Abstract
The development of a subgenomic replicon derived from the hepatitis C virus (HCV) strain Con1 enabled the study of viral RNA replication in Huh-7 cells. The level of replication of replicons, as well as full-length Con1 genomes, increased significantly by a combination of two adaptive mutations in NS3 (E1202G and T1280I) and a single mutation in NS5A (S2197P). However, these cell culture-adaptive mutations influenced in vivo infectivity. After intrahepatic transfection of chimpanzees, the wild-type Con1 genome was infectious and produced viral titers similar to those produced by other infectious HCV clones. Repeated independent transfections with RNA transcripts of a Con1 genome containing the three adaptive mutations failed to achieve active HCV infection. Furthermore, although a chimpanzee transfected with RNA transcripts of a Con1 genome with only the NS5A mutation became infected, this mutation was detected only in virus genomes recovered from serum at day 4; viruses recovered at day 7 had a reversion back to the original Con1 sequence. Our study demonstrates that mutations that are adaptive for replication of HCV in cell culture may be highly attenuating in vivo.
Hepatitis C virus (HCV) is an enveloped virus with a positive-strand RNA genome (1). Worldwide, more than 100 million people are persistently infected with HCV (2). Infected individuals are at increased risk of developing liver cirrhosis and hepatocellular carcinoma. In a subset of infected individuals it is possible to eliminate the infection by therapy with IFN and ribavirin (3, 4). Yet, for the development of vaccines or new therapeutics for HCV, it is of great importance to develop cell culture systems to replicate and propagate HCV. The recent construction of a subgenomic replicon, capable of replication in Huh-7 cells, might represent a first step in achieving this goal (5).
The HCV genome (≈9,600 nucleotides) has one ORF, which is flanked by UTRs. Translation produces a polyprotein of ≈3,000 amino acids that is cleaved into structural [core (C), viral envelope glycoproteins (E1 and E2)], p7, and nonstructural (NS2–5B) proteins by host or viral proteases (6, 7). The NS2 protein, with the N-terminal third of the NS3 protein, is a protease that mediates NS2/3 cleavage. The N-terminal part of the NS3 protein, with NS4A as a cofactor, has serine-protease activity and mediates the NS3/4A, 4A/4B, 4B/5A, and 5A/5B cleavages. The carboxy-terminal part of the NS3 protein contains an NTPase and an RNA-helicase. NS4B is a hydrophobic protein that induces the formation of a cytoplasmic membranous structure where all viral proteins are located and that most likely represents the site of RNA replication (8). The NS5A is a phosphorylated protein believed to be important for viral replication. A short region of the NS5A protein is thought to modulate the host IFN-mediated antiviral response (9). Mutations in this region, the IFN sensitivity-determining region, seems to be associated with the sensitivity of HCV genotype 1b viruses to IFN treatment. The protein NS5A also interacts with the IFN-induced cellular protein kinase, PKR (10). The interaction of the NS5A protein with PKR could represent a mechanism used by the virus to escape antiviral activity. The NS5B protein is an RNA-dependent RNA polymerase (RdRp).
A distinctive feature of HCV is its extensive genetic heterogeneity (11). HCV isolates were classified into six major genotypes and more than 100 subtypes. Sequences of different genotypes differ by ≈30%. Genotype 1, in particular subtype 1b, is the most common variant worldwide. Within infected individuals, HCV circulates as a quasispecies, the genome sequences of which typically differ by 1–2%. Finally, the virus has a capacity to evolve rapidly in an infected individual. Such mutations might enable HCV to replicate more efficiently or evade host immune responses.
The lack of a reliable cell culture system or of a small animal model to propagate HCV is a challenge for further studies. HCV can be propagated at low levels in human B and T cell lines (12). However, these replication systems are inefficient and results have been difficult to reproduce. The chimpanzee is the only animal that is both susceptible to HCV infection and available for study (13). Infectious cDNA clones have been generated for HCV strains of genotypes 1a (14–17), 1b (18–20), and 2a (21). Genomic RNA transcripts synthesized from full-length HCV clones were infectious when inoculated into the liver of chimpanzees. The course of HCV infection initiated by transfection of chimpanzees did not differ significantly from the infection observed in animals infected intravenously with the original virus (22, 23). Unfortunately, RNA transcripts infectious for chimpanzees have not been able to replicate in cell lines. Thus, the infectious clones have permitted studies of the importance of genetic components for replication in chimpanzees only (24–26). Therefore, it was an important advance when it was demonstrated that a subgenomic sequence of HCV strain Con1 (genotype 1b), consisting of the 5′ UTR, NS3-NS5B and the 3′ UTR as part of a selectable bi-cistronic construct can function as a self-replicating unit in Huh-7 cells (5). However, it is not known whether this viral sequence, when reconstituted into a full-length genome, is infectious for chimpanzees.
It was demonstrated that the efficiency of replication of the Con1-derived replicons in Huh-7 cells was increased by several orders of magnitude by adaptive mutations in NS3, NS5A, or NS5B (27, 28). Some of the most adaptive mutations are located at highly conserved serine residues (e.g., S2197P and S2204R/I) within a gene segment of NS5A upstream of the IFN sensitivity-determining region. However, a combination of two adaptive NS3 mutations (E1202G and T1280I) and a single adaptive mutation in NS5A (S2197P) resulted in the highest level of replication (29). In the present study, we determined whether such adaptive mutations had a similar effect on replication of the full-length Con1 genome in Huh-7 cells. Furthermore, we inoculated chimpanzees with recombinant HCV Con1 genomes to determine how the adaptive mutations affected replication in vivo.
Materials and Methods
Construction of Full-Length HCV Con1 Genomes.
Numbers refer to the nucleotide or the amino acid position of the Con1 isolate (European Molecular Biology Laboratory, AJ238799) (5). The pFK-Con1 plasmid containing the wild-type Con1 genome was described (5). pFK-Con1/5.1 was obtained by transfer of a SfiI fragment from a subgenomic replicon, which contained the E1202G, T1280I, and S2197P mutations (29), into pFK-Con1. Compared with pFK-Con1 this construct had five nucleotide changes (A3946G, C4180T, C6842T, C6926T, and T6930C) and three amino acid changes (E1202G, T1280I, and S2197P). Construction of pFK-Con1/NS5A was performed by digestion of pFK-Con1/5.1 with SalI (Invitrogen) and cloning the resulting fragment into the SalI-digested pFK-Con1. Compared with pFK-Con1 this construct had three nucleotide changes (C6842T, C6926T, and T6930C) and one amino acid change (S2197P). The pFK-Con1/D318N construct was a replication-deficient variant of pFK-Con1 with a single amino acid substitution, which changed the GDD motif of the NS5B polymerase active site to GND (29). After retransformation large-scale plasmid DNA was prepared as described (15), except that cultures were performed at 37°C. The sequences of the final DNA preparations of pFK-Con1, pFK-Con1/5.1, and pFK-Con1/NS5A were confirmed.
In Vitro Transcription, Electroporation, and Transient Replication Assays in Huh-7 Cells.
Plasmid DNA was digested with AseI and ScaI (New England Biolabs) and RNA transcripts were synthesized with T7 RNA polymerase (Promega) (5). Reactions were terminated by addition of 1.2 units of RNase-free DNase (Promega) per μg of DNA and incubation for 30 min at 37°C. After extraction with acidic phenol and chloroform, RNA was precipitated with isopropyl alcohol and dissolved in RNase-free water. For transient replication assays of full-length HCV genomes, 10 μg RNA transcripts were electroporated per 4 × 106 Huh-7 cells. The conditions for electroporation have been described (28). Six independent electroporations of the same transcript were pooled and seeded into parallel tissue culture dishes. Subsequently, cells were lysed at 4, 12, 24, 48, and 72 h, and processed for Northern or Western blotting. The Northern blot was performed as described (30) with 10 μg total RNA, except that the upper strip of the nylon membrane containing the HCV RNA was hybridized with a 32P-labeled negative-sense riboprobe complementary to NS5B and the 3′ UTR (nucleotides 8374–9440). The Western blot method was described (31). NS5B expression was monitored with monoclonal antibody 3B1.5.3 kindly provided by Darius Moradpour (Univ. of Freiburg, Freiburg, Germany) (30). β-Actin was detected with a mouse monoclonal antibody (Sigma).
Analysis of Recombinant HCV-Con1 Genomes in Chimpanzees.
The housing, maintenance, and care of the chimpanzees were in compliance with all relevant guidelines and requirements. To linearize plasmids pFK-Con1, pFK-Con1/5.1, or pFK-Con1/NS5A at the HCV 3′ end, DNA was digested first with AseI (New England Biolabs) at 37°C for 2 h in the ScaI buffer, and then ScaI (New England Biolabs) was added for another 2 h of digestion at 37°C. Digested DNA was phenol/chloroform-extracted and ethanol-precipitated. RNA was transcribed in vitro with T7 RNA polymerase from 10 μg of linearized plasmid as described (15). For each in vivo transfection, RNA transcripts from two transcription mixtures were percutaneously injected under ultrasonographic control into the liver of a naïve chimpanzee (18).
Serum samples were collected weekly or twice weekly from transfected chimpanzees. They were tested for HCV antibodies with the second-generation ELISA (Abbott) and for alanine aminotransferase (ALT) levels (Anilytics, Gaithersburg, MD). The presence of serum HCV-RNA was monitored in a reverse transcription (RT)-nested PCR (32) with primers for the 5′ UTR (33). The limit of detection by this assay is ≈10 genome equivalents per ml. In addition, HCV RNA was monitored by the HCV-Monitor Amplification kit version 2.0 (Roche Diagnostics). The lower detection limit by this test is 600 units/ml (≈500 genome equivalents per ml). When we tested aliquots of an International Units standard (34), containing 104 units/ml, in triplicate, the Monitor titers ranged from 104.1 to 104.2 units/ml. Monitor titers were in good agreement with those of the Bayer HCV RNA bDNA version 3.0 assay (R.E., unpublished data).
Serum samples collected from chimpanzees at days 3–4, 7, and 10–11 after transfections with Con1/5.1 or Con1/NS5A were tested also by RT-nested PCR with Con1-specific NS5A primers [I PCR: 6702S (TTCTTCACAGAAGTGGATGGGG) and 6999R (CATGACGGGTAGTGCATGTTGC); II PCR: 6765S (CGGGAGGAGGTCACATTCCTG) and 6993R (GGGTAGTGCATGTTGCCTTCAAG)]. The sensitivity of this assay for Con1 was equivalent to that of the 5′ UTR RT-nested PCR. NS5A amplicons obtained from the serum of chimpanzee 1614 after transfection with Con1/NS5A were cloned by the TOPO TA Cloning kit for Sequencing (Invitrogen) and sequenced. The entire ORF of virus recovered from serum of chimpanzees 1580 and 1614 at selected weeks was also amplified by RT-PCR with primers that are conserved among HCV genotype 1b strains (18) and sequenced directly to obtain consensus sequences.
Liver biopsies were collected weekly or twice weekly. For histological analysis liver biopsy specimens were fixed in 10% formalin solution. Paraffin-embedded liver biopsies collected from chimpanzees 1580 and 1614 were sectioned and stained with hematoxylin and eosin, and examined for necroinflammatory changes by using the following scoring criteria: 0, normal (no necrosis); 1+, mild (at least one focus of necrosis per ×10 field); 2+, mild–moderate (2–5 foci per ×10 field); 3+, moderate–severe (6–10 foci per ×10 field); 4+, severe (>10 foci per ×10 field).
Results
Transient Replication of Wild-Type (Con1) and Replicon-Derived (Cell Culture-Adapted) Full-Length HCV Genomes in Huh-7 Cells.
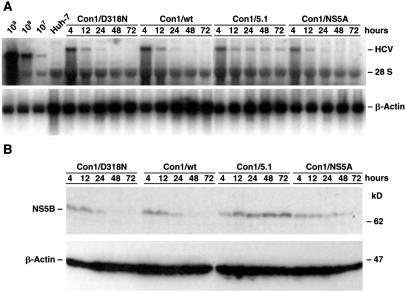
It was reported that a Con1-derived subgenomic replicon with E1202G and T1280I adaptive mutations in NS3 and a S2197P adaptive mutation in NS5A replicated more efficiently than replicons with individual NS3 or NS5A adaptive mutations (29). Because the ultimate goal of this study was to test the replication of full-length viral genomes carrying cell culture-adaptive mutations in chimpanzees, it was important to determine whether these adaptive mutations identified with subgenomic replicons had a similar effect on replication of full-length genomes in Huh-7 cells. Therefore, we performed transient replication assays with full-length genomes lacking heterologous sequences. Huh-7 cells were transfected with RNA transcripts of the wild-type genome (Con1), a variant (Con1/5.1) that contains the two mutations in NS3 and the mutation in NS5A, or a variant (Con1/NS5A) with only the single mutation in NS5A. Cells were harvested at 4, 12, 24, 48, and 72 h and replicating HCV RNA was detected by Northern blotting (Fig. 1A). The results show that Con1/5.1 replicated most efficiently. A quantitation of the blot by phosphoimaging, with β-Actin used to correct for the amount of RNA loaded in each lane, revealed that Con1/NS5A was less efficient than this highly adapted genome but slightly more efficient than the wild-type Con1 (data not shown). The adaptive phenotype of the NS5A mutation was further confirmed by Western blotting, which showed increased expression of NS5B for Con1/NS5A compared with the wild-type Con1 (Fig. 1B). Our study thus demonstrated that these NS3 and NS5A mutations that enhanced the efficiency of replication in the replicon system had a similar effect on replication of full-length genomes in cell culture.
Fig 1.
Transient replication of full-length HCV-Con1 genome and variant genomes with adaptive mutations. Huh-7 cells were transfected with the wild-type Con1 genome, with a replication-deficient variant of Con1 (Con1/D318N), or with two Con1 variants that contain cell culture-adaptive mutations [Con1/5.1 (E1202G, T1280I and S2197P) and Con1/NS5A (S2197P)]. Cells were harvested and replication was monitored by Northern and Western blot. (A) Detection of HCV RNA by Northern blot. Total RNA was prepared from harvested cells, and 10 μg was analyzed by using 32P-labeled riboprobes complementary to a sequence within the HCV NS5B and 3′ UTR (Upper) and β-Actin (Lower), respectively. As a reference, 10 μg of total RNA from control Huh-7 cells, as well as serial dilutions of in vitro transcripts of a subgenomic HCV replicon spiked with 2 μg of total RNA from naïve Huh-7 cells, were analyzed in parallel. (B) Detection of HCV protein by Western blot. Lysates, each corresponding to 5 × 105 cells, were loaded on a gel, and proteins were separated by electrophoresis and blotted. The upper part of the blot was probed with an HCV NS5B-specific monoclonal antibody, whereas the bottom part was incubated with antibodies specific for β-Actin.
The Wild-Type Con1 Genome Is Infectious for Chimpanzees.
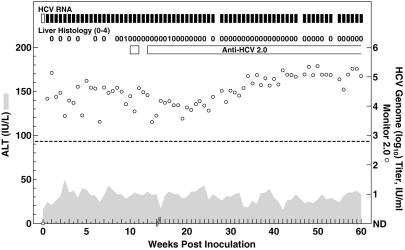
The first in vivo transfection with RNA transcripts synthesized in vitro from full-length wild-type Con1 genomes failed. Because this clone was readily infectious in the subsequent transfection in another naïve chimpanzee (see below), and because the original chimpanzee has since been infected with HCV, we believe that this failure was caused by technical difficulties in that particular experiment. When new RNA transcripts from the Con1 genomes were inoculated into the liver of chimpanzee 1580, the animal became infected; serum HCV RNA with a titer of 104-105 units/ml was detected during week 1 postinoculation (p.i.) and the animal remained HCV-RNA-positive throughout 60 weeks of follow-up (Fig. 2). The chimpanzee became anti-HCV-positive at week 10. ALT values remained normal, but necroinflammatory changes indicative of mild hepatitis were detected in liver biopsies at weeks 9 and 16.
Fig 2.
Intrahepatic inoculation of chimpanzee 1580 with RNA transcripts of the wild-type Con1 clone (pFK-Con1). Serum samples were tested for HCV-RNA by in-house RT-nested PCR with 5′ UTR primers and/or the Roche Monitor Test 2.0: filled rectangle, positive by RT-nested PCR and/or by Monitor; open rectangle, negative by RT-nested PCR in two independent assays; open circle, HCV Monitor titers; samples found to be below the detection limit of 600 units/ml (indicated by dashed line) are shown as not detected (ND). Seroconversion in the Abbott second-generation ELISA for anti-HCV is represented by a horizontal bar. Shaded area, serum levels of ALT. Liver Histology: necroinflammatory changes of liver biopsy samples graded as 0 (normal), 1 (mild), 2 (mild–moderate), 3 (moderate–severe), or 4 (severe).
We determined the consensus genome sequence (entire ORF) of viruses recovered from chimpanzee 1580 after transfection with Con1. During 1 year of follow-up 16 nucleotide changes (not shown), which resulted in amino acid changes at 11 positions, were identified (Table 1). The calculated mutation rate was 1.8 × 10−3 nucleotide substitutions per site per year and 3.7 × 10−3 amino acid substitutions per site per year. The sequence at week 1 was identical with the Con1 wild-type sequence; at week 8.5, a single nucleotide change that resulted in an amino acid change in NS2 (S1017N) was observed. Further analysis revealed that this mutation first appeared as a minor species during week 6, coinciding with a transient 1–2 log10 decrease in HCV titers. At week 28, this amino acid had changed again (N1017D) because of a mutation of a different nucleotide in the codon. The viral titers were ≈104 units/ml during weeks 15–25 p.i., but increased to ≈105 units/ml during the remainder of follow-up (Fig. 2). Several amino acid changes appeared to be emerging at week 28, and, by week 40, at 11 sites amino acid changes (in E1, E2, p7, NS2, NS3, and NS5A) seemed to have been fixed in the virus population, because no new mutations occurred between weeks 40 and 52.
Table 1.
Evolution of HCV-Con1 in chimpanzee 1580
| Week p.i.
|
E1 347 M
|
E2 430 N
|
p7 769 L
|
NS2 1017 S
|
NS3 | NS5A | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1177 V | 1444 F | 1498 I | 1568 I | 2356 E | 2362 S | 2375 D | |||||
| 1 | M | N | L | S | V | F | I | I | E | S | D |
| 2 | |||||||||||
| 3 | S | ||||||||||
| 4 | S | ||||||||||
| 5 | S | ||||||||||
| 6 | S | ||||||||||
| 6.5 | S/n | ||||||||||
| 7 | S/n | ||||||||||
| 8 | S/N | ||||||||||
| 8.5 | M | N | L | N/s | V | F | I | I | E | S | D |
| 28 | I/m | N/D | I/l | D | V | Y | I/V | I/M | E/g | N | D/g |
| 40 | I | D | I | D | A | Y | V | M | G | N | G |
| 52 | I | D | I | D | A | Y | V | M | G | N | G |
The consensus sequence of the entire ORF of viruses recovered from chimpanzee 1580 at weeks 1, 8.5, 28, 40, and 52 after transfection was determined. Only amino acid changes are given. The corresponding amino acids and their positions in the Con1 polyprotein sequence are shown on top. The differences from the wild-type Con1 sequence are indicated below. Dominant sequences are shown in capital letters; minor sequences are shown in lowercase letters. For sequential analysis of the NS2 position with a substitution identified at week 8.5, the consensus sequence of only a short genomic region was determined during weeks 3–8.
Entire polyprotein analyzed.
Variants of Wild-Type Con1 with Cell Culture-Adaptive Mutations Are Nonviable or Highly Attenuated in Chimpanzees.
Three independent transfections performed with three different RNA transcripts of the Con1/5.1 genome (E1202G, T1280I, and S2197P mutations) in two chimpanzees failed to achieve active HCV infection. After one such transfection, Con1-specific 5′ UTR sequences were amplified in RT-nested PCR (in one of two assays) from chimpanzee serum collected at day 3. However, this sample was negative in the NS5A RT-nested PCR assay. Also, all subsequent samples collected from this chimpanzee (weeks 1–24) were negative for HCV RNA in the 5′ UTR RT-nested PCR assay.
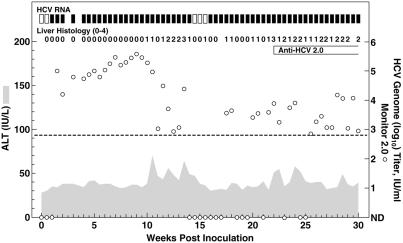
Chimpanzee 1614, transfected with RNA transcripts of the Con1/NS5A genome that contained only the single mutation in NS5A (S2197P), became infected (Fig. 3). The 5′ UTR RT-nested PCR assay was negative at day 4 (in two assays), but positive at days 7 and 11. The HCV titer at day 7 was <102 units/ml, but increased to ≈105 units/ml by day 11. The NS5A RT-nested PCR assay was positive at days 4 (in one of two assays), 7, and 11. Clonal analysis of these NS5A products revealed that the NS5A mutation was detected only in viruses recovered from chimpanzee serum at day 4 (28 of 28 clones), whereas viruses recovered at day 7 (52 of 52 clones from two independent amplifications) and at day 11 (19 of 19 clones) had reverted to the wild-type Con1 sequence. All clones analyzed at days 4, 7, and 11 contained the two silent NS5A mutations (C6842T and C6926T) found in Con1/NS5A but not in the Con1 sequence. Finally, the full-length ORF sequence of viruses recovered from this animal at weeks 6 and 9, respectively, was identical with the Con1 sequence except for the two silent NS5A mutations. These results suggest that the Con1 genome with the S2197P NS5A mutation was viable but highly attenuated in vivo.
Fig 3.
Intrahepatic inoculation of chimpanzee 1614 with RNA transcripts of pFK-Con1/NS5A, a variant of Con1 with a S2197P mutation in NS5A. The proline at position 2197 had reverted to the wild-type sequence (serine) in viruses recovered from chimpanzee serum at week 1. See also legend to Fig. 2.
After reversion to the wild-type Con1 sequence, the course of infection in chimpanzee 1614 was typical for acute hepatitis C (Fig. 3). Serum HCV titers increased to 105–106 units/ml during weeks 5–9 p.i. During weeks 10–15, virus titer decreased sharply to nondetectable levels during weeks 14 and 15. However, the animal remained infected with relatively low viral titers during weeks 16–30. HCV antibodies were detected from week 22. ALT values were elevated during weeks 10–14, 22–24, and 28. Moderate to severe necroinflammatory changes were observed in liver biopsies during weeks 10–15 and 20–30. Overall, the parent full-length genome, from which the first replicon was derived, was associated with typical HCV infections in chimpanzees. However, this genome, when modified with the NS3 and NS5A mutations that were adaptive for replication of full-length genomes in Huh-7 cells, had a highly attenuated replication phenotype in chimpanzees.
Discussion
It was reported that selectable subgenomic or full-length replicons of the HCV Con1 strain, with adaptive mutations in NS3 (E1202G, T1280I) and NS5A (S2197P), replicate very efficiently in Huh-7 cells (29, 31). The present study demonstrated that a full-length authentic Con1 genome without heterologous sequences but with these three mutations or with only the NS5A mutation also had enhanced replication capacity in Huh-7 cells. However, the replication in chimpanzees of recombinant genomes with the exact same adaptive mutations is apparently highly restricted. Although RNA transcribed from the wild-type Con1 cDNA clone (pFK-Con1) produced a typical HCV infection in a chimpanzee, RNA transcribed from a Con1 genome (pFK-Con1/5.1) with adaptive mutations in NS3 and NS5A could not produce an active HCV infection in chimpanzees. In addition, even though RNA transcribed from a Con1 genome (pFK-Con1/NS5A) with the single adaptive mutation in NS5A infected a chimpanzee, it apparently was highly attenuated; in all viruses recovered from chimpanzee serum at week 1, the proline had reverted back to the serine residue found in the wild-type Con1 sequence.
Compared with the recombinant wild-type Con1 genome, the two genomes with adaptive amino acid changes contained two synonymous nucleotide changes in NS5A. Whereas HCV genomes encoding for proline at position 2197 in Con1/NS5A mutated back to the wild-type sequence within the first week of infection in chimpanzee 1614, the two synonymous NS5A changes remained fixed in the viruses recovered during peak titers of 105-106 units/ml in this animal. No other compensating mutations were detected in the ORF of recovered viruses. These findings indicate that these NS5A mutations are silent in chimpanzees and that they did not affect the viability of Con1/5.1 or Con1/NS5A. Their presence proved that the HCV infection observed in chimpanzee 1614 after transfection with RNA transcripts of pFK-Con1/NS5A was not the result of contamination of the DNA preparation with the wild-type cDNA clone.
After one of the transfections with RNA transcripts of pFK-Con1/5.1, we did detect Con1-specific 5′ UTR sequences from chimpanzee serum, but only at day 3. HCV sequences were not detected thereafter. After transfection with RNA transcripts of pFK-Con1/NS5A, we did amplify the NS5A sequence with the adaptive mutation from chimpanzee serum, but only at day 4. The serum titers of these sequences were <10 units/ml. After day 4, only genomes that had reversion of the adaptive mutation to the wild-type sequence were detected. In previous experiments with transfection of chimpanzees with RNA transcription mixtures of defective genomes, we have not detected HCV RNA in chimpanzee serum during the first week. Therefore, we believe that the detection of Con1/5.1 and Con1/NS5A sequences represents true viral replication and not detection of input transcription mixtures. Thus, although these genomes with adaptive mutations were not capable of efficient replication in chimpanzees, they most likely replicated at low levels initially. The genome with three adaptive mutations could apparently not revert to wild-type, whereas the genome with only one mutation could.
The phenotype of wild-type Con1 infection in chimpanzees seemed similar to that of other infectious clones of HCV. In two chimpanzees, Con1 replicated to titers of 105-106 units/ml without acquiring substitutions in the virus polyprotein during the first 6 and 9 weeks, respectively. One chimpanzee had clear evidence of acute hepatitis. Finally, the virus apparently persisted in both animals. Thus, the Con1 sequence is fully functional and does not seem to be attenuated in vivo. Our study therefore indicates that the low efficiency of replication observed with the original Con1 subgenomic replicons (5, 27, 35, 36), was not due to defects in the NS3-NS5-coding region. Instead, it was due to an inherent limited capacity of these replicons to replicate in Huh-7 cells. The adaptive mutations, which enhance replication of Con1 in Huh-7 cells (27, 28), therefore most likely act at the level of interaction with host cell proteins. Thus, full-length wild-type Con1 genomes are fully capable of efficient HCV replication in vivo but not in Huh-7 cells and full-length Con1 genomes with adaptive mutations are capable of highly efficient replication in Huh-7 cells but are severely attenuated in vivo.
The NS5A protein is believed to play an important role in replication of HCV (12). A cluster of single mutations that promote replication of subgenomic Con1 in Huh-7 cells was identified in NS5A, in particular, in gene segments upstream of the IFN sensitivity-determining region (27, 28). Studies with subgenomic replicons suggest that genomes with the S2197P mutation are only moderately adapted, whereas genomes with other single NS5A mutations (S2204R/I) are more adapted (37). Therefore, we would predict that Con1 genomes with these or other NS5A adaptive mutations would also be highly attenuated in vivo. Recently, replicons were developed for HCV-N, another genotype 1b strain (35, 36). Replication of this strain was apparently not dependent on new adaptive mutations (36). However, efficient replication did depend on a unique amino acid insertion in the NS5A IFN sensitivity-determining region (36). RNA transcripts from a full-length clone of HCV-N were previously found to be infectious in a chimpanzee (19), but the infection was unusual, with low-titer viremia detectable only sporadically during weeks 3–14 p.i. Although this course of infection was originally believed by the authors to be the result of prior immunological priming, it is also possible, on the basis of our data, that the clone with the NS5A insertion was attenuated in vivo. Unfortunately, the NS5A sequence of viruses recovered from this animal was not analyzed. However, subsequent attempts to transfect RNA transcripts from this clone into other chimpanzees apparently failed to produce an active HCV infection (19, 36). It has not been determined whether the full-length HCV-N clone without the amino acid insertion in NS5A would more efficiently infect a chimpanzee. Thus, the in vivo attenuation observed with the HCV-N clone (19) could potentially be due to the effect of its nonconsensus amino acid sequence relative to its parent virus.
Many examples exist of other viruses in which cell culture-adaptive mutations result in attenuation in vivo. Among members of the related flaviviruses, the yellow fever 17D vaccine and candidate vaccines for Japanese encephalitis virus or dengue virus represent such attenuated viruses (38). However, the wild-type strains of these viruses grow efficiently in cell culture. Among the known hepatitis viruses only hepatitis A virus (HAV) routinely grows in cell culture. However, wild-type HAV grows poorly, and efficient growth of this positive-strand RNA virus is dependent on adaptive mutations in several gene regions (39). For example, adaptive mutations in the 2C protein are essential for efficient in vitro growth of wild-type, virulent HAV. However, adaptive mutations in this protein highly attenuate the virus for in vivo replication. Such attenuated viruses frequently develop back-mutations or compensating mutations in the 2C protein in vivo and revert to a virulent phenotype (40). Efficient in vitro growth of HAV is also dependent on an adaptive mutation in the 2B protein (2B: A216V) (39). Recently, it was shown, by using a replicon of a cell culture-adapted HAV, that this mutation in the 2B protein affects RNA replication in Huh-7 cells (41). In contrast to our finding with HCV, however, wild-type HAV genomes with this adaptive mutation seem to have a wild-type phenotype in the tamarin model (42). Thus, further studies are required to determine whether HCV genomes carrying other adaptive mutations are replication-competent in vivo. Relevant candidates for study might be adaptive mutations identified in the NS4B or NS5B proteins (28, 31), which might increase the efficiency of replication in Huh-7 cells by a different mechanism than the mutations identified within the NS5A protein.
Because the two chimpanzees that became infected with HCV in the present study were inoculated with RNA transcripts of recombinant genomes, they were infected with monoclonal Con1 viruses instead of the quasispecies present in a natural infection. As observed also in previous studies (16, 22, 23), the absence of a quasispecies during the early acute phase does not prevent chronicity since both animals became persistently infected. We found no evidence of changes within the hypervariable region 1 of E2 during the first year of follow-up in chimpanzee 1580, confirming that evolution of this region is not important for establishing a persistent HCV infection in chimpanzees (23, 25). Also, no change occurred in the hypervariable region 2, which has been identified among genotype 1b isolates (43). Furthermore, during the first several weeks of infection, when viral titers were as high as 105–106 units/ml, the dominant polyprotein sequence remained that of the wild-type Con1 sequence in both animals. The 11 amino acid changes that eventually occurred in Con1 viruses recovered from chimpanzee 1580 (Table 1) most likely represented selection after host immune responses or, alternatively, they represented random coselected mutations or second-site changes that compensated for decreased replicative fitness caused by other changes. Most adaptive mutations observed in the NS3 and NS5A proteins after replication of Con1 replicons in Huh-7 cells were at universally conserved positions (27, 28), whereas six of the seven mutations that evolved in these proteins in chimpanzee 1580 were at amino acid positions that varied even among different genotype 1b isolates (32). Only one in vivo change in NS3 (I1568M) was at a universally conserved position. Furthermore, the three NS5A mutations that evolved in the chimpanzee were all downstream of the cluster of mutations found to be adaptive for replication of Con1 in cell culture. Although we did not study the cellular immune responses to the virus in these animals, the following observations suggest that some mutations might have been the result of viral escape from the host immune responses. The first mutation to appear, in NS2 (S1017N), developed after a transient decrease in viral titer; this position also underwent a second change (N1017D), suggesting continuing immunological pressure. A change at this position (S1017N) was also observed during the acute phase in a chimpanzee infected with another genotype 1b strain (20). In addition, a mutation at the same position (G1017K) was recently reported to represent a cytotoxic T lymphocyte escape mutant in a chimpanzee infected with a genotype 1a strain (44). Furthermore, the F1444Y mutation, which was already fixed in the virus population at week 28, was within a cytotoxic T lymphocyte epitope in which escape mutants developed in two other infected chimpanzees (44). Thus, 2 of the 3 first mutations to appear in viruses recovered from chimpanzee 1580 might have been cytotoxic T lymphocyte escape mutants.
Our study demonstrated that Con1 genomes with cell culture-adaptive mutations were highly attenuated in vivo, which probably reflects host cell-specific interaction with HCV replication. The replicon system has become a widespread methodology to study HCV replication, virus-host interaction, and efficacy of antiviral drugs. Since most, if not all, replicons studied contain adaptive mutations specific for Huh-7 cells, our study has important implications for the interpretation of the biological relevance of findings in this in vitro system.
Acknowledgments
We thank Drs. R. H. Chanock, S. U. Emerson, and R. H. Purcell for critical reading of the manuscript. Analyses of HCV sequences were assisted by the Hepatitis database server (http://s2as02.genes.nig.ac.jp).
Abbreviations
HCV, hepatitis C virus
HAV, hepatitis A virus
p.i., postinoculation
E, envelope
NS, nonstructural
ALT, alanine aminotransferase
References
- 1.Houghton M. (1996) in Fields Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 1035–1058.
- 2.Cohen J. (1999) Science 285, 26-30. [DOI] [PubMed] [Google Scholar]
- 3.McHutchison J. G., Gordon, S. C., Schiff, E. R., Shiffman, M. L., Lee, W. M., Rustgi, V. K., Goodman, Z. D., Ling, M. H., Cort, S. & Albrecht, J. K. (1998) N. Engl. J. Med. 339, 1485-1492. [DOI] [PubMed] [Google Scholar]
- 4.Poynard T., Marcellin, P., Lee, S. S., Niederau, C., Minuk, G. S., Ideo, G., Bain, V., Heathcote, J., Zeuzem, S., Trepo, C. & Albrecht, J. (1998) Lancet 352, 1426-1432. [DOI] [PubMed] [Google Scholar]
- 5.Lohmann V., Korner, F., Koch, J., Herian, U., Theilmann, L. & Bartenschlager, R. (1999) Science 285, 110-113. [DOI] [PubMed] [Google Scholar]
- 6.Major M. E. & Feinstone, S. M. (1997) Hepatology 25, 1527-1538. [DOI] [PubMed] [Google Scholar]
- 7.Reed K. E. & Rice, C. M. (2000) Curr. Top. Microbiol. Immunol. 242, 55-84. [DOI] [PubMed] [Google Scholar]
- 8.Egger D., Wolk, B., Gosert, R., Bianchi, L., Blum, H. E., Moradpour, D. & Bienz, K. (2002) J. Virol. 76, 5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enomoto N., Sakuma, I., Asahina, Y., Kurosaki, M., Murakami, T., Yamamoto, C., Ogura, Y., Izumi, N., Marumo, F. & Sato, C. (1996) N. Engl. J. Med. 334, 77-81. [DOI] [PubMed] [Google Scholar]
- 10.Gale M., Blakely, C. M., Kwieciszewski, B., Tan, S. L., Dossett, M., Tang, N. M., Korth, M. J., Polyak, S. J., Gretch, D. R. & Katze, M. G. (1998) Mol. Cell. Biol. 18, 5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bukh J., Miller, R. H. & Purcell, R. H. (1995) Semin. Liver Dis. 15, 41-63. [DOI] [PubMed] [Google Scholar]
- 12.Bartenschlager R. & Lohmann, V. (2000) J. Gen. Virol. 81, 1631-1648. [DOI] [PubMed] [Google Scholar]
- 13.Alter H. J., Purcell, R. H., Holland, P. V. & Popper, H. (1978) Lancet 1, 459-463. [DOI] [PubMed] [Google Scholar]
- 14.Kolykhalov A. A., Agapov, E. V., Blight, K. J., Mihalik, K., Feinstone, S. M. & Rice, C. M. (1997) Science 277, 570-574. [DOI] [PubMed] [Google Scholar]
- 15.Yanagi M., Purcell, R. H., Emerson, S. U. & Bukh, J. (1997) Proc. Natl. Acad. Sci. USA 94, 8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong Z., Beaudet-Miller, M., Lanford, R. E., Guerra, B., Wright-Minogue, J., Skelton, A., Baroudy, B. M., Reyes, G. R. & Lau, J. Y. (1999) Virology 256, 36-44. [DOI] [PubMed] [Google Scholar]
- 17.Lanford R. E., Lee, H., Chavez, D., Guerra, B. & Brasky, K. M. (2001) J. Gen. Virol. 82, 1291-1297. [DOI] [PubMed] [Google Scholar]
- 18.Yanagi M., St. Claire, M., Shapiro, M., Emerson, S. U., Purcell, R. H. & Bukh, J. (1998) Virology 244, 161-172. [DOI] [PubMed] [Google Scholar]
- 19.Beard M. R., Abell, G., Honda, M., Carroll, A., Gartland, M., Clarke, B., Suzuki, K., Lanford, R., Sangar, D. V. & Lemon, S. M. (1999) Hepatology 30, 316-324. [DOI] [PubMed] [Google Scholar]
- 20.Thomson M., Nascimbeni, M., Gonzales, S., Murthy, K. K., Rehermann, B. & Liang, T. J. (2001) Gastroenterology 121, 1226-1233. [DOI] [PubMed] [Google Scholar]
- 21.Yanagi M., Purcell, R. H., Emerson, S. U. & Bukh, J. (1999) Virology 262, 250-263. [DOI] [PubMed] [Google Scholar]
- 22.Bukh J., Thimme, R., Govindarajan, S., Forns, X., Satterfield, W., Eder, E., Chang, K.-M., Yanagi, M., Emerson, S. U., Chisari, F. V. & Purcell, R. H. (2002) in Viral Hepatitis and Liver Disease, eds. Margolis, H. S., Alter, M. J., Liang, T. J. & Dienstag, J. L. (International Medical Press, Atlanta), pp. 336–340.
- 23.Major M. E., Mihalik, K., Fernandez, J., Seidman, J., Kleiner, D., Kolykhalov, A. A., Rice, C. M. & Feinstone, S. M. (1999) J. Virol. 73, 3317-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanagi M., St. Claire, M., Emerson, S. U., Purcell, R. H. & Bukh, J. (1999) Proc. Natl. Acad. Sci. USA 96, 2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forns X., Thimme, R., Govindarajan, S., Emerson, S. U., Purcell, R. H., Chisari, F. V. & Bukh, J. (2000) Proc. Natl. Acad. Sci. USA 97, 13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolykhalov A. A., Mihalik, K., Feinstone, S. M. & Rice, C. M. (2000) J. Virol. 74, 2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blight K. J., Kolykhalov, A. A. & Rice, C. M. (2000) Science 290, 1972-1974. [DOI] [PubMed] [Google Scholar]
- 28.Lohmann V., Korner, F., Dobierzewska, A. & Bartenschlager, R. (2001) J. Virol. 75, 1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krieger N., Lohmann, V. & Bartenschlager, R. (2001) J. Virol. 75, 4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pietschmann T., Lohmann, V., Rutter, G., Kurpanek, K. & Bartenschlager, R. (2001) J. Virol. 75, 1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietschmann T., Lohmann, V., Kaul, A., Krieger, N., Rinck, G., Rutter, G., Strand, D. & Bartenschlager, R. (2002) J. Virol. 76, 4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bukh J., Apgar, C. L., Engle, R., Govindarajan, S., Hegerich, P. A., Tellier, R., Wong, D. C., Elkins, R. & Kew, M. C. (1998) J. Infect. Dis. 178, 1193-1197. [DOI] [PubMed] [Google Scholar]
- 33.Bukh J., Purcell, R. H. & Miller, R. H. (1992) Proc. Natl. Acad. Sci. USA 89, 187-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saldanha J., Lelie, N. & Heath, A. (1999) Vox. Sang 76, 149-158. [DOI] [PubMed] [Google Scholar]
- 35.Guo J. T., Bichko, V. V. & Seeger, C. (2001) J. Virol. 75, 8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikeda M., Yi, M., Li, K. & Lemon, S. M. (2002) J. Virol. 76, 2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartenschlager R. & Lohmann, V. (2001) Antiviral Res. 52, 1-17. [DOI] [PubMed] [Google Scholar]
- 38.Burke D. S. & Monath, T. P. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 1043–1125.
- 39.Emerson S. U., Huang, Y. K. & Purcell, R. H. (1993) Virology 194, 475-480. [DOI] [PubMed] [Google Scholar]
- 40.Emerson S. U., Huang, Y. K., Nguyen, H., Brockington, A., Govindarajan, S., St. Claire, M., Shapiro, M. & Purcell, R. H. (2002) J. Virol. 76, 8551-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi M. & Lemon, S. M. (2002) J. Virol. 76, 1171-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emerson S. U., Lewis, M., Govindarajan, S., Shapiro, M., Moskal, T. & Purcell, R. H. (1992) J. Virol. 66, 6649-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hijikata M., Kato, N., Ootsuyama, Y., Nakagawa, M., Ohkoshi, S. & Shimotohno, K. (1991) Biochem. Biophys. Res. Commun. 175, 220-228. [DOI] [PubMed] [Google Scholar]
- 44.Erickson A. L., Kimura, Y., Igarashi, S., Eichelberger, J., Houghton, M., Sidney, J., McKinney, D., Sette, A., Hughes, A. L. & Walker, C. M. (2001) Immunity 15, 883-895. [DOI] [PubMed] [Google Scholar]