Abstract
Bifidobacteria are Gram-positive prokaryotes that naturally colonize the human gastrointestinal tract (GIT) and vagina. Although not numerically dominant in the complex intestinal microflora, they are considered as key commensals that promote a healthy GIT. We determined the 2.26-Mb genome sequence of an infant-derived strain of Bifidobacterium longum, and identified 1,730 possible coding sequences organized in a 60%–GC circular chromosome. Bioinformatic analysis revealed several physiological traits that could partially explain the successful adaptation of this bacteria to the colon. An unexpectedly large number of the predicted proteins appeared to be specialized for catabolism of a variety of oligosaccharides, some possibly released by rare or novel glycosyl hydrolases acting on “nondigestible” plant polymers or host-derived glycoproteins and glycoconjugates. This ability to scavenge from a large variety of nutrients likely contributes to the competitiveness and persistence of bifidobacteria in the colon. Many genes for oligosaccharide metabolism were found in self-regulated modules that appear to have arisen in part from gene duplication or horizontal acquisition. Complete pathways for all amino acids, nucleotides, and some key vitamins were identified; however, routes for Asp and Cys were atypical. More importantly, genome analysis provided insights into the reciprocal interactions of bifidobacteria with their hosts. We identified polypeptides that showed homology to most major proteins needed for production of glycoprotein-binding fimbriae, structures that could possibly be important for adhesion and persistence in the GIT. We also found a eukaryotic-type serine protease inhibitor (serpin) possibly involved in the reported immunomodulatory activity of bifidobacteria.
The human gastrointestinal tract (GIT) is colonized by a vast and diverse community of microbes that are essential to its functions. These microbes have evolved in concert with their host to occupy specific regions and niches in the GIT. A balanced, complex microflora is necessary for normal digestion and to maintain the homeostasis of intestinal ecosystem (1). This necessity is exemplified by the high incidence of GIT disorders after antimicrobial therapy, and the observation that germ-free mice have to consume 30% more calories to sustain body weight than do animals colonized with a natural flora (2). However, relatively little is known about the specific mechanisms of host–microbe interactions that play critical roles in GIT physiology.
The composition of human GIT microflora varies with age, diet, and location in the GIT. Although nonculture-based analyses suggest that the Bacteroides spp., bifidobacteria, enterococci, clostridia, and a few other groups dominate the colon, less than half of the intestinal microbiota has been cultured or described (3, 4). Bifidobacteria are obligate anaerobes in the Actinomycetales branch of the high–G+C Gram-positive bacteria that also includes the corynebacteria, mycobacteria, and streptomycetes. There are ≈32 species of bifidobacteria; a few have been isolated from human vagina and oral cavity, but the vast majority are from mammalian GITs (5). They are among the first colonizers of the sterile GITs of newborns and predominate in breast-fed infants until weaning, when they are surpassed by Bacteroides and other groups (6, 7). This progressive colonization is thought to be important for development of immune system tolerance, not only to GIT commensals, but also to dietary antigens (8); lack of such tolerance possibly leads to food allergies and chronic inflammation.
Although bifidobacteria represent only 3–6% of the adult fecal flora, their presence has been associated with beneficial health effects, such as prevention of diarrhea, amelioration of lactose intolerance, or immunomodulation (5). These correlations have led to widespread use of bifidobacteria as components of health-promoting foods. Nevertheless, there is only fragmentary information about the physiology, ecology, and genetics of any one species. To rapidly increase knowledge of bifidobacteria and the key role they play in the intestinal ecosystem, we determined the genome sequence of a Bifidobacterium longum strain isolated from infant feces. Bioinformatic analyses of the genome revealed many new insights. In particular, the predicted function of certain genes, including some that appeared to be duplicated or horizontally acquired, correlates well with the natural residence of this bacterium in the GIT. The genome information should catalyze exploration of the molecular mechanisms underlying key beneficial interactions in the GIT.
Genome Sequencing and Analysis
The genome sequence of B. longum NCC2705 was determined by shotgun-sequencing (GATC-Biotech, Constance, Germany). After achieving 8-fold coverage and <1 error per 105 nucleotides, ABI 3700 reads were assembled into 39 contigs with Seqman (DNAstar, Madison, WI). After manual editing, remaining ambiguities were resolved by additional sequencing. Gaps were closed by sequencing multiplex PCR products generated with oligonucleotides from contig extremities resulting in one large (2.26 Mb) and one small (3.6 kb) contig, which was 96% identical to plasmid pKJ36 (GenBank accession no. AF139129).
ORFs were identified with orpheus (Biomax Informatics, Martinsreid, Germany) and those with unlikely start codons or overlaps manually adjusted by using artemis (9). Criteria used for manually changing the start codon included presence of possible ribosome binding sites, GC frame plot analysis and alignments with similar ORFs from other organisms. Predicted amino acid sequences of ORFs were compared with public databases by using blastp and blastx (10); motif searches were performed by using hmmer on PFAM 5.4 (11). ORFs encoding products of <70 residues and with blastx expect values (E) > 10−3 were arbitrarily eliminated. When possible, predicted proteins were assigned an NCBI COG designation and functional category by Smith-Waterman (12) comparison to the COG database (13). Based on a manual evaluation of these results, a function or description was assigned to each ORF. tRNAs were identified by using trnascan-se (14) using default parameters and the prokaryotic covariance model. Secreted proteins were predicted by using signalp (15) and tmpred. Recent gene duplications, occurring since the split of Bifidobacterium from other high-G+C Gram-positive bacteria, were determined by comparing all B. longum ORFs to all ORFs in 54 genomes. When two or more ORFs were more similar to each other than to homologs from other species, their sequences and those of their homologs were collected. Where n > 3, phylogenetic trees were generated and parsed for the subcluster of only B. longum genes.
Results and Discussion
General Genome Characteristics.
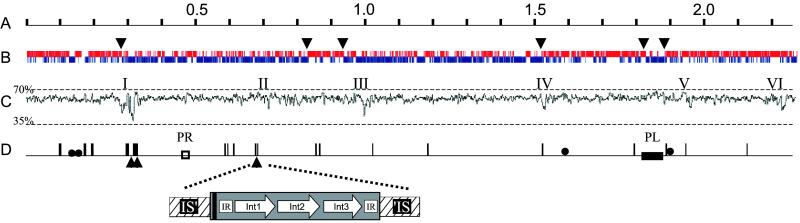
B. longum NCC2705 has a 2,256,646-bp, 60% G+C chromosomal replicon containing 4 nearly identical rrn operons, 57 tRNAs, 16 intact insertion sequence (IS) elements, as well as possible prophage and integrated plasmid remnants (Fig. 1). Eight of the IS elements constitute new members of five known families, including one rare IS607-type. We also found many related IS-elements that were interrupted or fragmented. In silico reconstruction of IS fragments indicated that many originated from intact elements, strongly suggesting substantial genome rearrangement. The genome harbored 14 integrase/recombinase genes, 9 of which were organized in 3 unusual and nearly identical structures. Each is composed of three different contiguous integrases flanked by two 97-bp perfect inverted repeats and a 20-bp palindrome; all were inserted at the same sequence and position in slightly different IS3-type elements (Fig. 1D). Although these integrases are in a phage integrase family (PFAM, PF00589), the flanking repeats and palindrome suggest that they may represent a new type of mobile genetic element (M. Chandler, personal communication). A search of GenBank for similar elements identified only a few, all in the genomes of 3 rhizobia.
Fig 1.
Linear map of the B. longum chromosome. (A) Scale in Mb. (B) Coding regions by strand. Upper and lower lines represent plus and minus strand ORFs, respectively. Arrows indicate transition points in cumulative GC skew from ORILOC. (C) G+C content with scale (window = 1,000). Roman numerals mark regions where G+C is 2.5 SD units below average. (D) Intact (two IS3-type, five IS21-type, three IS30-type, five IS256-type, and one IS607-type) as well as partial IS elements are represented by vertical lines; boxes mark possible prophage remnant (PR) and integrated plasmid (PL). Filled circles represent rRNA operons. Positions of the 3 nearly identical copies of the potentially new type of mobile genetic element in B. longum are indicated by triangles and an expanded view of one shown. The three different integrases (Int) are represented by arrows; the interrupted IS3-type element containing them is hatched. Black bar, 20-bp palindrome; IR, 97-bp perfect inverted repeat.
Total GC-skew analysis (9, 16) did not identify any clear origin of replication (OriC), but cumulative GC-skew at the third codon position using oriloc (17) showed reversal in 6 regions (Fig. 1B). Region 2 (located at ≈0.8 Mb, Fig. 1B) contains parB, parA, gid, rnpA, rpmH, dnaA, dnaN, recF, gyrB, gyrA, and 4 putative DnaA binding motifs in exact synteny with the confirmed OriC region of Mycobacterium tuberculosis (18). Unlike many other bacterial genomes, the position of the major coding strand shift (located at ≈0.28 Mb, Fig 1B) is distant from this region, again suggesting major genomic rearrangement.
We identified 1,730 probable coding regions comprising 86% of the genome. A specific or general function was assigned to 1,225 (71%) of them; 1,346 (78%) were attributed to a COG family. No functional description could be assigned to 505 genes (29%). Of these, 389 (22%) had no similarity (E > 10−2) to any predicted protein in public databases indicating either that they were either specific to B. longum or that they remain to be identified in other genomes. When the best blastp hits for each predicted protein (cutoff E > 10−2) were parsed by organism, we found that 34% of the ORFs had best hits from Streptomyces coelicolor, 9.3% from M. tuberculosis, and 3.8% from Corynebacterium glutamicum. This finding confirms assignment of B. longum to the Actimomycetales group. Surprisingly, a disparate number of best hits were found in genomes of phylogenetically distant genera that include GIT inhabitants. For example, 5.3% of the best hits were from Clostridia, 4.0% from Streptococcus, and 1.9% from Escherichia coli, whereas <0.7% were found in any other single prokaryotic genome.
Predicted Biosynthetic Capabilities.
Although bifidobacteria have been studied for over a century, lack of genetic tools and uniformity among studied isolates have prevented a comprehensive and coherent view of their biosynthetic capabilities. Our genome analysis supports and extends previous studies about vitamin and amino acid production (5), and more importantly reveals some previously undescribed physiological characteristics that may relate to their habitat. Moreover, because many bifidobacteria are often difficult to culture in defined media, the predicted metabolic abilities from genome analysis should facilitate development of new methods for growth and isolation.
B. longum has genes for synthesis of at least 19 amino acid from NH4 and major biosynthetic precursors (phosphoenolpyruvate, oxaloacetate, oxoglutarate, and fumarate) provided by its partial Krebs cycle that lacks fumarase, oxoglutarate dehydrogenase, and malate dehydrogenase. As in other high–G+C Gram-positive bacteria, all of the biosynthetic genes for tryptophan biosynthesis are present except TrpF (phosoribosylanthranilate isomerase), suggesting that an unrecognized orthologous replacement has occurred in this group. Asparagine synthetases (AsnA/AsnB) and asparaginyl–tRNA synthetase are absent indicating that B. longum exclusively uses the gatABC/asparaginyl–tRNA-dependent route to produce asparagine from aspartate (19). Also missing is the widespread sulfate/sulfite assimilation pathway involving adenosine-5′-phosphosulfate (APS) kinase, ATP sulfurylase, serine acetyl transferase, and cysteine synthetase (20). However, in B. longum cysteine biosynthesis and sulfur assimilation may be accomplished by an atypical pathway involving its homologs of cystathionine γ-synthase (EC 4.2.99.9), cystathionine β-synthetase (EC 4.2.1.22), and cystathionine γ-lyase (EC 4.4.1.1) using succinylhomoserine and the H2S or methanethiol produced by other colonic microflora as substrates (21).
B. longum has all homologs needed for biosynthesis of pyrimidine and purine nucleotides from glutamine. There are 2 homologs each of PyrE and PyrF, and 3 of PyrD, more than most prokaryotic genomes. Homologs of most enzymes needed for synthesis of folic acid, thiamin, and nicotinate are present, whereas all those for riboflavin, biotin, cobalamin, pantothenate, and pyridoxine are missing. Inability to make some vitamins and dependence on H2S or methanethiol for Cys and Met biosynthesis probably limits the ecological range of B. longum, unless this organism harbors totally novel biosynthetic pathways we failed to detect. Unlike the vast majority of sequenced prokaryotes, B. longum lacks an individual acyl carrier protein and has a 3,100-residue multifunctional type I fatty acid synthetase (FAS) instead of the multipolypeptide type II FAS. A similar type I FAS is found only in Mycobacterium, Corynebacterium, and Brevibacterium.
Predicted Energy Metabolism Typical of a Microaerotolerant Anaerobe.
B. longum has no aerobic or anaerobic respiratory components confirming it is a strict fermentative anaerobe. It is moderately aerotolerant, and as such has homologs of enzymes that repair oxidative damage. Although previous work showed that B. longum has NADH oxidase, NADH peroxidase, and low superoxidase dismutase activities for minimizing the toxicity of active oxygen species (22), we only found an NADH-oxidase homolog. However, we did find three other predicted proteins that reverse oxidative damage to proteins and lipids: thiol peroxidase, alkyl hydroperoxide reductase (ahpC), and peptide methionine sulfoxide reductase.
Homologs of all enzymes needed for fermentation of glucose, fructose, or gluconate to lactate and acetate are present. This includes the characteristic xylulose 5-phosphate/fructose-6-phosphate phosphoketolase, and all other components of the “fructose-6-phosphate shunt” (5, 20, 23), including a partial Embden–Meyerhoff pathway. Homologs of enzymes needed to feed fructose, galactose, NAc-glucosamine, NAc-galactosamine, arabinose, xylose, ribose, sucrose, lactose, cellobiose, melibiose, gentobiose, maltose, isomaltose, raffinose, and mannose, but not fucose, into the fructose-6-phosphate shunt are present. This finding corroborates and extends previous results showing that B. longum ferments a wide variety of sugars (5).
Like some other anaerobes, B. longum may ferment amino acids by using its homologs of 2-hydroxyacid dehydrogenase, serine dehydratase, threonine aldolase, and other predicted deaminases and dehydratases. B. longum has >20 predicted peptidases that could provide amino acids from proteinaceous substrates in the GIT, or in the vagina, where carbohydrates are less abundant. Among the >25 predicted ATP-binding cassette (ABC)-type transporter systems, several appear to be specific for oligopeptides or amino acids. There are 4 recently duplicated long chain fatty acyl–CoA synthetases (EC 6.2.1.3) predicted in the genome, more than any other prokaryote, except S. coelicolor and another GIT-inhabitant, B. fragilis. These could play a role in fatty acid utilization.
Genomic Adaptation for Utilization of a Diversity of Complex Carbohydrates.
Bifidobacteria colonize the lower GIT, an environment poor in mono- and disaccharides because they are consumed by the host and microflora in the upper GIT. Although past work showed that B. longum utilizes a variety of plant-derived dietary fibers, such as arabinogalactans and gums (24, 25), the genome sequence suggests that this ability is much more extensive than previously anticipated, reflecting its adaptation to a special colonic niche. The genome contains a plethora of predicted proteins assigned to COGs in the carbohydrate transport-metabolism category, >8.5% of the total predicted proteins. This is 30% more than E. coli, Enterococcus faecium, L. lactis, B. halodurans, and B. subtilis, and twice the number for M. leprae and D. radiodurans. Numerous assignments were to COGs related to oligosaccharide hydrolysis and uptake such as COG3534 (α-l-arabinofuranosidases), COG1472 (β-glucosidase-related glycosidases), COG1501 (glycosyl hydrolase family 31), COG395 (sugar permeases), and COG1653 (solute binding proteins). B. longum has >40 predicted glycosyl hydrolases whose predicted substrates cover a wide range of di-, tri-, and higher order oligo saccharides (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). Based on amino acid sequence identity of >35% and >75% overlap with biochemically validated homologs, predicted activities include 2 xylanases, 9 arabinosidases, 2 α-galactosidases, neopullanase, isomaltase, maltase, inulinase (β-fructofuranosidase), 4 β-galactosidases, 3 β-glucosidases, 3 hexosaminidases, and 3 α-mannosidases. To take full advantage of these enzymes and minimize crossfeeding of competitors, B. longum has 8 high-affinity MalEFG-type oligosaccharide transporters (27), more than in any other published prokaryotic genome. These likely help B. longum compete for uptake of structurally diverse oligosaccharides (degree of polymerization <8) released from plant fibers.
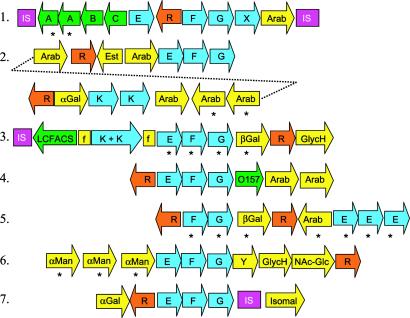
Interestingly, many of the glycosyl hydrolases and oligosaccharide transporters are organized in >7 clusters that display a conserved modular architecture (Fig. 2). Each cluster consists of (i) a LacI-type, sugar-responsive repressor, (ii) an ABC-type MalEFG oligosaccharide transporter (26), and (iii) 1–6 genes encoding various types of glycosyl hydrolases. For example, cluster 2 (Fig. 2) has 5 predicted arabinosidases and a rare α-galactosidase homolog (BL1761; Table 1 and below) implying that its function is to release arabinose and galactose from internalized fragments of arabinogalactans and arabinoxylans. These fragments are probably generated by extracellular enzymes such as endoarabinosidase BL0183 and endoxylanase BL1544 acting on larger, hemicellulosic plant fibers in the GIT. Interestingly, the only other genome sequences with large numbers of arabinosidases and oligosaccharide transporters are those of the colonic inhabitants E. faecium and B. fragilis. Some are in clusters analogous to those shown in Fig. 2, thus suggesting that these features may be shared among some GIT-inhabiting microbes.
Fig 2.
Oligosaccharide utilization gene clusters. Genes are represented by arrows. IS, insertion sequence; F and G, MalF-type and MalG-type permease subunits of ABC transporter, respectively; E and K, MalE-type solute binding protein and MalK-type ATP-binding protein of ABC transporter, respectively; R, LacI-type repressor; Arab, arabinosidase; βGal, β-galactosidase; αMan, α-mannosidase; αGal, α-galactosidase; GlycH, glycosyl hydrolase of unknown specificity; Isomal, isomaltase; NAc-Glc, N-acetyl glucosamindase; O157, ORF with homolog only in E. coli O157; X and Y, unique hypothetical proteins; LCFACS, long chain fatty acyl CoA synthetase; Est, possible xylan esterase; A, L. lactis phage infection protein homolog; B, oxidoreductase; C, phosphoglycerate mutase; f, fragment of AraE permease. Asterisks mark recent gene duplications.
The extent of B. longum's metabolic adaptation to the GIT environment is further highlighted by cluster 6 (Fig. 2). In addition to the MalEFG-type oligosaccharide transporter and a glycosyl hydrolase, this cluster contains three α-mannosidases and an endo-NAc glucosaminidase (Table 1), which are more commonly found in eukaryotes, where they remove the N-linked Man8-NAcGlc2 chains of glycoproteins (27). Adjacent to cluster 6 are genes encoding a peptidase, an oligopeptide permease system, and enzymes needed to direct mannose and NAc-glucosamine into fermentation pathways. Thus, cluster 6 may function in catabolism of galactomannan-rich plant gums, that among major GIT inhabitants, are fermented only by B. longum (24, 28) or more interestingly, in catabolism of glycans, glycoconjugates, or glycoproteins produced by epithelial cells of the colon. In support of this Hooper et al. (29) showed in vivo that the GIT commensal Bacteroides thetaiotamicron induced mouse intestinal epithelium to produce specific glycans (oligosaccharides), which were metabolized by specialized bacterial enzymes for its own nutritional benefit, possibly enhancing colonization. Thus, it is clear that the gene clusters shown in Fig. 2 provide B. longum with access to a diverse spectrum of substrates, either from the host's diet or perhaps from the host itself. It is noteworthy that several of these clusters have recently duplicated genes and adjacent IS elements, implying that B. longum is under strong pressure to evolve catabolic diversity to cope with nutritional competition.
Several B. longum glycosyl hydrolases have their best homologs in the eukaryota. One of these, BL0177, is a predicted α-galactosidase or galactomannanase. Although a protein with 60% similarity is predicted in B. halodurans, all other strong homologs in >100 genomes searched are from diverse plants, and a few other eukaryotes, including Homo sapiens. Another example is BL1761, a predicted β-1,3 exoglucanase that is >50% similar to characterized enzymes found only in Saccharomyces cerevisiae, Candida albicans, Schizosacchromyces pombe, and C. dificile. Although the unusual prokaryotic distribution of BL0177 and BL1761 could result from the limited availability of complete genome sequences of GIT inhabitants, it nonetheless strongly suggests a special role for them in adaptation of B. longum to its GIT niche. Another noteworthy protein is BL0739, which shows 50% similarity to several eukaryotic-type UDP-glucose pyrophosphorylases (EC 2.7.7.9), but very little to prokaryotic enzymes. Indeed, no homologs of BL0739 were detected in >100 prokaryotic genomes. This enzyme, which was apparently purified from Bifidobacterium bifidum, is unique in that it can use glucose-1-PO4, galactose-1-PO4, and to a lesser extent xylose-1-PO4 as substrates (30), thus expanding its versatility.
Possible Adaptation of Transcriptional Regulation to Substrate Variability in the Colon.
In contrast to most prokaryotes, B. longum appears to predominantly use negative transcriptional control of gene expression; nearly 70% of its predicted transcriptional regulators appear to be repressors. It is thought that repressors allow a quicker and more stringent response to environmental change, and consistent with B. longum's need to adapt to wide substrate fluctuation in the GIT, we found an overabundance of repressors in the TetR or LacI families. Of >100 bacterial genomes analyzed, none had more LacI-type regulators than B. longum. This richness of repressors seems to be expanding, because several show clear evidence of recent duplication. All 22 of the predicted LacI-type repressors have a periplasmic sugar binding motif (PFAM PF00532), suggesting that they are sugar-responsive. Over half are in the modular gene clusters for oligosaccharide utilization (Fig. 2), and thus presumably control expression of catabolic genes in response to the available saccharides. B. longum also has four predicted NagC/XylR family repressors often associated with sugar metabolism.
Besides repressors we found a few other regulators: one alternate σ factor [heat shock sigH (σ24)], a LexA SOS-response regulator, 7 two-component systems, 5 LysR-type activators, and an AraC homolog. Interestingly, we found 2 WhiB-type regulators most often associated with control of mycellial development of Streptomyces. In contrast to many other bacterial genomes, there are only a few MarR-type and GntR-type regulators. B. longum has 3 MerR-type regulators, a Fur homolog, and a LuxS homolog, which is the synthetase for a recently identified extracellular quorum sensing and communication molecule, AI-2 (31).
Extracellular Components Possibly Involved in Host Interactions.
Extracytoplasmic proteins and structures play critical roles in establishing and maintaining interactions between a microbe and its environment. In bifidobacteria these could mediate important functions affecting the host, such as adhesion, nutrient availability, immune system modulation, or pathogen inhibition. To identify such proteins, we used SignalP (15) to identify ≈200 proteins with probable Sec-type signal peptides. Of these, 59 were apparently surface-associated lipoproteins (PROSITE PS00013), including 26 solute binding proteins of ABC transporter systems. In agreement with the extensive ability of B. longum to scavenge nutrients from extracellular polymers, several enzymes for polymer fragmentation were predicted to be secreted: 2 endoxylanases, 2 endoarabinosidases, an arabinogalactan β-galactosidase, a N-acetyl β-glucosaminidase, a M23/M27 peptidase, and a carboxypeptidase homolog. Additionally, 8 putative secreted proteins displayed a clear Gram-positive cell-surface anchor motif (PFAM PF00746). Two of these (BL0420 and BL0421) were very large and possibly involved in attachment to or degradation of xylan/hemicellulose.
The most intriguing protein with a cell-surface anchor motif is BL0675. It is 30% identical to FimA(P), the major component of the type 2 glycopropotein-binding fimbriae of the oral cavity inhabitant Actinomyces naeslundii (32, 33). Fimbriae have never been described or proposed for bifidobacteria, yet they are of great potential significance because they are cell-surface filaments that can mediate microbial adhesion to and colonization of epithelial, mucosal, or other host cell surfaces (34, 35). The finding of homologs for other A. naeslundii fimbrial biogenesis components in an apparent operon with BL0675, further supports the proposal that B. longum makes fimbriae for attachment. These additional cell-surface components include BL0676, 31% identical to a fimbrial-associated sortase-like protein of A. naeslundii (34), BL0674, a 262-kDa protein with a repetitive glycine-rich sequence characteristic of some Rickettsial cell-surface proteins, and BL0486 a predicted prepilin peptidase with 35% identity to the product of orfC, the fourth gene in the fimbrial biogenesis operon of A. naeslundii (36). Preliminary electron micrographs have revealed fimbriae-like structures on the surface of B. longum (M. Rouvet, personal communication), but it remains to be determined if they act like fimbriae, and if they possibly contribute to attachment or retention in the GIT.
Of all predicted secreted proteins, BL0108 is most remarkable because it displays identity to proteins from the serpin family of protease inhibitors found predominantly in mammals. In >100 genomes searched, we found prokaryotic homologs of BL0108 only in the heterocyst-forming cyanobacterium Nostoc sp. PCC7120; however, unlike what is found in B. longum, this serpin homolog is not predicted to be secreted and is adjacent to a gene encoding a probable target protease. In eukaryotes, serpins control key steps in physiological regulatory cascades by inhibiting specific proteases (36). In some cases serpins play important roles in immune system evasion during pathogenesis, as in the case of a myxoma virus serpin that modulates the inflammatory response of its host (37).
Horizontal Gene Acquisitions Contributing to Physiological Specialization of B. longum.
Analysis of G+C content, dinucleotide bias (38), and codon preference identified >6 genomic regions that appear to contain recently acquired foreign genes (Fig. 1C). Region I was of particular interest as it is very large (42-kb) and appears to be a hotspot for genome evolution because it contains 4 truncated IS-elements, 3 pairs of recently duplicated genes (BL0213/BL0216; BL0205/BL0215; BL0237/BL0249), and 2 of the novel integrase-IS3 elements (Fig. 1D). Moreover, most predicted proteins in Region I are related to production of exopolysaccharides, which often are important molecules in host–microbe interactions. Two contiguous genes in Region I (BL0235–BL0236) have a clear and recent streptococcal origin as they show 63% ungapped DNA sequence identity to the contiguous cps2F–cps2T with ≈20% lower GC-content and encoding rhamnosyl transferases of Streptococcus salivarius. Phylogenetic tree building with BL0235 and BL0236 (and other proteins) in Region I supports recent acquisition from a Streptococcus. The collective function of Region I could be biosynthesis of a teichoic acid-linked rhamnose-containing exopolysaccharide because it apparently contains genes encoding (i) six glycosyl transferases; (ii) a rhamnose biosynthesis pathway; (iii) a putative 3-protein polysaccharide export system; (iv) homologs of uncharacterized exopolysaccharide-related proteins; (v) TagD glycerol-3-phosphate cytidylyltransferase; and (vi) a protein associated with incorporation of phophorylcholine into lipopolysaccharides or (lipo)teichoic acids.
Regions II and VI also display codon utilization and dinucleotide frequencies quite different from the B. longum average, which together with the presence of a phage-family integrase at the border of Region VI, are again suggestive of acquisition by horizontal gene transfer. Both these regions contain genes encoding two different types of restriction-modification systems, one of which is highly homologous to the Sau3A system. Region III contains 11 genes, but the product of only one (BL0821) displays homology to a known protein, a thioredoxin-dependent thiol peroxidase that reverses oxidative damage (39). Region IV is adjacent to multiple IS elements and contains 6 genes of unknown function, as well as a gene whose product is similar to AbiL protein involved in phage resistance (40).
Lastly, Region V contains two xylanase homologs (BL1543/BL1544). Consistent with a recent acquisition of this region is the fact that of >20 different B. longum isolates surveyed, none fermented xylan (24). Hence, the presence of these xylanase genes could be a defining characteristic of NCC2705.
Conclusions
The lack of genetic tools and the diversity of the studied isolates has impeded development of a comprehensive molecular understanding of bifidobacteria. At present, <50 nonredundant bifidobacterial proteins are in GenBank. Therefore, our analysis of the complete genome sequence of B. longum NCC2705 with 1,730 predicted proteins represents a major step forward in bifidobacterial biology. It especially provides insight into the physiological and ecological specialization of B. longum in relation to its GIT environment and sheds light on its interactions with its host.
Our most striking observation was that B. longum has an excessive number of genes associated with oligosaccharide metabolism, comprising >8% of the genome. The amplification of some of these by gene duplication (Table 1), and apparent horizontal acquisition of others suggests that B. longum has been subjected to a strong environmental pressure to amplify the level and diversity of its metabolic capabilities, perhaps in response to competition for varied substrates in the GIT ecosystem. The apparent absence of pectinases, cellulases, and α- and β-amylases in B. longum contrasts sharply with its numerous other glycosyl hydrolases. These sometimes novel glycosyl hydrolases appear to attack a wide spectrum of heterogenous, less common linkages found in plant polymers such as hemicelluloses, arabinogalactans, arabinoxylans, gums, inulins, galactomannans, and branched starches (limit dextrins). This observation substantiates previous studies of nutrient utilization by bifidobacteria that foreshadowed this extensive ability (25, 41). The persistence of B. longum in the colon may result from its adaptation to catabolize the substrates that are poorly digested by the host or other GIT microorganisms, which instead focus on utilization of sugars and the more abundant uniform polymers like pectins and linear starch. Interestingly, previous work showed that high molecular weight carbohydrate concentrations were lower in the colon than in the upper located illeum (42), implying that complex carbohydrates are largely broken down in the colon. Consistent with this proposal, B. longum also has numerous high-affinity MalEFG-type oligosaccharide transporters, but only one PTS-type sugar transporter, the more common type of carbohydrate transporter in E. faecium, E. coli, and other less dominant GIT bacteria. An additional possible manifestation of competitive adaptation of B. longum is the impressive number of LacI- and TetR-type repressors that likely control expression of its many gene clusters for oligosaccharide catabolism. These negative regulators could facilitate a rapid response to fluctuations in nutrient type and availability in the GIT.
Bifidobacteria dominate the GITs of breast-fed infants, but less so those of formula-fed infants (6, 7). Besides lactose, human milk contains >80 diverse oligosaccharides, which constitute >20% of its total carbohydrate (43). A significant number of these (e.g., lacto-N-fucopentaoses, sialylated-lactoses, and other NAc-hexose-rich oligosaccharides) have uncommon structures and thus pass intact into the colon (44), where they could be selective substrates for B. longum and its arsenal of unusual glycosyl hydrolases. In agreement with this hypothesis, the genome of Lactobacillus johnsonii, an inhabitant of the more mono- and disaccharide-rich ileum, has far fewer catabolic genes for oligosaccharides. Another complex and perhaps selective substrate for B. longum is the abundant mucin coating of the colon. Although most bifidobacteria do not extensively degrade mucin in vitro (25), the presence of the prokaryotically-rare α-mannosidases, an endo-NAc glucosaminidase, and other enzymes suggest that B. longum may partially break down mucin glycoproteins or glycans to generate nutrients and enhance selective colonization.
Bifidobacteria can attach to human cells or mucins in vitro (5, 45, 46), but the situation in vivo is unclear. Strong adherence to the surface of the colon would provide a marked advantage for colonization by B. longum because it would decrease excretion in faeces. Relevant to this, we found evidence for the existence of fimbriae and a teichoic acid-linked surface polysaccharide, both of which could mediate attachment to the surface of the colon (34, 35, 47, 48).
From limited clinical studies, it is widely claimed that bifidobacterial probiotics promote GIT homeostasis and health because of antidiarrheal, immunomodulating, and possibly anticarcinogenic properties (5). However, the physiological mechanisms underlying these observations are unknown. The B. longum genome sequence will stimulate the generation and testing of hypotheses that can dissect the molecular basis of these and other important host-commensal interactions. For example, the B. longum sequence suggested it has a eukaryotic-like protease inhibitor that could alter host physiology, such as its immune response, by interfering with a regulatory serine protease. We also found that B. longum lacks the major prokaryotic DNA recombination pathway encoded by recBCD. This discovery may explain the very low homologous recombination frequencies observed with B. longum (F.A. and M.D., unpublished data) and more importantly, implies that supplying recBCD in trans could greatly improve recombination frequency, facilitating site-directed gene knockout strategies to test the roles of many B. longum genes in GIT colonization.
In addition to this complete genome analysis of a Gram-positive GIT commensal, full comparisons to genomes of other GIT commensals will undoubtedly give more insight into the molecular physiology of microflora-GIT interactions, and how various members of the GIT consortium adapt to different niches. This information will lead to a better understanding of how diet, probiotics, and other factors influence the intestinal ecosystem to affect human and animal health.
Note Added in Proof.
While this work was under review, a partial sequence of another B. longum genome was made publicly available at www.jgi.doe.gov/JGI_microbial/html/.
Supplementary Material
Acknowledgments
We thank J. R. Neeser, B. Mollet, and A. Pfeifer for their unwavering support. We are grateful to D. Söll, A. Mercenier, and B. German for valuable suggestions to improve this manuscript. We acknowledge S. Blum, E. Schiffrin, and J. Medrano for helpful discussions. We thank E. Moret for comments on vitamin biosynthesis. We also thank G. Bothe, T. Pohl, and the GATC sequencing team.
Abbreviations
GIT, gastrointestinal tract
E, expect value
IS, insertion sequence
References
- 1.Simon G. L. & Gorbach, S. L. (1986) Dig. Dis. Sci. 31, 147S-162S. [DOI] [PubMed] [Google Scholar]
- 2.Wostmann B. S., Larkin, C., Moriarty, A. & Bruckner-Kardoss, E. (1983) Lab. Anim. Sci. 33, 46-50. [PubMed] [Google Scholar]
- 3.Suau A., Bonnet, R., Sutren, M., Godon, J. J., Gibson, G. R., Collins, M. D. & Dore, J. (1999) Appl. Environ. Microbiol. 65, 4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marteau P., Pochart, P., Dore, J., Bera-Maillet, C., Bernalier, A. & Corthier, G. (2001) Appl. Environ. Microbiol. 67, 4939-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biavati B. & Mattarelli, P. (2001) in The Prokaryotes, eds. Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K. H. & Stackebrandt, E. (Springer, New York), pp. 1–70.
- 6.Favier C. F., Vaughan, E. E., De Vos, W. M. & Akkermans, A. D. (2002) Appl. Environ. Microbiol. 68, 219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harmsen H. J., Wildeboer-Veloo, A. C., Raangs, G. C., Wagendorp, A. A., Klijn, N., Bindels, J. G. & Welling, G. W. (2000) J. Pediatr. Gastroenterol. Nutr. 30, 61-67. [DOI] [PubMed] [Google Scholar]
- 8.Hooper L. V. & Gordon, J. I. (2001) Science 292, 1115-1118. [DOI] [PubMed] [Google Scholar]
- 9.Rutherford K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A. & Barrell, B. (2000) Bioinformatics 16, 944-945. [DOI] [PubMed] [Google Scholar]
- 10.Altschul S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bateman A., Birney, E., Durbin, R., Eddy, S. R., Howe, K. L. & Sonnhammer, E. L. (2000) Nucleic Acids Res. 28, 263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith T. F. & Waterman, M. S. (1981) J. Mol. Biol. 147, 195-197. [DOI] [PubMed] [Google Scholar]
- 13.Tatusov R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., Kiryutin, B., Galperin, M. Y., Fedorova, N. D. & Koonin, E. V. (2001) Nucleic Acids Res. 29, 22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen H., Engelbrecht, J., Brunak, S. & von Heijne, G. (1997) Protein Eng. 10, 1-6. [DOI] [PubMed] [Google Scholar]
- 16.Lobry J. R. (1996) Mol. Biol. Evol. 13, 660-665. [DOI] [PubMed] [Google Scholar]
- 17.Frank A. C. & Lobry, J. R. (2000) Bioinformatics 16, 560-561. [DOI] [PubMed] [Google Scholar]
- 18.Qin M. H., Madiraju, M. V. & Rajagopalan, M. (1999) Gene 233, 121-130. [DOI] [PubMed] [Google Scholar]
- 19.Min B., Pelaschier, J. T., Graham, D. E., Tumbula-Hansen, D. & Soll, D. (2002) Proc. Natl. Acad. Sci. USA 99, 2678-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs G. (1999) in Biology of the Prokaryotes, eds. Lengler, J. W., Drews, G. & Schlegel, H. G. (Springer, New York), pp. 116–123.
- 21.Suarez F. L., Furne, J., Springfield, J. & Levitt, M. D. (1999) Am. J. Gastroenterol. 94, 208-212. [DOI] [PubMed] [Google Scholar]
- 22.Shimamura S., Abe, F., Ishibashi, N., Miyakawa, H., Yaeshima, T., Araya, T. & Tomita, M. (1992) J. Dairy Sci. 75, 3296-3306. [DOI] [PubMed] [Google Scholar]
- 23.Meile L., Rohr, L. M., Geissmann, T. A., Herensperger, M. & Teuber, M. (2001) J. Bacteriol. 183, 2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salyers A. A., West, S. E., Vercellotti, J. R. & Wilkins, T. D. (1977) Appl. Environ. Microbiol. 34, 529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crociani F., Alessandrini, A., Mucci, M. M. & Biavati, B. (1994) Int. J. Food Microbiol. 24, 199-210. [DOI] [PubMed] [Google Scholar]
- 26.Boos W. & Shuman, H. (1998) Microbiol. Mol. Biol. Rev. 62, 204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabral C. M., Liu, Y. & Sifers, R. N. (2001) Trends Biochem. Sci. 26, 619-624. [DOI] [PubMed] [Google Scholar]
- 28.Salyers A. A., Vercellotti, J. R., West, S. E. & Wilkins, T. D. (1977) Appl. Environ. Microbiol. 33, 319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooper L. V., Xu, J., Falk, P. G., Midtvedt, T. & Gordon, J. I. (1999) Proc. Natl. Acad. Sci. USA 96, 9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee L., Kimura, A. & Tochikura, T. (1979) J. Biochem. (Tokyo) 86, 923-928. [DOI] [PubMed] [Google Scholar]
- 31.Chen X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L. & Hughson, F. M. (2002) Nature 415, 545-549. [DOI] [PubMed] [Google Scholar]
- 32.Yeung M. K., Donkersloot, J. A., Cisar, J. O. & Ragsdale, P. A. (1998) Infect. Immun. 66, 1482-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeung M. K. (1999) Crit. Rev. Oral Biol. Med. 10, 120-138. [DOI] [PubMed] [Google Scholar]
- 34.Li T., Johansson, I., Hay, D. I. & Stromberg, N. (1999) Infect. Immun. 67, 2053-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubiet M., Ramphal, R., Weber, A. & Smith, A. (2000) Infect. Immun. 68, 3362-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silverman G. A., Bird, P. I., Carrell, R. W., Church, F. C., Coughlin, P. B., Gettins, P. G., Irving, J. A., Lomas, D. A., Luke, C. J., Moyer, R. W., et al. (2001) J. Biol. Chem. 276, 33293-33296. [DOI] [PubMed] [Google Scholar]
- 37.Macen J. L., Upton, C., Nation, N. & McFadden, G. (1993) Virology 195, 348-363. [DOI] [PubMed] [Google Scholar]
- 38.Karlin S. (1998) Curr. Opin. Microbiol. 1, 598-610. [DOI] [PubMed] [Google Scholar]
- 39.Jeong W., Cha, M. K. & Kim, I. H. (2000) J. Biol. Chem. 275, 2924-2930. [DOI] [PubMed] [Google Scholar]
- 40.Deng Y. M., Liu, C. Q. & Dunn, N. W. (1999) J. Biotechnol. 67, 135-149. [DOI] [PubMed] [Google Scholar]
- 41.Van Laere K. M., Hartemink, R., Bosveld, M., Schols, H. A. & Voragen, A. G. (2000) J. Agric. Food Chem. 48, 1644-1652. [DOI] [PubMed] [Google Scholar]
- 42.Vercellotti J. R., Salyers, A. A., Bullard, W. S. & Wilkins, D. (1977) Can. J. Biochem. 55, 1190-1196. [DOI] [PubMed] [Google Scholar]
- 43.Newburg D. S. & Neubauer, S. H. (1995) in Handbook of Milk Composition, ed. Jensen, R. C. (Academic, London), pp. 273–349.
- 44.Engfer M. B., Stahl, B., Finke, B., Sawatzki, G. & Daniel, H. (2000) Am. J. Clin. Nutr. 71, 1589-1596. [DOI] [PubMed] [Google Scholar]
- 45.Juntunen M., Kirjavainen, P. V., Ouwehand, A. C., Salminen, S. J. & Isolauri, E. (2001) Clin. Diagn. Lab. Immunol. 8, 293-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Re B., Sgorbati, B., Miglioli, M. & Palenzona, D. (2000) Lett. Appl. Microbiol. 31, 438-442. [DOI] [PubMed] [Google Scholar]
- 47.Tsukioka Y., Yamashita, Y., Oho, T., Nakano, Y. & Koga, T. (1997) J. Bacteriol. 179, 1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stinson M. W., Nisengard, R. J. & Bergey, E. J. (1980) Infect. Immun. 27, 604-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.