolog (POH1), which confers pleiotropic P-glycoprotein-independent resistance to paclitaxel, doxorubicin, 7-hydroxystaurosporine, and UV light in mammalian cells (47); subunits X, Y, and Z, implicated in substrate specificity of the proteasome and its modulation by extracellular stimuli such as IFN-γ (48–52); as well as the p97 subunit of 26S proteasome, a polypeptide identical to the type-1 tumor necrosis factor receptor-associated protein-2 (53). This coordinated up-regulation of multiple genes encoding proteasome subunits triggered by PS-341 likely represents a stress response, similar to that reported in S. cerevisiae (21).
PS-341 Increases Transcription of Stress Proteins in MM.1S Cells.
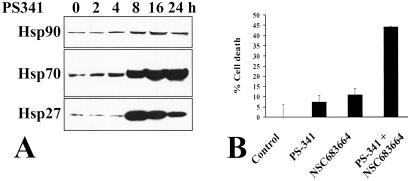
Our transcriptional profiling studies also showed that PS-341 induces a pronounced and global increase in hsp transcripts, including hsp90, hsp70, and hsp40 families; hsp28; hsp apg-1; and mitochondrial hsp75 (Fig. 1). These changes are among the most pronounced responses to PS-341 observed in our transcriptional analysis and likely reflect a stress response, which would be consistent with the well documented role of these molecular chaperones in conferring protection against therapeutic agents (54). Immunoblotting confirmed PS-341-induced up-regulation of hsp27, -70, and -90 (Fig. 3A). The functional significance of up-regulation of hsps, in general, and, in particular, hsp90, in conferring a protective effect against PS-341 is confirmed by the fact that inhibitors of the hsp90-chaperoning function (55, 56), such as the geldanamycin analog NSC683664, sensitized MM cells to PS-341-mediated apoptosis (Fig. 3B).
Fig 3.
Increased expression of stress-response proteins in apoptosis induced by proteasome inhibition. (A) Immunoblotting confirms PS-341 induces time-dependent increase in levels of hsp90, hsp70, and hsp27. (B) The hsp90 inhibitor NSC683664 (100 nM) sensitizes MM.1S cells to a subtoxic concentration of PS-341 (5 nM).
Discussion
Proteasome inhibitors constitute a class of anticancer agents with highly encouraging preliminary clinical evidence of activity, particularly against MM, the second most commonly diagnosed hematologic malignancy in the United States. Even when administered as a single agent, PS-341 offers disease stabilization or objective responses (even complete ones), in ≈80% of MM patients (13, ∗∗) who previously failed all available conventional therapies and several other investigational anti-MM agents (e.g., thalidomide and IMiDs) (13, ∗∗). This exciting result prompted us to characterize the molecular sequelae of PS-341 treatment of MM cells, and define molecular pathways that mediate its proapoptotic activity or that may, conversely, attenuate its effect; potentiation of the former mechanisms and/or neutralization of the latter ones may improve the antitumor activity of proteasome inhibitor-based therapies.
Because of the pivotal role of the proteasome in cellular physiology, it could have been expected that proteasome inhibitors would induce nonspecific accumulation of undegraded substrates, cause massive and indiscriminate transcriptional shutdown, and prove too toxic for clinical use. Nevertheless, Fleming et al. demonstrated that only a limited number of genes is involved in the PS-341-induced sequelae in S. cerevisiae (21). By using oligonucleotide microarray-based transcriptional profiling of PS-341-treated MM cells, we now found that proteasome inhibition induces, in human tumor cells, a specific coordinated pattern of transcriptional events consistent with its proapoptotic effect, e.g., down-regulation of transcripts involved in key growth/survival signaling pathways (e.g., the IGF-1-receptor pathway), and up-regulation of molecules implicated in proapoptotic pathways (e.g., caspase cascade).
The down-regulation, by proteasome inhibition, of transcripts implicated in growth signaling, and in particular of IGF-1 and key mediators of its signaling cascade, are consistent with our previous data, which demonstrate that IGF-1 mediates resistance to dexamethasone and to anticancer therapies (e.g., Apo2L/TRAIL) by Akt signaling (26). Because IGF-1 is present in vivo both in the serum and the bone marrow microenvironment where MM cells primarily reside, our current findings may explain the clinical observation of the, at least additive, in vivo anti-MM effect of PS-341 with dexamethasone (∗∗): by counteracting the expression of IGF-1 and its signaling mediators, PS-341 can block not only the proliferative effect of this cascade, but also abrogate its ability to confer resistance against dexamethasone. Furthermore, in view of our recent findings that the IGF-1/Akt axis reduces the sensitivity of MM cells to TRAIL/Apo2L (26), our current data may also explain why PS-341 has a synergistic effect with TRAIL/Apo2L, in in vitro experiments in the presence of IGF-1-containing serum (27). The PS-341 sensitivity of MM cells was reduced in the presence vs. absence of IGF-1, whereas overexpression of Akt (the key mediator of IGF-1-induced anti-apoptotic signaling), in a constitutively active form, also decreased the degree of proteasome inhibitor-induced apoptosis. These pieces of information taken together suggest that the IGF-1/Akt axis and the proteasome inhibition constitute two opposing forces, which tend to promote cell survival vs. apoptosis, respectively, and that, from a therapeutic standpoint, the antitumor activity of PS-341 can be enhanced, instead of using increased PS-341 doses, by inhibition of IGF-1 and its downstream signaling.
Several key proteins in pathways regulating transcription and growth/apoptosis are known substrates for proteasomal degradation, such as IκB (36), c-myc (38, 57, 58), and JNK (42). However, the precise link between inhibition of proteasome activity and induction of apoptosis is was unknown. Our data now confirm that PS-341 induces dual (caspase 8 and 9) apoptotic signaling, consisting of early cytoplasmic release of mitochondrial cytochrome c and caspase-9 activation; followed by independent activation of the Fas/caspase-8 pathway. Cytochrome c binds to Apaf-1 to form the “apoptosome” that activates caspase-9 and in turn cleaves and activates downstream executioner caspases (59, 60). Cytochrome c release is facilitated by decreased levels of antiapoptotic members of the Bcl-2 family, such as Bcl-2 or A1/Bfl-1, which inhibit the proapoptotic Bax. PS-341 stabilizes the NF-κB inhibitor IκB (36), and we now show that it induces rapid decrease in NF-κB activity and protein levels of several apoptosis inhibitors, including the mitochondrial proteins Bcl-2 and A1 (thus facilitating cytoplasmic cytochrome c release); and the caspase-9 inhibitor XIAP (further enhancing caspase-9 activity). NF-κB is necessary for MM cell survival (14); and, conversely, specific inhibition of its activity down-regulates Bcl-2 and A1 expression, induces cytochrome c release into the cytoplasm, and activates caspase-9 (but not caspase-8) (14). PS-341-induced apoptosis in MM cells is, therefore, mediated, at least in part, by a Bcl-2/A1-cytochrome c–caspase-9 pathway triggered by NF-κB inhibition. The importance of this pathway is confirmed by the early PS-341-induced cleavage of caspase-9 and the partial protection conferred by the caspase-9 inhibition. Also, overexpression of Bcl-2 modestly attenuates PS-341 sensitivity and delays the kinetics of PS-341-induced cell death; whereas a cell-permeable peptide that specifically prevents BH3 domain-mediated interaction between proapoptotic and antiapoptotic members of the Bcl-2 family (37) sensitizes MM.1S cells to PS-341.
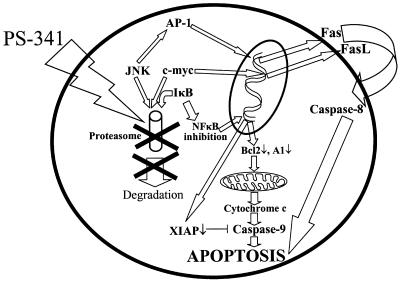
Moreover, caspase-8 inhibition has a significant protective effect in our model, confirming the importance of this second, independent apoptotic pathway. Because caspase-8 is the apical caspase of apoptosis induced by death ligands, such as tumor necrosis factor-α, FasL, and TRAIL (35), the ability of PS-341 to up-regulate Fas and down-regulate expression of caspase-8 inhibitors, such as FLIP and cIAP-2, could facilitate its enzymatic activity. Indeed, we have already reported that PS-341 sensitizes MM.1S cells to tumor necrosis factor-α (36) and TRAIL/Apo2L (27), and we now confirm a similar effect on Fas-mediated apoptosis. Fas up-regulation in our model was dependent on the JNK/c-Jun/AP-1 axis, because AP-1 has been reported to regulate Fas expression (41). Treatment with PS-341 stabilizes JNK and increases c-Jun phosphorylation and DNA-binding activity of the transcription factor AP-1, correlating with increased protein levels of Fas. The JNK inhibitor SP600125 partially blocks PS341-induced Fas up-regulation and apoptosis. These data are in agreement with Meriin et al. (42), who described a crucial role for JNK, a proteasome substrate, in apoptosis induced by the proteasome inhibitor MG132 in U937 cells, because it was suppressed by a dominant-negative SEK1 mutant and a JNK antisense oligonucleotide. Taken together, these data suggest that JNK activity contributes to PS-341-induced apoptosis. In our study, FasL expression is also up-regulated in PS-341-treated cells. FasL expression is regulated by the transcription factor c-myc (40), a well known substrate for the ubiquitin-proteasome degradation pathway (38, 57, 58). In human glioma cells treated with proteasome inhibitors lactacystin and acetyl-leucinyl-leucinyl-norleucinal, up-regulation of FasL expression has also been attributed to accumulation of c-myc (61). In this study, we similarly demonstrate that FasL up-regulation in PS-341-treated cells was associated with increased levels of c-myc. The functional significance of the up-regulation of Fas and FasL in our model is confirmed by using a soluble decoy Fas construct, which protected against PS-341-induced apoptosis. Our data, therefore, suggest a dual apoptotic mechanism induced by PS-341 in MM cells: one involving the mitochondrial/caspase-9 axis, and another mediated by JNK/Fas/caspase-8. The apoptotic pathways triggered by PS-341 are schematically summarized in Fig. 4.
Fig 4.
Schematic diagram of apoptotic pathways activated by PS-341 inhibition.
Finally, we demonstrate that PS-341 triggers transcription of proteasome subunits and molecular chaperones of the hsp family, representing a stress response. It is likely that PS-341-treated tumor cells attempt to compensate for the loss of proteasome activity by synthesizing new proteasomes and new chaperones. Although these cytoprotective responses are insufficient to rescue MM cells from apoptosis, they do support the notion that proteasome activity is required for cell survival and, importantly, further suggest that agents inhibiting the activity of molecular chaperones may block this protection and enhance the antitumor activity of proteasome inhibitors. Indeed, we found that a specific inhibitor of the chaperoning function of hsp90 (22, 55, 56), sensitizes MM cells to subtoxic concentrations of PS-341. This sensitizing effect can be attributed to the fact that hsp90 inhibitors suppress a wide constellation of antiapoptotic protective cellular pathways, thereby rendering cells more sensitive to various proapoptotic stimuli. Specifically, we have previously found that inhibitors of the hsp90 chaperone can induce apoptosis of even drug-resistant MM cells, which is preceded by down-regulation of several kinases implicated in tumor cell growth/survival (including IGF-1R, Akt, IκB kinase-α) as well as intracellular antiapoptotic proteins such as FLIP, XIAP, and cIAP-2,†† suggesting that hsp90 inhibition increases the PS-341 sensitivity of MM cells by means of multifactorial molecular mechanisms. On one hand, hsp90 inhibitors abrogate IGF-1R/Akt signaling, thus neutralizing its attenuating effect on PS-341 sensitivity. On the other hand, hsp90 inhibitors can potentiate certain key proapoptotic sequelae of PS-341, e.g., PS-341 suppresses the proteasomal degradation of phosphorylated IκB, whereas hsp90 inhibitors suppress IκΒ phosphorylation by down-regulating IκB kinase-α.†† Thus, both these classes of antitumor agents block NF-κB activation; however, because they target separate distinct levels of regulation of NF-κB activity, combined use of subtoxic concentrations of these drugs may achieve a synergistic inhibitory effect on NF-κB and cell survival. The strong synergistic interaction between the hsp90 inhibitor and PS-341 confirms the functional significance of the up-regulation of hsps, in general, and, in particular, hsp90, as a protective mechanism against PS-341-induced apoptosis and provides the framework for combination treatments that will include hsp90 inhibitors in an effort to augment clinical efficacy and overcome clinical refractoriness to PS-341.
In summary, the current study characterized the molecular sequelae of PS-341 treatment in MM, the prototypic disease model of antitumor activity of this proteasome inhibitor, and defined apoptotic pathways triggered by this anticancer agent. Our studies show that PS-341 induces down-regulation of proliferative pathways, up-regulation of apoptotic signaling, up-regulation of the proteasome/ubiquitin pathway, and activation of stress kinases. Specifically, we identify a dual apoptotic mechanism, with a pivotal role for the mitochondria/cytochrome c/caspase-9 in one arm and the JNK/death receptor-activated caspase-8 in the other. Overexpression of Bcl-2 and Akt activity may attenuate PS-341 effects, suggesting that these molecules may predict for response to PS-341 treatment and, at least in part, account for emergence of resistance in relapsing patients. Combining PS-341 with agents that inhibit Bcl-2 and Akt expression or function (e.g., antisense oligonucleotides) could, therefore, improve clinical outcome. Similarly, coupling PS-341 with agents which block the chaperoning function of hsp90 can also enhance their efficacy. These studies therefore not only shed light into mechanisms of action of proteasome inhibitors against MM cells, but also suggest therapeutic strategies to overcome potential clinical resistance to this emerging class of antitumor agents.
Supplementary Material
Acknowledgments
This work was supported by the Multiple Myeloma Research Foundation (N.M. and C.S.M.), Lauri Strauss Leukemia Foundation (N.M. and C.S.M.), National Institutes of Health Grants RO-1 50947 and PO-1 78378, National Institutes of Health Grant R24 DK58739 (to T.A.L.), the Leukemia and Lymphoma Society Scholar in Translational Research Award (to N.C.M.), and the Doris Duke Distinguished Clinical Research Scientist Award (to K.C.A.).
Abbreviations
MM, multiple myeloma
hsp, heat-shock protein
IGF-1, insulin-like growth factor 1
MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
This paper was submitted directly (Track II) to the PNAS office.
Richardson, P. G., Berenson, J., Irwin, D., Jagannath, S., Traynor, A., Rajkumar, V., Alsina, M., Kuter, D., Srkalovic, G., Siegel, S., et al. (2001) Blood 98, 774a (abstr.).
Mitsiades, C. S., Mitsiades, N., Poulaki, V., Akiyama, M., Treon, S. P. & Anderson, K. C. (2001) Blood 98, 377a (abstr.).
References
- 1.Ciechanover A., Finley, D. & Varshavsky, A. (1984) J. Cell. Biochem. 24, 27-53. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A., Finley, D. & Varshavsky, A. (1984) Cell 37, 57-66. [DOI] [PubMed] [Google Scholar]
- 3.Varshavsky A. (1997) Trends Biochem. Sci. 22, 383-387. [DOI] [PubMed] [Google Scholar]
- 4.Varshavsky A., Bachmair, A., Finley, D., Gonda, D. K. & Wunning, I., (1989) Bio/Technology 13, 109–143. [PubMed]
- 5.Johnson E. S., Bartel, B., Seufert, W. & Varshavsky, A. (1992) EMBO J. 11, 497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldberg A. L. (1990) Semin. Cell. Biol. 1, 423-432. [PubMed] [Google Scholar]
- 7.Goldberg A. L., Elledge, S. J. & Harper, J. W. (2001) Sci. Am. 284, 68-73. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg A. L. & Rock, K. (2002) Nat. Med. 8, 338-340. [DOI] [PubMed] [Google Scholar]
- 9.Kisselev A. F. & Goldberg, A. L. (2001) Chem. Biol. 8, 739-758. [DOI] [PubMed] [Google Scholar]
- 10.Adams J., Palombella, V. J., Sausville, E. A., Johnson, J., Destree, A., Lazarus, D. D., Maas, J., Pien, C. S., Prakash, S. & Elliott, P. J. (1999) Cancer Res. 59, 2615-2622. [PubMed] [Google Scholar]
- 11.Hideshima T., Richardson, P., Chauhan, D., Palombella, V. J., Elliott, P. J., Adams, J. & Anderson, K. C. (2001) Cancer Res. 61, 3071-3076. [PubMed] [Google Scholar]
- 12.LeBlanc R., Catley, L. P., Hideshima, T., Lentzsch, S., Mitsiades, C. S., Mitsiades, N., Neuberg, D., Goloubeva, O., Pien, C. S., Adams, J., et al. (2002) Cancer Res. 62, 4996-5000. [PubMed] [Google Scholar]
- 13.Richardson P. G., Barlogie, B., Berenson, J., Traynor, A., Singhal, S., Jagannath, S., Irwin, D., Rajkumar, V., Srkalovic, G., Alsina, M., et al., (2002) American Society of Clinical Oncology Annual Meeting Proceedings (Lippincott Williams & Wilkins, Baltimore), Vol. 21, pp. 11a. [Google Scholar]
- 14.Mitsiades N., Mitsiades, C. S., Poulaki, V., Chauhan, D., Richardson, P. G., Hideshima, T., Munshi, N. C., Treon, S. P. & Anderson, K. C. (2002) Blood 99, 4079-4086. [DOI] [PubMed] [Google Scholar]
- 15.Auphan N., DiDonato, J. A., Rosette, C., Helmberg, A. & Karin, M. (1995) Science 270, 286-290. [DOI] [PubMed] [Google Scholar]
- 16.Ramdas J. & Harmon, J. M. (1998) Endocrinology 139, 3813-3821. [DOI] [PubMed] [Google Scholar]
- 17.Wissink S., van Heerde, E. C., vand der Burg, B. & van der Saag, P. T. (1998) Mol. Endocrinol. 12, 355-363. [DOI] [PubMed] [Google Scholar]
- 18.Scheinman R. I., Cogswell, P. C., Lofquist, A. K. & Baldwin, A. S., Jr. (1995) Science 270, 283-286. [DOI] [PubMed] [Google Scholar]
- 19.Mitsiades N., Mitsiades, C. S., Poulaki, V., Chauhan, D., Richardson, P. G., Hideshima, T., Munshi, N. C., Treon, S. P. & Anderson, K. C. (2002) Blood 99, 4525-4530. [DOI] [PubMed] [Google Scholar]
- 20.Hideshima T., Chauhan, D., Richardson, P., Mitsiades, C., Mitsiades, N., Hayashi, T., Munshi, N., Dang, L., Castro, A., Palombella, V., et al. (2002) J. Biol. Chem. 277, 16639-16647. [DOI] [PubMed] [Google Scholar]
- 21.Fleming J. A., Lightcap, E. S., Sadis, S., Thoroddsen, V., Bulawa, C. E. & Blackman, R. K. (2002) Proc. Natl. Acad. Sci. USA 99, 1461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basso A. D., Solit, D. B., Munster, P. N. & Rosen, N. (2002) Oncogene 21, 1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitsiades N., Yu, W. H., Poulaki, V., Tsokos, M. & Stamenkovic, I. (2001) Cancer Res. 61, 577-581. [PubMed] [Google Scholar]
- 24.Mitsiades N., Mitsiades, C. S., Poulaki, V., Anderson, K. C. & Treon, S. P. (2002) Blood 99, 2162-2171. [DOI] [PubMed] [Google Scholar]
- 25.Ogawa M., Nishiura, T., Oritani, K., Yoshida, H., Yoshimura, M., Okajima, Y., Ishikawa, J., Hashimoto, K., Matsumura, I., Tomiyama, Y. & Matsuzawa, Y. (2000) Cancer Res. 60, 4262-4269. [PubMed] [Google Scholar]
- 26.Mitsiades, C. S., Mitsiades, N., Poulaki, V., Richardson, P. G., Schlossman, R., Akiyama, M., Chauhan, D., Hideshima, T., Munshi, N. C., Treon, S. P. & Anderson, K. C. (2002) Oncogene, in press. [DOI] [PubMed]
- 27.Mitsiades C. S., Treon, S. P., Mitsiades, N., Shima, Y., Richardson, P., Schlossman, R., Hideshima, T. & Anderson, K. C. (2001) Blood 98, 795-804. [DOI] [PubMed] [Google Scholar]
- 28.Hitoshi Y., Lorens, J., Kitada, S. I., Fisher, J., LaBarge, M., Ring, H. Z., Francke, U., Reed, J. C., Kinoshita, S. & Nolan, G. P. (1998) Immunity 8, 461-471. [DOI] [PubMed] [Google Scholar]
- 29.Grishin A. V., Azhipa, O., Semenov, I. & Corey, S. J. (2001) Proc. Natl. Acad. Sci. USA 98, 10172-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollander M. C., Sheikh, M. S., Yu, K., Zhan, Q., Iglesias, M., Woodworth, C. & Fornace, A. J., Jr. (2001) Int. J. Cancer 96, 22-31. [DOI] [PubMed] [Google Scholar]
- 31.Hollander M. C., Zhan, Q., Bae, I. & Fornace, A. J., Jr. (1997) J. Biol. Chem. 272, 13731-13737. [DOI] [PubMed] [Google Scholar]
- 32.Sheikh M. S., Hollander, M. C. & Fornance, A. J., Jr. (2000) Biochem. Pharmacol. 59, 43-45. [DOI] [PubMed] [Google Scholar]
- 33.Jourdan M., Ferlin, M., Legouffe, E., Horvathova, M., Liautard, J., Rossi, J. F., Wijdenes, J., Brochier, J. & Klein, B. (1998) Br. J. Haematol. 100, 637-646. [DOI] [PubMed] [Google Scholar]
- 34.Chauhan D., Hideshima, T., Rosen, S., Reed, J. C., Kharbanda, S. & Anderson, K. C. (2001) J. Biol. Chem. 276, 24453-24456. [DOI] [PubMed] [Google Scholar]
- 35.Mitsiades N., Poulaki, V., Mitsiades, C. S., Koutras, D. A. & Chrousos, G. P. (2001) Trends Endocrinol. Metab. 12, 384-390. [DOI] [PubMed] [Google Scholar]
- 36.Hideshima T., Chauhan, D., Schlossman, R., Richardson, P. & Anderson, K. C. (2001) Oncogene 20, 4519-4527. [DOI] [PubMed] [Google Scholar]
- 37.Degterev A., Lugovskoy, A., Cardone, M., Mulley, B., Wagner, G., Mitchison, T. & Yuan, J. (2001) Nat. Cell Biol. 3, 173-182. [DOI] [PubMed] [Google Scholar]
- 38.Flinn E. M., Busch, C. M. & Wright, A. P. (1998) Mol. Cell. Biol. 18, 5961-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hueber A. O., Zornig, M., Lyon, D., Suda, T., Nagata, S. & Evan, G. I. (1997) Science 278, 1305-1309. [DOI] [PubMed] [Google Scholar]
- 40.Brunner T., Kasibhatla, S., Pinkoski, M. J., Frutschi, C., Yoo, N. J., Echeverri, F., Mahboubi, A. & Green, D. R. (2000) J. Biol. Chem. 275, 9767-9772. [DOI] [PubMed] [Google Scholar]
- 41.Li X. R., Chong, A. S., Wu, J., Roebuck, K. A., Kumar, A., Parrillo, J. E., Rapp, U. R., Kimberly, R. P., Williams, J. W. & Xu, X. (1999) J. Biol. Chem. 274, 35203-35210. [DOI] [PubMed] [Google Scholar]
- 42.Meriin A. B., Gabai, V. L., Yaglom, J., Shifrin, V. I. & Sherman, M. Y. (1998) J. Biol. Chem. 273, 6373-6379. [DOI] [PubMed] [Google Scholar]
- 43.Saito A., Watanabe, T. K., Shimada, Y., Fujiwara, T., Slaughter, C. A., DeMartino, G. N., Tanahashi, N. & Tanaka, K. (1997) Gene 203, 241-250. [DOI] [PubMed] [Google Scholar]
- 44.Yokota K., Kagawa, S., Shimizu, Y., Akioka, H., Tsurumi, C., Noda, C., Fujimuro, M., Yokosawa, H., Fujiwara, T., Takahashi, E., et al. (1996) Mol. Biol. Cell 7, 853-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hori T., Kato, S., Saeki, M., DeMartino, G. N., Slaughter, C. A., Takeuchi, J., Toh-e, A. & Tanaka, K. (1998) Gene 216, 113-122. [DOI] [PubMed] [Google Scholar]
- 46.Kominami K., DeMartino, G. N., Moomaw, C. R., Slaughter, C. A., Shimbara, N., Fujimuro, M., Yokosawa, H., Hisamatsu, H., Tanahashi, N., Shimizu, Y., et al. (1995) EMBO J. 14, 3105-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spataro V., Toda, T., Craig, R., Seeger, M., Dubiel, W., Harris, A. L. & Norbury, C. (1997) J. Biol. Chem. 272, 30470-30475. [DOI] [PubMed] [Google Scholar]
- 48.Akiyama K., Kagawa, S., Tamura, T., Shimbara, N., Takashina, M., Kristensen, P., Hendil, K. B., Tanaka, K. & Ichihara, A. (1994) FEBS Lett. 343, 85-88. [DOI] [PubMed] [Google Scholar]
- 49.Aki M., Shimbara, N., Takashina, M., Akiyama, K., Kagawa, S., Tamura, T., Tanahashi, N., Yoshimura, T., Tanaka, K. & Ichihara, A. (1994) J. Biochem. (Tokyo) 115, 257-269. [DOI] [PubMed] [Google Scholar]
- 50.Akiyama K., Yokota, K., Kagawa, S., Shimbara, N., Tamura, T., Akioka, H., Nothwang, H. G., Noda, C., Tanaka, K. & Ichihara, A. (1994) Science 265, 1231-1234. [DOI] [PubMed] [Google Scholar]
- 51.Hisamatsu H., Shimbara, N., Saito, Y., Kristensen, P., Hendil, K. B., Fujiwara, T., Takahashi, E., Tanahashi, N., Tamura, T., Ichihara, A. & Tanaka, K. (1996) J. Exp. Med. 183, 1807-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaczynska M., Goldberg, A. L., Tanaka, K., Hendil, K. B. & Rock, K. L. (1996) J. Biol. Chem. 271, 17275-17280. [DOI] [PubMed] [Google Scholar]
- 53.Tsurumi C., Shimizu, Y., Saeki, M., Kato, S., Demartino, G. N., Slaughter, C. A., Fujimuro, M., Yokosawa, H., Yamasaki, M., Hendil, K. B., et al. (1996) Eur. J. Biochem. 239, 912-921. [DOI] [PubMed] [Google Scholar]
- 54.Lee D. H. & Goldberg, A. L. (1998) Mol. Cell. Biol. 18, 30-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Munster P. N., Srethapakdi, M., Moasser, M. M. & Rosen, N. (2001) Cancer Res. 61, 2945-2952. [PubMed] [Google Scholar]
- 56.Munster P. N., Basso, A., Solit, D., Norton, L. & Rosen, N. (2001) Clin. Cancer Res. 7, 2228-2236. [PubMed] [Google Scholar]
- 57.Gregory M. A. & Hann, S. R. (2000) Mol. Cell. Biol. 20, 2423-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bahram F., von der Lehr, N., Cetinkaya, C. & Larsson, L. G. (2000) Blood 95, 2104-2110. [PubMed] [Google Scholar]
- 59.Li P., Nijhawan, D., Budihardjo, I., Srinivasula, S. M., Ahmad, M., Alnemri, E. S. & Wang, X. (1997) Cell 91, 479-489. [DOI] [PubMed] [Google Scholar]
- 60.Zou H., Li, Y., Liu, X. & Wang, X. (1999) J. Biol. Chem. 274, 11549-11556. [DOI] [PubMed] [Google Scholar]
- 61.Tani E., Kitagawa, H., Ikemoto, H. & Matsumoto, T. (2001) FEBS Lett. 504, 53-58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.