Abstract
The TCL1 protooncogene is overexpressed in many mature B cell lymphomas, especially from AIDS patients. To determine whether aberrant expression promotes B cell transformation, we generated a murine model in which a TCL1 transgene was overexpressed at similar levels in both B and T cells. Strikingly, transgenic mice developed Burkitt-like lymphoma (BLL) and diffuse large B cell lymphoma (DLBCL) with attendant Bcl-6 expression and mutated JH gene segments at a very high penetrance beginning at 4 months of age. In contrast, only one mouse developed a T cell malignancy at 15 months, consistent with a longer latency for transformation of T cells by TCL1. Activation of premalignant splenic B cells by means of B cell antigen receptor (BCR) engagement resulted in significantly increased proliferation and augmented AKT-dependent signaling, including increased S6 ribosomal protein phosphorylation. Transgenic spleen cells also survived longer than wild-type spleen cells in long-term culture. Together these data demonstrate that TCL1 is a powerful oncogene that, when overexpressed in both B and T cells, predominantly yields mature B cell lymphomas.
The TCL1 (T cell leukemia 1) protooncogene is expressed in CD3−CD4−CD8− precursor T cells and is extinguished at the CD4+CD8+ stage of thymocyte development (1). In B cells, TCL1 is first expressed in pro-B cells and remains high in naive mantle zone B cells of peripheral lymphoid tissues (1–4). Down-regulation of TCL1 expression in follicle center centroblasts and centrocytes is followed by gene extinction in post-germinal center (GC) memory B cells and plasma cells (4, 5).
Continued high-level TCL1 expression, because of chromosomal rearrangements, was implicated in mature peripheral T cell malignancies (6, 7). Polyclonal and oligoclonal T cell expansions preceded clonal outgrowth by many years, suggesting that additional lesions were required for transformation (8, 9). Supporting this tumorigenic mechanism, transgenic mice expressing TCL1-family-member proteins exclusively in T cells developed polyclonal T cell expansions before the evolution of clonal malignancies at 15 to 20 months (10, 11). Overexpression of TCL1, or MTCP1 (mature T cell proliferation 1), in mouse T cells did not affect B cell development or produce B cell lymphomas. These findings indicate that aberrant expression of TCL1 or MTCP1 in T cells perturbs T cell homeostasis through cell autonomous pathways without inducing premalignant or malignant changes in bystander B cells.
About 15% of AIDS patients develop aggressive B cell non-Hodgkin lymphoma (AIDS-NHL) (12, 13). Most AIDS-NHL originate from GC or post-GC B cells, but the early events leading to AIDS-NHL remain poorly defined (13, 14). Diffuse large B cell lymphoma (DLBCL) is the most prevalent type of AIDS-NHL, and these tumors generally lack consistent genetic and/or viral tumor-promoting alterations. Recently, abundant TCL1 expression was shown in a high percentage of AIDS-NHL of post-GC origin (3, 4). This discovery led us to postulate that TCL1 dysregulation could contribute to, and possibly initiate, B cell malignancies, especially in individuals with impaired immunity. To test our hypothesis we generated transgenic mice that abnormally express TCL1 throughout both B and T cell development. This model has allowed for a direct comparison of the strength of TCL1 transforming capacity in B cells as compared with T cells.
Materials and Methods
Generation of TCL1 Transgenic Mice.
A 381-bp human TCL1 (hTCL1) coding sequence obtained by RT-PCR from an AIDS-related B cell lymphoma was inserted into the vector pEμ-B29, creating pEμ-B29-TCL1 (3). The construct was injected into eggs from (C57BL/6 × C3H)F1 females mated to C57BL/6 males, and three transgenic founders were obtained.
Western Blot.
Ten to 15 μg of protein per sample was separated by SDS/PAGE and Western blotted with anti-TCL1 sera by standard techniques (4). Spleen cells were fractionated with anti-B220-coated microbeads (clone RA3–6B2, PharMingen) by using a magnetic bead sorting system (Miltenyi Biotec, Auburn, CA). B and T cell-enriched populations were >85% and >95% pure, respectively, by flow cytometry (data not shown).
Histologic Examination and TCL1 Immunohistochemistry.
Tissue sections were stained with hematoxylin and eosin (H&E). Bone marrow (BM), blood smears, and ascites fluid were Wright–Giemsa stained. Paraffin sections were stained with antisera to TCL1, Bcl-6, and terminal deoxynucleotidyltransferase (3, 4, 15). Tumor diagnoses used criteria generated by an international panel of experts in mouse and human hematopathology. Flow cytometry and molecular studies provided complementary and confirmatory data for each diagnosis. The closest counterpart of each transgenic mouse diagnosis described here to the new World Health Organization human lymphoma classification is shown in Table 1.
Table 1.
World Health Organization lymphoma classifications
| Mouse | Human |
|---|---|
| Follicular B cell lymphoma (FBL) | Follicular lymphoma |
| Diffuse large B cell lymphoma (DLBCL) | DLBCL |
| Histiocyte-associated (HA) | Histiocyte-rich |
| T cell-rich (TR) | TR |
| Burkitt-like lymphoma (BLL) | BLL |
Flow Cytometry.
Suspensions from blood, BM, spleen, lymph node, or thymus were depleted of red cells, blocked with anti-FcγII/III receptor antibody, and surface-stained with phycoerythrin- or FITC-conjugated antibodies from PharMingen. For triple staining, streptavidin conjugated to energy-coupled dye (ECD; Immunotech, Westbrook, ME) was used. Data were analyzed by using fcs express (De Novo Software, Thornhill, ON, Canada).
Clonality Determination.
IgH, T cell antigen receptor (TCR)-β, and TCR-γ rearrangements were determined by high-fidelity PCR using primers as described, followed by 1.2% agarose gel electrophoresis (16). Genomic DNA was isolated by using a DNeasy Tissue kit (Qiagen, Chatsworth, CA). Gel-purified PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen) and sequenced with an ABI 377XL automated DNA sequencer.
Adoptive Transfer.
Ascites fluid from mouse K7 was collected and injected i.p. into three 5-month-old wild-type (C57BL/6 × C3H)F1 males. In addition, mouse K7 ascites cells were passaged for >3 months in RPMI medium 1640 supplemented with 10% FCS and antibiotics at 37°C in 5% CO2.
In Vitro Proliferation and Survival Assays.
Suspensions from lymph nodes, spleen, and thymus were depleted of red cells and macrophages and cultured at 1 × 105 cells per well in 96-well plates in RPMI medium 1640 with 10% FCS. For proliferation assays, cells were cocultured with the indicated amounts of anti-IgM F(ab′)2 (Jackson ImmunoResearch), or Escherichia coli lipopolysaccharide (LPS; Calbiochem). At 48 h, cells were pulsed with 1 μCi (1 Ci = 37 GBq) of [3H]thymidine for 12 h and harvested onto glass fiber filters, and cpm was determined. For survival assays, cells were incubated for the indicated number of days and viability was determined by the addition of annexin V and propidium iodide, followed by flow cytometry.
AKT, S6 Ribosomal Protein, and ERK1 and ERK2 Phosphorylation.
Spleen cells were washed and resuspended in RPMI medium 1640 without serum. After incubation for 30 min, cells were stimulated with 20 μg/ml anti-IgM F(ab′)2. At 0, 2 (or 3), 10, and 60 min after stimulation, cells were centrifuged and washed, followed by lysis with Nonidet P-40 in the presence of protease and phosphatase inhibitors. Lysates were fractionated by SDS/PAGE, immunoblotted with anti-AKT (9272), anti-phosphoserine(473)-AKT (9270), anti-phosphoserine(235/236)-S6 (2211), anti-phosphothreonine(202)/tyrosine(204)-p44/42 MAP kinase (9101), anti-p44/42 MAP kinase (9100), and anti-actin antibodies (Cell Signaling Technology, Beverly, MA).
Results
Transgenic Mice Express TCL1 in B Cells and T Cells at Equal Levels.
The genetic background, timing, and strength of TCL1 expression that might cause B cell tumors in mice have not been defined. Because broad developmental expression of TCL1 could be critical for inducing B cell malignancies, an expression construct was created by using the B29 minimal promoter, coupled to the IgH intronic enhancer (17, 18). This construct was designed to yield similar levels of TCL1 in B cells and T cells. We performed this study in a (C57BL/6 × C3H)F1 background because this mixed strain is permissive for both B and T cell tumor formation (19).
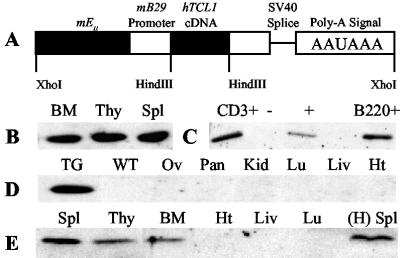
Mice expressing a hTCL1 cDNA were generated with a 2.5-kb pEμ–B29-TCL1 construct (Fig. 1A). Two independent founders, designated K and P, were used to generate lines for study. Three-month-old hTCL1 mice showed abundant TCL1 protein expression in lymphoid tissues, whereas no expression was detected in nonlymphoid tissues (Fig. 1 B, D, and E). The level of TCL1 expression in BM, thymus, and spleen was similar within hTCL1 lines and within purified B cells and T cells (Fig. 1 B and C and Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org).
Fig 1.
pEμ-B29-TCL1 transgenic mice express TCL1 only in lymphoid tissues. (A) A 2.5-kb XhoI fragment of the pEμ-B29-TCL1 construct was isolated and used in egg injections. (B–E) Detection of TCL1 protein in 3-month-old heterozygotes. Fifteen micrograms of protein was immunoblotted with TCL1 antiserum. Membranes were stained with Ponceau S before immunoblotting to ensure equal protein loading in each lane (data not shown). (B) TCL1 expression in K line lymphoid tissues. (C) TCL1 expression in magnet-activated cell sorting (MACS)-sorted B (B220+) and T (CD3+) cells from K line spleens. (D) TCL1 expression is absent from nonlymphoid tissues from K line mice. (E) TCL1 expression in lymphoid and nonlymphoid tissues from P line mice and the spleen from a 6-month-old H line female founder that died before successful breeding.
Mild Lymphocytosis in Young hTCL1 Mice.
Compared with WBC counts in normal littermates (70 × 104 per ml), WBC counts were slightly higher in the K line (117 × 104 per ml; P = 0.026) and higher in the P line (178 × 104 per ml; P < 0.001) hTCL1 mice (Table 2). Intact lymphoid structures were seen with H&E-stained sections from thymus, spleen, and lymph nodes (data not shown). Spleen weights were similar for wild-type and hTCL1 mice. The ratios of splenic B to T cells and the numbers of mature IgM+B220+ B cells and CD4+ or CD8+ T cells were similar between mice (Fig. 2 and data not shown). No evidence of B cell activation, as reflected in cell size or level of CD5, CD44, or CD69 expression, was detected in K line spleen cells (data not shown). Thymocyte subpopulations were indistinguishable between wild-type and hTCL1 mice (Fig. 2 and data not shown). Flow cytometric analysis of wild-type and hTCL1 BM indicated no differences in lymphoid lineage development (data not shown). Overall, the data indicate that TCL1 expression in early and mature lymphocytes resulted in a modest increase of WBC counts in P line mice and no detectable effect on lymphoid organ morphology in hTCL1 mice up to 3 months of age (Fig. 8, which is published as supporting information on the PNAS web site).
Table 2.
WBC counts and spleen weights for wild-type and TCL1 transgenic mice
| Type | Age, months | No. | WBC counts, ×104 per ml | Range | Spleen weight, g | Range |
|---|---|---|---|---|---|---|
| WT | 1–3 | 9 | 70 ± 17.7 | 50–95 | 0.09 ± 0.02 | 0.07–0.12 |
| WT | 4–6 | 6 | 150 ± 61 | 110–220 | 0.10 ± 0.01 | 0.08–0.12 |
| WT | 7–13 | 8 | 113 ± 50.9 | 50–200 | 0.15 ± 0.04 | 0.10–0.20 |
| Line K +/− | 1–3 | 10 | 117 ± 55 | 80–226 | 0.11 ± 0.02 | 0.09–0.14 |
| Line K +/− | 4–6 | 11 | 1,088 ± 1,129 | 90–2460 | 0.21 ± 0.14 | 0.09–0.43 |
| Line K +/− | 7–13 | 20 | 624 ± 1,063 | 40–3400 | 1.17 ± 1.06 | 0.15–3.70 |
| Line P +/− | 1–3 | 5 | 178 ± 73 | 110–255 | 0.12 ± 0.02 | 0.10–0.14 |
| Line P +/− | 4–6 | 5 | 250 ± 178 | 93–450 | — | — |
| Line P +/− | 12,13 | 2 | 2,600 ± 1,400 | 1,260–4,000 | 2.30 ± 1.00 | 1.28–3.35 |
| Tumor injection | 6 weeks post | 3 | 1576 ± 202 | 1,300–1,780 | 2.60 ± 0.36 | 2.10–2.90 |
Mice were grouped by type and age. WBC counts and spleen weights were determined by standard methods. WT, wild-type; K +/−, line K heterozygotes; P +/−, line P heterozygotes; Tumor injection, 5-month-old (C57BL/6 × C3H)F1 wild-type male mice injected with 107 K7 ascites cells i.p., and necropsied at 6 weeks after injection.
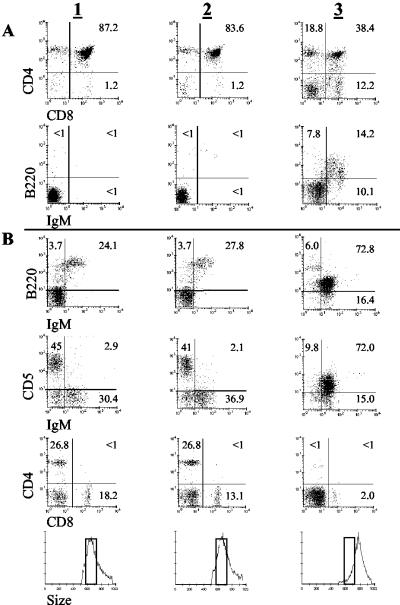
Fig 2.
Older hTCL1 mice develop IgM+B220loCD5lo monotypic B cell expansions. (A and B) Flow cytometric analyses of thymocytes (A) and spleen cells (B) from 6-week-old wild-type (column 1), 6-week-old K line (column 2), and malignant 12-month-old K10 (column 3) hTCL1 mice are shown. (Bottom) Forward-angle light scatter shows that monotypic IgM+B220loCD5lo B cells are significantly larger than 6-week-old wild-type and hTCL1 spleen cells. Wild-type 12-month-old spleen cell and thymocyte marker and size profiles were similar to those for younger wild-type and hTCL1 mice (data not shown).
Lymphoproliferative Disorder in Older hTCL1 Mice.
In all, 34 K line mice were generated, of which 30 were killed after becoming visibly ill at 12.4 ± 3.4 months of age. Seventeen of these mice were necropsied and demonstrated spleen weights averaging a 7.8-fold increase over wild-type spleen weights (Table 3, which is published as supporting information on the PNAS web site). Two mice became ill at 4 months with WBC counts that were 7- to 12-fold higher than WBC counts observed in wild-type mice. Most hTCL1 mice appeared ill between 7 and 13 months and, on average, had a 5.5-fold increased WBC count compared with that of wild-type mice. Two 6-month-old P line mice had WBC counts >300 × 104 per ml and became visibly ill at 9 and 12 months of age (Table 2). Wild-type mice never appeared ill and WBC counts never exceeded 200 × 104 per ml during this study.
Wright–Giemsa-stained blood smears from ill hTCL1 mice demonstrated numerous lymphocytic blasts (data not shown). Necropsies of hTCL1 mice demonstrated variable splenomegaly, lymphadenopathy, and macroscopic lesions of the liver, lung, kidney, and intestines (Fig. 3). Microscopic examination confirmed that these lesions were multiorgan, TCL1-positive lymphocytic infiltrates. Most hTCL1 mice had splenic and nodal effacement because of a diffusely infiltrative process. BM and thymic infiltrates were also observed in several cases. Periportal, perivascular, and sinusoidal liver infiltrates were common, as was involvement of other solid organs. Collectively, the data indicate that these mice developed lymphoproliferative disease significantly more rapidly and more frequently than transgenic mice expressing either TCL1 or MTCP1 exclusively in T cells (10, 11).
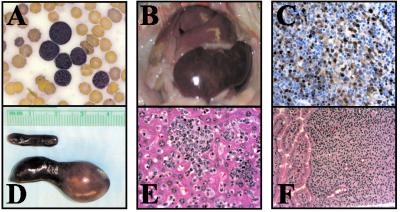
Fig 3.
Appearance of BLL in 11-month-old mouse K7. (A) Wright–Giemsa-stained peripheral blood smear showing monotypic lymphoblastoid B cells. (B) Massively enlarged spleen and liver infiltrated with tumor cells. (C) TCL1 immunohistochemical stain of spleen. (D) Size comparison between K7 (lower) and wild-type (upper) age-matched K line spleens. (E and F) BLL infiltrating the liver (E) and small intestine (F).
Clonal B Cell Malignancies Beginning at 4 Months of Age.
hTCL1 mice with lymphoproliferative disorders contained polyclonal expansions of B and T cells (data not shown). However, some mice had splenomegaly with monotypic B cell populations as determined by flow cytometry. Results from mouse K10 are shown, whereas data for additional mice are summarized in Table 3. The spleen from K10 contained about 75% IgM+B220loCD5lo B cells (Fig. 2). These cells did not express Mac-1 or Gr-1, and T cells constituted <5% of total spleen cells. This monotypic B cell population also accounted for 15% of the total lymphocytes in the thymus. By forward scatter plot analysis, these cells were larger than spleen cells from wild-type or unaffected hTCL1 mice.
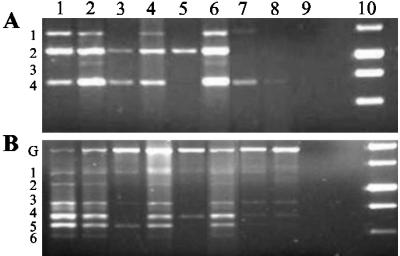
To determine whether these monotypic expansions were clonal, PCR analysis of rearranged TCR and Ig diversity and junction regions was performed (Fig. 4 and data not shown). Clonal or oligoclonal gene rearrangements were detected for IgD–J junctions in 9 of 16 mice tested (Table 3). Spleen cells from 7 mice, and both liver and spleen cells from mouse K7, showed clonal Ig rearrangements. Two mice demonstrated oligoclonal Ig rearrangements and mouse K8 demonstrated oligoclonal TCR bands in addition to a DJH2 rearrangement. Studies of Ig gene rearrangements in the remaining 7 mice revealed, in most cases, evolving oligoclonal or clonal expansions, detected by changes in band intensities, and associated with monomorphic B cell populations detected by flow cytometry.
Fig 4.
Clonal B cell expansions detected by the PCR analysis of Ig and TCR genomic DNA. (A and B) Detection of Ig DJH1 to DJH4 rearrangements (A) and a survey of TCR DJβ2.1 to DJβ2.6 gene rearrangements (B). Lane 1, wild-type thymus; lane 2, wild-type spleen; lane 3, K4 spleen; lane 4, K14 spleen; lane 5, K8 spleen; lane 6, K12 spleen; lane 7, K7 liver; lane 8, K7 spleen; lane 9, water blank; lane 10, 1-kb DNA ladder.
Histopathologic analyses revealed the occurrence of different lymphoma types, almost always derived from mature B cells. These included DLBCL with histiocyte-associated (HA) and T cell-rich (TR) features, Burkitt-like lymphomas (BLLs), and follicular lymphoma (20, 21). Confirmation that these tumors were of mature B cell origin was performed by immunohistochemical analyses. All three lymphoma types exhibited strong Bcl-6-positive and terminal deoxynucleotidyltransferase-negative staining (Fig. 9, which is published as supporting information on the PNAS web site). Furthermore, each tumor type demonstrated somatic hypermutation in JH region sequence analysis (Table 4, which is published as supporting information on the PNAS web site). Tumors showed ongoing mutations both within and outside of mutational “hot spots,” indicating a lack of antigen-driven selective pressure as seen in malignant transformation. Combined, the data are consistent with the generation of GC-derived B cell lymphomas of multiple types.
Demonstration of Complete B Cell Transformation.
Tumor ascites developed in three hTCL1 mice between 10 and 12 months of age (Fig. 10 and Table 3, which are published as supporting information on the PNAS web site). Ascites cells from mouse K7 were grown in culture for >3 months and chosen for extended analysis. Immunoblotting showed abundant TCL1 protein in tumor cells, whereas flow cytometric analysis indicated that the cells were IgM+B220loCD5lo. Surface staining for Mac-1, Gr-1, CD3, CD4, and CD8 was negative (data not shown). To demonstrate transformation, isolated ascites cells were injected i.p. into three syngeneic wild-type males. At 6 weeks after injection all three mice became visibly ill. Immediately before killing, WBC counts were 10- to 20-fold higher than in age-matched mice. At necropsy each mouse showed abundant ascites and enlarged spleens (Table 2). Histologic examination of the spleens demonstrated diffuse effacement by an infiltrative lymphocytic process (data not shown). Fluorescence-activated cell sorter (FACS) analysis demonstrated retention of the initial IgM+B220loCD5lo surface phenotype (Fig. 10). Clonality studies demonstrated retention of the DJH4 Ig rearrangement (data not shown). These results indicate that aberrant expression of TCL1 within the B lineage induced transformation of peripheral B cells as evidenced by the establishment of a TCL1-positive clonal B cell line.
Splenic B Cells from Young hTCL1 Mice Exhibit Increased Proliferation.
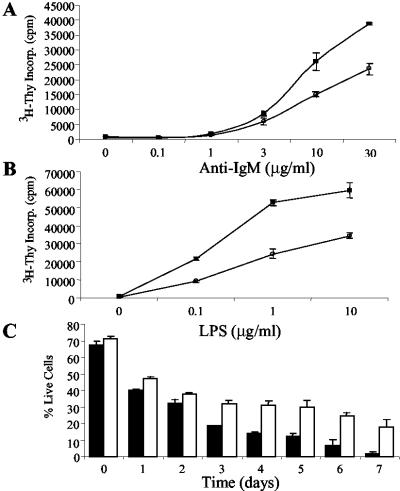
Prior in vitro and transformed cell studies suggest that TCL1 and AKT interact (22, 23). In addition, AKT activation plays a key role in immune receptor-mediated lymphocyte proliferation and survival (24–26). We therefore evaluated the response of nontransformed B cells from hTCL1 mice and age-matched controls to B cell antigen receptor (BCR) engagement and to treatment with LPS. Splenic B cells proliferated strongly and in a dose-dependent response to LPS or anti-IgM treatments (Fig. 5 A and B). The proliferative response was consistently 2- to 3-fold higher than wild-type B cells with both anti-IgM and LPS stimulations. In preliminary studies, splenic T cells from hTCL1 mice responded more strongly and in a dose-dependent manner to TCR stimulation or Con A treatments than did wild-type T cells (K.K.H. and M.A.T., unpublished work). Combined, these data indicate that TCL1 augments B cell (and T cell) proliferation through immune receptor stimulation.
Fig 5.
TCL1 expression induces a dose-dependent proliferative response and increased survival in extended culture. (A and B) Premalignant 6- to 8-week-old hTCL1or wild-type spleen cells were treated with the indicated amounts of anti-IgM (A) or LPS (B) and assayed at 60 h for [3H]thymidine incorporation. Results from wild-type (•) and hTCL1 (▪) mice are shown. Data are representative of six independent experiments. (C) Spleen cells were cultured for the indicated number of days, stained with annexin V and propidium iodide, and assayed for viability by flow cytometry. The percentage of viable wild-type (black bars) and hTCL1 (white bars) cells detected at each time is shown. Data are representative of three independent experiments.
TCL1 Protects Transgenic Spleen Cells from Apoptosis.
Isolated hTCL1 spleen cells showed increased survival in week-long culture compared with spleen cells from age-matched controls (Fig. 5C). The percentage of input B and T cells was equivalent to the percentage of each cell type remaining viable in culture after 7 days as determined by flow analysis (data not shown). The data argue that aberrant TCL1 expression protects both B and T cells from cell death to the same extent. Combined with our preliminary T cell stimulation results, the data suggest that TCL1-induced B and T cell expansions likely result from similar mechanisms (10, 11).
AKT and S6 Phosphorylation Are Elevated in hTCL1 Mice.
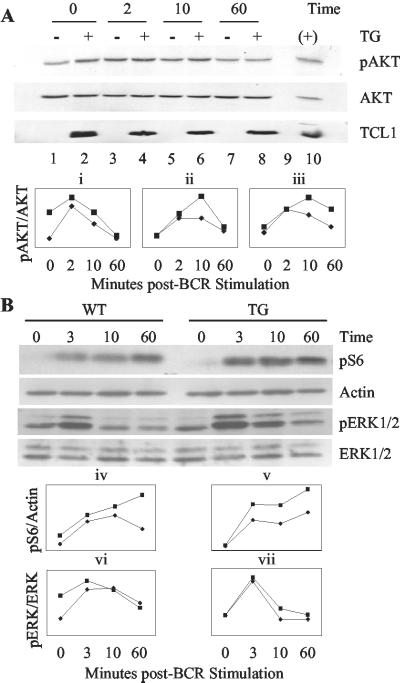
We next evaluated the role for TCL1 overexpression in AKT-dependent immune receptor signaling. Nontransformed spleen cells from hTCL1 and wild-type mice were stimulated by BCR engagement (Fig. 6A). Stimulation with anti-IgM resulted in AKT phosphorylation that peaked between 2 and 10 min in both hTCL1 and wild-type spleen cells. Phospho-AKT levels in hTCL1 spleen cells were equal to or higher than wild type at all time points tested, with the mean being ≈25% higher than wild-type spleen cells. Some assays showed phospho-AKT levels that were up to 90% higher in hTCL1 compared with that of wild-type spleen cells. Occasional hTCL1 mice contained spleen cells with elevated phospho-AKT levels before stimulation by BCR ligation. These results indicate that AKT phosphorylation levels ranged narrowly between assays but were consistently higher in hTCL1 B cells and that TCL1 did not significantly alter the kinetics of AKT activation through BCR stimulation. Notably, the increased activation of AKT seen here in B cells appears roughly equivalent to the level of augmented AKT activation in PTEN-null T cells (27).
Fig 6.
BCR stimulation induces higher AKT Ser-473 and S6 ribosomal protein phosphorylation in hTCL1 vs. wild-type spleen cells. Six- to 8-week-old hTCL1 (▪) or wild-type (♦) spleen cells were stimulated with 20 μg/ml anti-IgM F(ab′)2 for 2 (or 3), 10, and 60 min, harvested, and immunoblotted. (A Upper) Three representative Western blots for AKT Ser-473 phosphorylation (pAKT), total AKT, and TCL1 from one experiment. (Lower) Densitometry results of three independent stimulation experiments in which the line graphs represent the ratio of Ser-473-phosphorylated AKT to total AKT measured. (B Upper) Immunoblots of Ser-235/236 S6 ribosomal protein phosphorylation (pS6), an actin loading control, phospho-Thr-202/Tyr-204 ERK1 and ERK2 (pERK1 and pERK2) and total ERK1 and ERK2 from a single stimulation experiment. (Lower) Densitometry results for pS6 to actin (iv and v) and pERK1 and pERK2 to total ERK1 and ERK2 ratios (vi and vii) for two independent stimulation experiments.
The phosphorylation status of the ribosomal protein S6 was determined as an indicator of augmented AKT activation by TCL1 (Fig. 6B). For comparison, the phosphorylation of ERK1 and ERK2 was determined because ERK signaling proteins are activated by RAS-dependent, AKT-independent pathways (28). Anti-IgM-stimulated splenic B cells showed a time-dependent increase in S6 phosphorylation that peaked after 10 min. hTCL1 B cells demonstrated higher levels of S6 phosphorylation at each time point assayed. By contrast, the anti-IgM dependent increase in ERK1 and ERK2 phosphorylation was equivalent in hTCL1 and wild-type B cells. Together, these results implicate TCL1 overexpression in the augmented activation of PI3K pathway-dependent AKT and p70S6 kinases exclusive of the ERK activation pathway. These results suggest at least one potential mechanism for TCL1 activity in promoting peripheral B cell malignancies.
Discussion
The findings we provide here demonstrate that TCL1 is a powerful B cell oncogene. TCL1 overexpression in two independent transgenic lines promoted high-frequency and rapid development of multiple mature B cell lymphoma types. The tumorigenic mechanism depends, at least in part, on enhanced signaling through phosphatidylinositol 3-kinase, resulting in increased AKT and p70S6 kinase activity.
Premalignant TCL1-positive splenic B and T cells were not immortalized and, although they demonstrated increased survival, did not indefinitely grow in culture. By contrast, transformed ascites B cells were readily established as cell lines and induced identical tumors after transplantation. These findings agree with our prediction that inappropriate TCL1 levels provide a survival and proliferative advantage to premalignant mature B cells (3, 4). The B cell tumors observed here were classified as follicular B cell lymphoma (FBL), BLL, or DLBCL, consistent with a GC or post-GC B cell origin. They also consistently had an activated, IgM+B220loCD5lo mature B cell phenotype, demonstrated somatic hypermutation, and strongly expressed Bcl-6. Notably, CD5 expression in these tumors is not indicative of a chronic lymphocytic leukemia/low-grade lymphoma (CLL/SLL) or mantle cell lymphoma as it is in humans, and more likely represents expression of an activation marker because over 80% of mouse B cell lymphomas of all types express CD5 (21). Also, B cell CLL/SLL is Bcl-6 negative in humans and mice.
Despite similar expression of the TCL1 transgene in both mature T and B cell populations, a striking observation was the rapid rate, high frequency, and essentially exclusive development of B cell tumors. This malignancy rate far exceeded the rate and frequency of T cell tumors observed in previous TCL1 or MTCP1 transgenic T cell models at older ages (10, 11). Notably, this predilection for B lymphomas occurred despite the fact that both transgenic B and T cells (K.K.H. and M.A.T., unpublished work) proliferated strongly in response to immune receptor stimulation and showed increased survival in extended culture compared with that of wild-type spleen cells. The mechanisms responsible for the enhanced oncogenicity of TCL1 in B cells compared with T cells were not identified. However, the observed results suggest some testable models. The preference for B cell tumor formation could be because of the generation of secondary mutations that occur more frequently in B cells vs. T cells, consistent with our finding of evolving somatic mutations in Ig genes of several tumor lines (Table 4). An attractive source for such mutations would be errors in the GC hypermutation events that drive antibody affinity maturation. Such errors occur at moderate frequencies in B cell development and result from mistakes in double-strand break repair during somatic hypermutation and class switching, leading to point mutations, translocations, and probably additional types of chromosomal aberrations (29–31). The lack of a similar error-prone mechanism for TCR diversification, coupled with the absence of tumors from pre-GC stages of B cell development, is consistent with this proposal. Another testable hypothesis is that the early appearance of B cell lymphomas may preclude the appearance of T cell lymphomas that appear only with longer latencies. Consistent with this idea, mouse K11 had a polyclonal T cell expansion and might have developed a T cell neoplasm had it not developed a clonal B cell malignancy that resulted in morbid illness at 12 months. In addition, one mouse, K16, did not develop a B cell lymphoma but, rather, had a CD8+ T cell malignancy at 15 months. The absence of complementing mutations that contribute to early development of B cell malignancy may underlie the development of a T cell tumor in this mouse at a later age.
The molecular mechanisms by which dysregulated TCL1 induces lymphoid malignancies are emerging. Recently, TCL1 was shown to associate in a multimeric complex with AKT serine/threonine kinase family members (22, 23, 32). This association resulted in a dose-dependent augmentation of AKT activation and suggested that TCL1-induced activation of AKT might be critical for TCL1-induced neoplasia. Activated AKT was previously shown to increase cell survival and proliferation through the phosphorylation of numerous substrates in distinct signaling pathways (33–37). Conflicting results, however, have been reported from in vitro studies with regard to TCL1 modulation of AKT targets. Overexpressed TCL1 lead to an increased phosphorylation of AKT target substrates FKHR, GSK3-β, and BAD in some studies, whereas others indicated no effect on the AKT target substrates p70S6 kinase, BAD, IκB, and NUR77 (22, 23, 38, 39). Therefore, it has remained unresolved as to whether dysregulated TCL1 expression in premalignant lymphoid development had an effect on AKT activity and target substrates. These findings also created some uncertainty about a potential mechanism for TCL1-induced tumor formation.
The present study in nontransformed B lymphocytes shows that TCL1 augments phosphorylation of an AKT downstream target substrate, S6 ribosomal protein, and is consistent with the notion that hyperactivation of the phosphatidylinositol 3-kinase pathway is at least part of the mechanism for initiating transformation. It has been suggested that overactive p70S6 kinase may have oncogenic potential, possibly by affecting S6-regulated protein translation and cell size (40, 41). However, one study reported no effect on p70S6 kinase activity with TCL1 overexpression in 293 T cells (23). The disparity between results reported here and those of Pekarsky et al. (23) could indicate a cell type or transformation-state dependence on the outcome of TCL1 overexpression in regulating p70S6 kinase activity. Membrane-linked AKT has been shown to directly activate p70S6 kinase, whereas PDK1 phosphorylates p70S6 kinase when AKT is cytosol-restricted (42–44). Recently, we have shown that AKT and TCL1 interact optimally at the cytoplasmic membrane in transformed B cells (45). Combined, these colocalization results and the sequential activation of AKT followed by p70S6 kinase (Fig. 6) suggest a model in which TCL1-mediated hyperactivation of AKT directly increases p70S6 kinase activity. Additional support for this direct model is provided by the studies of Laine et al. (32), which showed that TCL1 does not interact directly with PDK1 (22, 32). However, it still remains possible that TCL1 has AKT-dependent and AKT-independent pathway activation effects, potentially through direct interactions with p70S6 kinase or additional PDK1 target substrate molecules.
Finally, B cells from patients with AIDS are chronically stimulated by antigens and develop into aggressive mature B-NHL at a very high rate. Many of these tumors contain aberrantly high levels of TCL1 expression. Secondary mutations required for malignancy could be generated by somatic hypermutation in HIV-altered GC reactions. This situation is similar to the high rate of mature B cell lymphomas detected in the TCL1 transgenic model presented here and predicts that TCL1-induced B cell neoplasia may be dependent on faulty DNA repair reactions and cell selection mechanisms in the GC. One compelling test of this prediction is to analyze mice that dysregulate TCL1 but fail to form GCs.
Note
During review of this manuscript, an independent study was published on the generation of CD5+ B cell chronic lymphocytic leukemia by TCL1 overexpression in B cells of transgenic mice (46).
Supplementary Material
Acknowledgments
We thank Riccardo Dalla-Favera for the Bcl-6 antibody, Owen Witte for the pEμ-B29 vector, and Charles Sawyers, Ken Dorshkind, Enca Montecino-Rodriguez, and Shane Smith for valuable advice. We also thank Peter Shintaku, Katherine Williams, Foaad Hanna (Specialty Labs, Santa Monica, CA), and members of the Teitell laboratory for technical assistance. This study was supported by the Lymphoma Research Foundation, the American Cancer Society, the Jonsson Comprehensive Cancer Foundation, the University of California Los Angeles AIDS Institute Research Award, an Amgen/University of California BioStar Award (S98-35), and the National Institutes of Health (Grants T32CA09056, GM08042, CA09120, HD37091, CA81140, CA74929, CA85841, and GM40185).
Abbreviations
DLBCL, diffuse large B cell lymphoma
BLL, Burkitt-like lymphoma
TCR, T cell antigen receptor
BCR, B cell antigen receptor
GC, germinal center
NHL, non-Hodgkin lymphoma
BM, bone marrow
LPS, lipopolysaccharide
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Virgilio L., Narducci, M. G., Isobe, M., Billips, L. G., Cooper, M. D., Croce, C. M. & Russo, G. (1994) Proc. Natl. Acad. Sci. USA 91, 12530-12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pekarsky Y., Hallas, C., Isobe, M., Russo, G. & Croce, C. M. (1999) Proc. Natl. Acad. Sci. USA 96, 2949-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teitell M., Damore, M. A., Sulur, G. G., Turner, D. E., Stern, M. H., Said, J. W., Denny, C. T. & Wall, R. (1999) Proc. Natl. Acad. Sci. USA 96, 9809-9814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Said J. W., Hoyer, K. K., French, S. W., Rosenfelt, L., Garcia-Lloret, M., Koh, P. J., Cheng, T. C., Sulur, G. G., Pinkus, G. S., Kuehl, W. M., et al. (2001) Lab. Invest. 81, 555-564. [DOI] [PubMed] [Google Scholar]
- 5.Narducci M. G., Pescarmona, E., Lazzeri, C., Signoretti, S., Lavinia, A. M., Remotti, D., Scala, E., Baroni, C. D., Stoppacciaro, A., Croce, C. M. & Russo, G. (2000) Cancer Res. 60, 2095-2100. [PubMed] [Google Scholar]
- 6.Pekarsky Y., Hallas, C. & Croce, C. M. (2001) Oncogene 20, 5638-5643. [DOI] [PubMed] [Google Scholar]
- 7.Pekarsky Y., Hallas, C. & Croce, C. M. (2001) J. Am. Med. Assoc. 286, 2308-2314. [DOI] [PubMed] [Google Scholar]
- 8.Narducci M. G., Virgilio, L., Isobe, M., Stoppacciaro, A., Elli, R., Fiorilli, M., Carbonari, M., Antonelli, A., Chessa, L., Croce, C. M., et al. (1995) Blood 86, 2358-2364. [PubMed] [Google Scholar]
- 9.Thick J., Metcalfe, J. A., Mak, Y. F., Beatty, D., Minegishi, M., Dyer, M. J., Lucas, G. & Taylor, A. M. (1996) Oncogene 12, 379-386. [PubMed] [Google Scholar]
- 10.Gritti C., Dastot, H., Soulier, J., Janin, A., Daniel, M. T., Madani, A., Grimber, G., Briand, P., Sigaux, F. & Stern, M. H. (1998) Blood 92, 368-373. [PubMed] [Google Scholar]
- 11.Virgilio L., Lazzeri, C., Bichi, R., Nibu, K., Narducci, M. G., Russo, G., Rothstein, J. L. & Croce, C. M. (1998) Proc. Natl. Acad. Sci. USA 95, 3885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raphael M. M., Audouin, J., Lamine, M., Delecluse, H. J., Vuillaume, M., Lenoir, G. M., Gisselbrecht, C., Lennert, K. & Diebold, J. (1994) Am. J. Clin. Pathol. 101, 773-782. [DOI] [PubMed] [Google Scholar]
- 13.Knowles D. M. (1997) Semin. Diagn. Pathol. 14, 67-82. [PubMed] [Google Scholar]
- 14.Gaidano G., Carbone, A. & Dalla-Favera, R. (1998) Am. J. Pathol. 152, 623-630. [PMC free article] [PubMed] [Google Scholar]
- 15.Said J. W., Pinkus, J. L., Shintaku, I. P., deVos, S., Matsumura, F., Yamashiro, S. & Pinkus, G. S. (1998) Mod. Pathol. 11, 1-5. [PubMed] [Google Scholar]
- 16.Kawamoto H., Ikawa, T., Ohmura, K., Fujimoto, S. & Katsura, Y. (2000) Immunity 12, 441-450. [DOI] [PubMed] [Google Scholar]
- 17.Omori S. A. & Wall, R. (1993) Proc. Natl. Acad. Sci. USA 90, 11723-11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malone C. S., Omori, S. A. & Wall, R. (1997) Proc. Natl. Acad. Sci. USA 94, 12314-12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yukawa K., Kikutani, H., Inomoto, T., Uehira, M., Bin, S. H., Akagi, K., Yamamura, K. & Kishimoto, T. (1989) J. Exp. Med. 170, 711-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hori M., Xiang, S., Qi, C. F., Chattopadhyay, S. K., Fredrickson, T. N., Hartley, J. W., Kovalchuk, A. L., Bornkamm, G. W., Janz, S., Copeland, N. G., et al. (2001) Blood Cells Mol. Dis. 27, 217-222. [DOI] [PubMed] [Google Scholar]
- 21.Kovalchuk A. L., Qi, C. F., Torrey, T. A., Taddesse-Heath, L., Feigenbaum, L., Park, S. S., Gerbitz, A., Klobeck, G., Hoertnagel, K., Polack, A., et al. (2000) J. Exp. Med. 192, 1183-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laine J., Kunstle, G., Obata, T., Sha, M. & Noguchi, M. (2000) Mol. Cell 6, 395-407. [DOI] [PubMed] [Google Scholar]
- 23.Pekarsky Y., Koval, A., Hallas, C., Bichi, R., Tresini, M., Malstrom, S., Russo, G., Tsichlis, P. & Croce, C. M. (2000) Proc. Natl. Acad. Sci. USA 97, 3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craxton A., Otipoby, K. L., Jiang, A. & Clark, E. A. (1999) Adv. Immunol. 73, 79-152. [DOI] [PubMed] [Google Scholar]
- 25.Gold M. R., Scheid, M. P., Santos, L., Dang-Lawson, M., Roth, R. A., Matsuuchi, L., Duronio, V. & Krebs, D. L. (1999) J. Immunol. 163, 1894-1905. [PubMed] [Google Scholar]
- 26.Genot E. M., Arrieumerlou, C., Ku, G., Burgering, B. M., Weiss, A. & Kramer, I. M. (2000) Mol. Cell. Biol. 20, 5469-5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki A., Yamaguchi, M. T., Ohteki, T., Sasaki, T., Kaisho, T., Kimura, Y., Yoshida, R., Wakeham, A., Higuchi, T., Fukumoto, M., et al. (2001) Immunity 14, 523-534. [DOI] [PubMed] [Google Scholar]
- 28.Gudermann T. (2001) Novartis Found. Symp. 239, 68-79. [DOI] [PubMed] [Google Scholar]
- 29.Pasqualucci L., Neumeister, P., Goossens, T., Nanjangud, G., Chaganti, R. S., Kuppers, R. & Dalla-Favera, R. (2001) Nature 412, 341-346. [DOI] [PubMed] [Google Scholar]
- 30.Pasqualucci L., Migliazza, A., Fracchiolla, N., William, C., Neri, A., Baldini, L., Chaganti, R. S., Klein, U., Kuppers, R., Rajewsky, K. & Dalla-Favera, R. (1998) Proc. Natl. Acad. Sci. USA 95, 11816-11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen H. M., Peters, A., Baron, B., Zhu, X. & Storb, U. (1998) Science 280, 1750-1752. [DOI] [PubMed] [Google Scholar]
- 32.Laine J., Kunstle, G., Obata, T. & Noguchi, M. (2002) J. Biol. Chem. 277, 3743-3751. [DOI] [PubMed] [Google Scholar]
- 33.Datta S. R., Brunet, A. & Greenberg, M. E. (1999) Genes Dev. 13, 2905-2927. [DOI] [PubMed] [Google Scholar]
- 34.Testa J. R. & Bellacosa, A. (2001) Proc. Natl. Acad. Sci. USA 98, 10983-10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collado M., Medema, R. H., Garcia-Cao, I., Dubuisson, M. L., Barradas, M., Glassford, J., Rivas, C., Burgering, B. M., Serrano, M. & Lam, E. W. (2000) J. Biol. Chem. 275, 21960-21968. [DOI] [PubMed] [Google Scholar]
- 36.Muise-Helmericks R. C., Grimes, H. L., Bellacosa, A., Malstrom, S. E., Tsichlis, P. N. & Rosen, N. (1998) J. Biol. Chem. 273, 29864-29872. [DOI] [PubMed] [Google Scholar]
- 37.Sonenberg N. & Gingras, A. C. (1998) Curr. Opin. Cell Biol. 10, 268-275. [DOI] [PubMed] [Google Scholar]
- 38.Pekarsky Y., Hallas, C., Palamarchuk, A., Koval, A., Bullrich, F., Hirata, Y., Bichi, R., Letofsky, J. & Croce, C. M. (2001) Proc. Natl. Acad. Sci. USA 98, 3690-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Künstle G., Laine, J., Pierron, G., Kagami, S.-i., Nakajima, H., Hoh, F., Roumestand, C., Stern, M.-H. & Noguchi, M. (2002) Mol. Cell. Biol. 22, 1513-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barlund M., Forozan, F., Kononen, J., Bubendorf, L., Chen, Y., Bittner, M. L., Torhorst, J., Haas, P., Bucher, C., Sauter, G., Kallioniemi, O. P. & Kallioniemi, A. (2000) J. Natl. Cancer Inst. 92, 1252-1259. [DOI] [PubMed] [Google Scholar]
- 41.Podsypanina K., Lee, R. T., Politis, C., Hennessy, I., Crane, A., Puc, J., Neshat, M., Wang, H., Yang, L., Gibbons, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dufner A. & Thomas, G. (1999) Exp. Cell Res. 253, 100-109. [DOI] [PubMed] [Google Scholar]
- 43.Dufner A., Andjelkovic, M., Burgering, B. M., Hemmings, B. A. & Thomas, G. (1999) Mol. Cell. Biol. 19, 4525-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rintelen F., Stocker, H., Thomas, G. & Hafen, E. (2001) Proc. Natl. Acad. Sci. USA 98, 15020-15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.French S. W., Shen, R. R., Koh, P. J., Malone, C. S., Mallick, P. & Teitell, M. A. (2002) Biochemistry 41, 6376-6382. [DOI] [PubMed] [Google Scholar]
- 46.Bichi R., Shinton, S. A., Martin, E. S., Koval, A., Calin, G. A., Cesari, R., Russo, G., Hardy, R. R. & Croce, C. M. (2002) Proc. Natl. Acad. Sci. USA 99, 6955-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.