Abstract
The HIV-1 Rev protein is an essential regulator of the HIV-1 mRNA expression that promotes the export of unspliced and partially spliced mRNA. The export receptor for the leucine-rich nuclear export signal (NES) of Rev has recently been recognized as CRM1. We identified a low molecular weight compound PKF050-638 as an inhibitor of HIV-1 Rev. This drug inhibits in a dose-dependent fashion Rev-dependent mRNA expression in a cellular assay for Rev function. We show that PKF050-638 is an inhibitor of the CRM1-mediated Rev nuclear export. By using a quantitative in vitro CRM1-NES cargo-binding assay, we could demonstrate that PKF050-638 disrupts CRM1-NES interaction. This mode of action is confirmed in cell culture because the drug reversibly interferes with the colocalization of CRM1 and Rev in the nucleolus of the cell. In addition, we prove that the inhibition is through direct interaction of the compound with Cys-539 of CRM1. These effects are similar to those of the known CRM1 inhibitor leptomycin B and suggest that the inhibitory effect of the compound is caused by binding to CRM1 at a similar site. The compound displayed strict structural requirements for its activity, as its enantiomer was inactive in all assays tested. These results show that we identified a drug that interferes with the CRM1-mediated nuclear export of Rev through inhibition of the CRM1-NES complex formation. The reversibility of its binding to CRM1 and its availability through chemical synthesis could make it useful for studying CRM1-mediated export pathways.
The Rev protein is an essential factor for HIV replication and promotes the export of unspliced or partially spliced mRNA responsible for the production of the viral structural proteins. Rev is an 18-kDa protein that has been shown to shuttle continuously between the nucleus and the cytoplasm (1, 2). Nuclear import of Rev is mediated by its nuclear localization signal (NLS) embedded in the RNA-binding domain that binds a unique RNA stem-loop structure termed the Rev responsive element (RRE). The function of the Rev NLS in the context of Rev is apparently multimerization dependent. Mutants defective in multimerization do not accumulate in the nucleus. Instead, these mutants are localized throughout the cell, although their NLS is completely intact (3, 4). Nuclear export of Rev is mediated by its leucine-rich nuclear export signal (NES) and is known to use the CRM1 export factor to export the viral RNA from the nucleus to the cytoplasm. CRM1 is a nuclear export receptor for proteins carrying the leucine-rich NES (5–7).
Nucleocytoplasmic transport is mediated largely by the superfamily of transport receptors that interact with nuclear pore complexes (NPCs), share an N-terminal RanGTP-binding motif and are related to importin β (8, 9). CRM1 binds its cargo in the nucleus, translocates it to the cytoplasm, releases the cargo, and returns back to the nucleus. The RanGTPase cycle is key to promoting the directionality of this transport (reviewed in refs. 10 and 11). Export receptors bind their cargo with much higher affinity in the presence of RanGTP (5, 12). The export receptor/cargo/RanGTP complex is translocated through the nuclear pore. When this complex encounters RanGAP on the cytoplasmic side of the NPC, RanGTP is hydrolyzed to RanGDP, and the complex disassembles, releasing the export cargo into the cytoplasm (12–14). The nuclear transport factor 2 binds RanGDP in the cytoplasm and delivers Ran back into the nucleus (15, 16), where RanGEF exchanges its GDP with GTP.
Recently, direct binding of Rev to CRM1 has been shown in vitro (17). Footprinting experiments revealed a more detailed picture of the regions of Rev and CRM1 involved in complex formation. Within Rev, the region Leu-64 to Arg-80 was protected by CRM1, whereas Rev specifically interacts with residue Asp-716 and the neighborhood of Lys-810 of CRM1 (17). RanGTP binding to CRM1 is expected to be mediated by a region near the N terminus (8, 9). A comparison of leucine-rich NESs of several proteins revealed that the Rev NES has a relatively low affinity for CRM1 (18).
The Streptomyces cytotoxin leptomycin B (LMB) has been identified as an inhibitor of the CRM1-mediated nuclear export (19). Its effect is direct because LMB inactivates CRM1 by covalent modification at Cys-539 (5, 20). In Schizosaccharomyces pombe, resistance to LMB maps to the crm1 gene (21). LMB has been shown to inhibit the Rev/CRM1/RanGTP complex formation in vitro (17). The use of LMB in the study of nuclear export pathways has been hampered by the variability of the quality of LMB production lots.
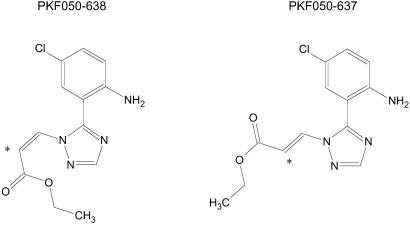
Here, we describe the chemical structure (Fig. 1), and we address the mechanism of action of a chemically synthesized low molecular weight inhibitor of Rev function.
Fig 1.
Chemical formulae of PKF050-638 and its inactive enantiomer PKF050-637. *, Asymmetric carbon.
Materials and Methods
Plasmids and Cells.
pBrev-GFP, pBrev14-GFP, and pBrev38-GFP plasmids produce fusion proteins of Rev, Rev14, and Rev38, respectively, fused to the enhanced version of the GFP emitting green light (GFPsg25; ref. 22). In the Rev14 mutant, amino acids 14–16 (RTV) are mutated to EED (23). To obtain the mutant pBrev38-GFP, the amino acids 38–44 were deleted. pBrev-blue fluorescent protein (BFP) expresses the Rev protein fused to the enhanced BFP emitting blue light (22). hCRM1-GFP was a kind gift of Barbara Felber (National Cancer Institute, Frederick, MD). pTat-GFP-NES expresses a Tat-GFP hybrid protein containing the Rev NES (24).
pCMV-Luc contains the firefly luciferase reporter gene under control of the CMV immediate early promoter. pEF-Rev expresses the HIV-1 Rev protein from the EF1-α promoter. pCMVgagLucRRE contains the firefly luciferase reporter gene fused to the p17 gag sequences and the RRE, flanked by the HIV-1 major splice sites, and driven by the CMV promoter.
HeLa and HLtat cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) FCS. Jurkat cells were grown and maintained in RPMI medium 1640 supplemented with 10% (vol/vol) FCS.
Rev Function Assay.
The Rev-dependent expression of HIV-1 was monitored as described (D.D., unpublished work). Briefly, Jurkat cells were cotransfected with pCMVgagLucRRE and pEFRev. Jurkat cells (10 million) were suspended in 200 μl of medium, and 16 μg of plasmid DNA was electroporated (260 V, 1050 μF, and indefinite resistance) into the cells. Electroporated cells (100,000) were incubated in the presence of different concentrations of test compounds in 96-well plates. Rev-dependent expression was measured by quantification of reporter gene activity 24 h after transfection. Luciferase activity was monitored by adding 100 μl of luciferase reagent containing lysis buffer (LucLite; Packard, Belgium) to the same volume of cells according to the user's manual. The IC50 value was calculated as being the inhibitor concentration that reduces reporter gene expression by 50%. Specificity was tested by measuring inhibition of luciferase expression from the Rev-independent pCMVLuc plasmid. Toxicity of the test compounds to the cells was measured according to a tetrazolium-based viability assay (CellTiter 96 Aqueous Cell Proliferation Assay; Promega, Belgium).
Microscopy of Living Cells.
HLtat cells were seeded onto coated 50-mm glass-bottom microwell dishes (MatTek), cultured in phenol red-free DMEM [supplemented with 10% (vol/vol) FCS], and transfected 24 h later with the various plasmids by using Superfect (Qiagen, Chatsworth, CA), according to the manufacturers manual. One day later, the cells were washed with PBS and treated with the appropriate concentrations of drug in DMEM (10% FCS) without phenol red. After 2 h of incubation, the cells were analyzed with a Zeiss LSM 410 Micro System in the confocal mode. Nomarski images were made by using a 543-nm green laser and appropriate polarized lenses. For GFP excitation, an argon/krypton laser at 488-nm wavelength was used, whereas a UV laser at 364-nm wavelength was used to excite BFP. Emitted fluorescence was detected with a 510- to 540-nm band-pass filter for GFP or a 400- to 440-nm band-pass filter for BFP.
Yeast Growth.
The CRM1T539C (containing Cys at position 539 instead of Thr) strain was kindly provided by Michael Rosbash (Howard Hughes Medical Institute, Waltham, MA) (25). Yeasts were grown in rich media at 30°C. Growth curves were made by using cultures of 100–350 μl in 100-well plates in a Bioscreen C apparatus (Labsystems, Zellik, Belgium). For this purpose, stationary phase cultures were diluted to an OD600 of 0.1 (photometer length of 1 cm) in the presence or absence of different concentration of drugs. Growth was followed over a period of 12–24 h with a 30-s shaking period every minute and OD600 measurements every 30 min.
RanGAP Hydrolysis Assay.
The hydrolysis of RanGTP to RanGDP in the presence of NES and CRM1 was analyzed as described (18). Briefly, RanGTP was loaded with [γ-32P]GTP (Amersham Pharmacia) in the presence of 10 mM EDTA. The reaction was stopped with 20 mM MgCl2 followed by gel filtration on a Bio-Spin 6 column (Bio-Rad) equilibrated with Ran buffer [0.1 M Tris·HCl, pH 7.5/0.5 M NaCl/10 mM MgCl2/2 mM DTT/0.25 μg/μl BSA/10% (vol/vol) glycerol]. Reaction mixtures containing ≈200 pM Ran[γ-32P]GTP, 0–200 nM CRM1, and 40 μM NS2 NES peptide (CVDEMTKKFGTLTIHDTEK) or protein kinase inhibitor (PKI) peptide (CELALKLAGLDIN) in 40 μl of reaction buffer (30 mM Tris·HCl, pH 7.5/90 mM NaCl/6 mM MgCl2/1.5 mM GTP/0.1 μg/μl BSA/1% glycerol) were assembled on ice. After a 20-min incubation, 25 nM of Rna1p (RanGAP from Saccharomyces cerevisiae) was added in 10 μl of reaction buffer and immediately placed at 25°C for exactly 2 min. Reactions were stopped by adding 1 ml of charcoal suspension [7% (wt/vol) charcoal/10% (vol/vol) ethanol/0.1 M HCl/10 mM KH2PO4], and the mixture was centrifuged for 5 min. Release of [32P]phosphate was determined by scintillation counting in 0.7 ml of the supernatant.
Results
PKF050-638 Specifically Inhibits Rev-Dependent Luciferase Reporter Gene Expression.
To identify new anti-HIV Rev inhibitors, we analyzed a series of small molecules by using a cell-based assay for Rev function. This assay is based on the expression of a Rev-dependent luciferase reporter gene in the Jurkat T cell line (D.D., unpublished work). Briefly, the cells are cotransfected with a Rev-dependent luciferase gene and then incubated with different concentrations of test compounds. The Rev-dependent luciferase gene is flanked by splice sites under the control of the RRE. Inhibitors of the Rev function cause a dose-dependent inhibition of Rev-dependent luciferase expression. The specificity of the compounds on Rev function was tested on a Rev-independent luciferase gene. The compound LMB, which is reported to be a Rev inhibitor, inhibited the Rev-dependent expression of the luciferase reporter gene at an IC50 value of 0.5 nM (Table 1), a result that is compatible with previous reports (19). By this assay, we identified a compound, PKF050-638 (Fig. 1), inhibiting Rev function in several independent experiments. The IC50 value for PKF050-638 in Jurkat cells was 0.04 μM, whereas its enantiomer PKF050-637 showed hardly any activity (Table 1). The specificity of the compounds was determined in parallel experiments with a Rev-independent luciferase reporter gene construct. The maximum concentration tested was 3.3 μM.
Table 1.
Inhibitory effect on the Rev-dependent luciferase expression
| Inhibition of Rev-mediated expression IC50, μM | Inhibition of Rev-independent expression IC50, μM | Toxicity CC50, μM | |
|---|---|---|---|
| LMB | 0.0005 | >0.125 | >0.125 |
| PKF050-638 | 0.04 | 1.90 | 3.10 |
| PKF050-637 | 1.98 | >3.30 | >3.30 |
Concentration of inhibitor required for 50% inhibition of Rev-dependent luciferase expression in transfected Jurkat cells.
Concentration of inhibitor required for 50% inhibition of luciferase expression from the CMV promoter in transfected Jurkat cells.
Concentration of inhibitor required for 50% inhibition of cell proliferation as measured by a tetrazolium-based viability assay.
PKF050-638 Inhibits Nuclear Export of Rev.
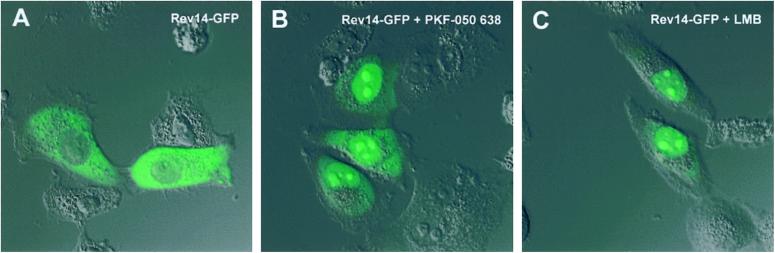
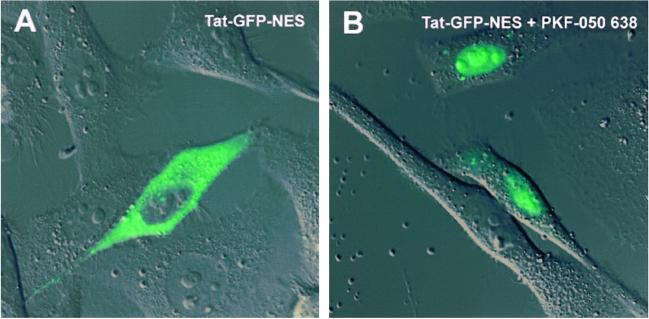
WT Rev accumulates in the nucleolus and shuttles between the nucleolus and the cytoplasm. Upon treatment of the cells with actinomycin D, Rev translocates from the nucleus to the cytoplasm. The effect of actinomycin D on Rev export is reversible; upon relief of actinomycin D from the cells, Rev steady-state localization becomes nucleolar again (not shown). PKF050-638 has no effect on the nucleolar localization of Rev-GFP (data not shown). PKF050-638 applied in addition to actinomycin D was able to prevent redistribution of Rev to the cytoplasm. These data indicated that PKF050-638 prevented the actinomycin D-induced export of Rev to the cytoplasm. To verify this result, we used two previously characterized cytoplasmic Rev mutants. As shown, the Rev mutants Rev14-GFP and Rev38-GFP (3) are localized predominantly in the cytoplasm (Fig. 2A) and are defective in their nuclear retention but not in nuclear import. They are continuously shuttling between the nucleus and the cytoplasm. Upon treatment with 5 μM PKF050-638 for 2 h, the Rev14-GFP mutant accumulated in the nucleus, indicating that its export to the cytoplasm was greatly inhibited (see Fig. 2B). This inhibitor of export was similar to what has been observed with LMB (Fig. 2C). The effect on nuclear retention of the Rev38-GFP mutant was similar (data not shown). An additional experiment indicated that the NES of Rev is the only part of Rev required for export inhibition by PKF050-638. A Tat-GFP protein fused to the Rev NES has been shown to localize in the cytoplasm (Fig. 3A) but is able to shuttle between the cytoplasm and the nucleus. Tat itself only has a nuclear import signal, and WT Tat is, therefore, localized in the nucleus. The ability of Tat-GFP-NES to shuttle between nucleus and cytoplasm, therefore, signifies the acquisition of a CRM1-mediated mode of nuclear export, associated with the NES of Rev (24). When cells expressing Tat-GFP-NES were incubated with 5 μM PKF050-638, the Tat-GFP-NES protein was retained in the nucleus of the cells, clearly demonstrating inhibition of NES-dependent nuclear export (Fig. 3B). These data suggest that the target region of the drug is the NES of Rev, either directly or indirectly.
Fig 2.
Effect of PKF050-638 on the localization of Rev14-GFP in living cells. HLtat cells were transfected with a plasmid expressing Rev14-GFP, and 24 h later, they were treated with PKF050-638 or LMB. Two hours after the addition of drugs, the cells were analyzed by confocal laser scan microscopy. Rev14-GFP localized in the cytoplasm but is able to shuttle between cytoplasm and nucleus (A). Addition of 5 μM PKF050-638 (B) or 50 nM LMB (C) resulted in predominantly nuclear/nucleolar localization of the mutant.
Fig 3.
Effect of PKF050-638 on the Rev NES in living cells. (A) Tat-GFP-NES accumulates in the cytoplasm of cells. (B) Treatment of the cells with 5 μM PKF050-638 induces nuclear retention of Tat-GFP-NES within 2 h.
The Effect of PKF050-638 Is Reversible.
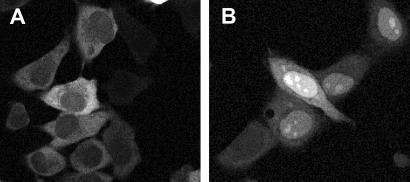
We tested whether the inhibition of export by PKF050-638 seen in Fig. 2 could be restored upon removal of the compound. Therefore, we incubated pRev14-GFP-transfected cells with 7.5 μM PKF050-638 or 100 nM LMB for 3 to 4 h. The concentrations used, 200-fold the respective compound IC50, showed approximately the same nuclear localization of Rev14-GFP after 4 h of incubation for both compounds. In cells incubated with PKF050-638 or LMB, most of the Rev14-GFP was localized in the nucleus and nucleolus as seen in Fig. 2, whereas in untreated cells, Rev14-GFP was still in the cytoplasm. The cells were washed three times with PBS to remove the drugs, and new medium was added. After overnight incubation, cells were monitored for Rev14-GFP localization by fluorescence microscopy as shown in Fig. 4. Rev14-GFP in cells incubated with PKF050-638 was almost completely translocated in the cytoplasm, whereas in cells incubated with LMB, Rev14-GFP was still localized in the nucleoli for >50% of the cells (Fig. 4B). The percentage of cells with GFP remaining in the nucleoli was determined after image analysis by confocal microscopy. Whereas 50% of LMB-treated cells contained Rev14-GFP in the nucleoli, only 14% of cells incubated with PKF050-638 had any detectable Rev14-GFP in the nucleolus. Therefore, in contrast to LMB-treated cells, the majority of PKF050-638-treated cells were able to reverse completely the inhibitory effects of the drug.
Fig 4.
Reversible effect of PKF050-638 on the localization of Rev14-GFP in living cells. Four hours after drug treatment, the drugs were washed out, and cells were analyzed by confocal scan microscopy after overnight incubation. Rev14-GFP in untreated localizes in the cytoplasm as shown in Fig. 2A. (A) Cells incubated with 7.5 μM PKF050-638 and washed resulted in predominant relocalization of the Rev14-GFP in the cytoplasm, whereas (B) in most cells treated with 100 nM LMB and washed, the Rev14-GFP stayed in the nucleoli. Before addition of compound (0 h), the Rev14-GFP in all cells was cytoplasmatic (0% of cells showed GFP in nucleoli). After the addition of either compound, all of the cells had Rev14-GFP in the nucleoli. The cells then were washed, and in 50% of cells incubated with LMB, Rev14-GFP still localized in the nucleoli, whereas Rev14-GFP could be detected in the nucleolus in only 14% of cells incubated with PKF050-638.
Inhibition of the Rev/hCRM1 Interaction in Vivo.
To examine the effect of PKF050-638 on the association of hCRM1 and Rev in mammalian cells, we cotransfected HeLa cells with the BFP-tagged Rev protein (22) and the GFP-tagged hCRM1 (26). hCRM1 was localized at the nuclear rim as well as within the nucleus (Fig. 5A), in agreement with published data (9, 26), whereas Rev localized predominantly to the nucleoli (Fig. 5D). Importantly, in the cells that expressed Rev (compare Fig. 5 A and D), a significant fraction of hCRM1 was found in the Rev-containing nucleoli. Such colocalization was observed in all Rev-expressing cells, as reported (27). Upon treatment with PKF050-638, this Rev-dependent hCRM1 nucleolar localization was abolished after 60 min (Fig. 5 B and C), whereas no effect on hCRM1 distribution was observed in cells that did not produce detectable amounts of Rev. PKF050-638 did not affect the Rev nucleolar distribution (Fig. 5F). Taken together, these results suggest that PKF050-638 is interfering with the CRM1-mediated nuclear export machinery.
Fig 5.
Colocalization of hCRM1-GFP with Rev-BFP in HeLa cells. HeLa cells were cotransfected with phCRM1-GFP and pRev-BFP and analyzed by fluorescence microscopy. For GFP, emitted fluorescence was detected with a 510- to 540-nm band-pass filter (Upper), and BFP expression was analyzed with a 400–440 nm band-pan filter (Lower). Rev-BFP is localized into nucleoli (Nu in D), whereas hCRM1-GFP is found at the nuclear rim of the cells not expressing Rev-BFP (NE in A). In cells coexpressing Rev-BFP and hCRM1-GFP, the latter colocalizes with Rev-BFP in the nucleoli (Nu in A). Thirty and 60 minutes after the addition of 7.5 μM PKF050-638, the colocalization of hCRM1-GFP with Rev-BFP disappeared (B and C), whereas Rev-BFP stays in the nucleoli (E and F).
Effect on CRM1-NES-RanGTP Complex Formation.
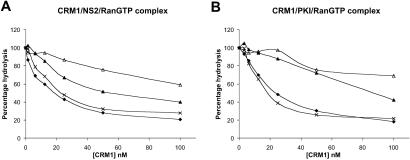
Because CRM1 is the export receptor for leucine-rich NESs (5), exemplified by the Rev NES, we tested whether PKF050-638 disrupts CRM1-NES binding. It has been reported that a complex of CRM1-NES-RanGTP is formed and is important for export of the NES-containing cargo to the cytoplasm (5). To measure the effect of the compounds on the CRM1-NES-RanGTP complex formation, we used a system developed to study interactions between RanGTP and its interaction partners (28), such as members of the importin β family (8, 9). This assay is termed the RanGTP hydrolysis assay and uses the ability of RanGAP to stimulate Ran's GTPase activity when RanGTP is unbound but not when it is in a complex with an importin β family member (e.g., CRM1), thereby allowing quantification of unbound RanGTP (28). Forty μM of NES peptide from the NS2 protein of minute virus of mice or 200 μM of PKI NES peptide was incubated with 200 pM [γ-32P]GTP-loaded Ran in the presence of different concentrations of recombinant CRM1, in the absence or presence of 1 μM LMB, or 330 μM of either PKF050-638 or PKF050-637. Subsequently, 25 nM of Rna1p (the S. pombe RanGAP) was added, and the released [32P]phosphate was measured. In the absence of CRM1 or NES, RanGTP is hydrolysed to RanGDP, whereas in the presence of both CRM1 and NES, no or little RanGTP hydrolysis occurs. As shown in Fig. 6A, LMB and PKF050-638 were both able to disrupt the CRM1-NS2-RanGTP interaction, whereas PKF050-637 had no effect.
Fig 6.
Quantitative analysis of NES-CRM1 affinity in vitro using the RanGAP hydrolysis assay. Inhibitory effect of LMB (1 μM; ▵), PKF050-638 (330 μM; ▴), PKF050-637 (330 μM; ×), and no drug (♦) on CRM1-NS2 (A) or CRM1-PKI (B) binding as measured from CRM1-dependent protection of Rna1p-stimulated GTP hydrolysis on Ran, as a function of increasing concentrations of CRM1. In all series, NS2 or PKI peptides are present at 40 and 200 μM, respectively.
Defining the Sensitive Region of CRM1.
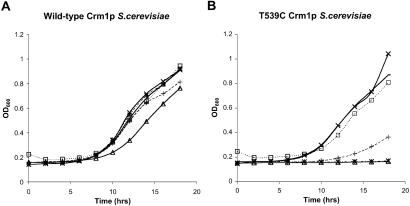
The experiments above suggested a direct interaction of PKF050-638 with CRM1, as is the case for LMB. To study this interaction, we compared the inhibitory effect of PKF050-638 with the effect of LMB on WT type Crm1p and the Crm1pT539C mutant in S. cerevisiae. It has been shown that in contrast to vertebrates and S. pombe, LMB is not toxic for S. cerevisiae (29). LMB interacts directly with S. pombe and human CRM1, whereas S. cerevisiae Crm1p has no apparent affinity for LMB (5, 20, 25). Crm1p is a highly conserved protein, and a single Thr to Cys change at position 539 (Thr-539→C) is sufficient to render S. cerevisiae Crm1p fully LMB susceptible, indicating that Crm1p is the only relevant target of LMB and suggesting an essential role for Cys-539 in Crm1p binding (25). We tested the effect of LMB, PKF050-638, and its enantiomer PKF050-637 on cell growth of WT S. cerevisiae (LMB resistant) and S. cerevisiae-expressing Crm1pT539C (LMB susceptible). Fig. 7 shows the growth curves of the WT S. cerevisiae (Fig. 7A) and the T539C Crm1p mutated S. cerevisiae (Fig. 7B) in the absence and presence of LMB or different concentrations of the Rev inhibitor PKF050-638 and its inactive enantiomer PKF050-637. At 240 and 48 μM, PKF050-638 inhibited the growth of the WT S. cerevisiae weakly, whereas it completely inhibited the growth of the Crm1pT539C mutant. In contrast, its enantiomer PKF050-637 did not affect the growth of either the WT or the mutated S. cerevisiae. At the concentration tested, LMB also inhibited the mutant Crm1pT539C yeast and not the WT. These results indicated that, similar to LMB, PKF050-638 interacts specifically with a CRM1 region proximate to the Cys at position 539.
Fig 7.
Analysis of the effect of PKF050-638 on WT S. cerevisiae (A) and a Crm1pT539C-expressing strain (B). Overnight cultures of the two strains were diluted until OD600 ≈ 0.1 and were grown at 30°C in the presence of 80 nM LMB (*), different concentrations PKF050-638 (▵, 240 μM; +, 48 μM) and PKF050-637 (□, 240 μM; −, 48 μM), or with no drug (×). The evolution of the optical density at 600 nm is plotted.
Discussion
Nuclear export of proteins containing a leucine-rich NES is mediated by a specific NES receptor known as CRM1 (5, 6, 30). This protein interacts directly with the NES in a RanGTP-dependent manner. The HIV Rev protein, a prototype NES-family member, facilitates the export of the viral RNAs by means of the CRM1 pathway. Rev binds to CRM1 through its NES. LMB has been identified as a Rev function inhibitor, acting as an inhibitor of the CRM1-mediated nuclear export (19).
We identified a low molecular weight compound, PKF050-638 (Fig. 1), as a specific inhibitor to the HIV-1 Rev function. This drug dose-dependently inhibits the Rev-dependent mRNA expression. PKF050-638 is active in the micromolar range with a selectivity index of 76, whereas LMB acts in the nanomolar range with a greatly varying toxicity, depending on the production lot. Although PKF050-638 displays cellular toxicity, a selective window of activity can be defined. Interestingly, its enantiomer, PKF050-637, is 50 times less active against Rev function (Table 1).
By using Rev-GFP fusion proteins, we could visualize Rev nuclear export and its inhibition by PKF050-638 (Fig. 2). This effect was clearly reversible in the majority (86%) of the cells, whereas the inhibitory effect of LMB could be reversed only in half of the cells (Fig. 4) within 16 h. The inhibitory effect was directed against the Rev NES, because the compound also promoted nuclear retention of a Tat-NES-GFP fusion protein carrying the Rev NES (Fig. 3).
These findings prompted us to investigate whether the compound disrupts Rev-CRM1 binding by monitoring the effect of PKF050-638 on the colocalization of CRM1 and Rev in human cells (27). One hour after addition of drug to the cells, the Rev-mediated nucleolar colocalization of CRM1 was completely disrupted, suggesting that PKF050-638 inhibits the interaction of Rev with CRM1.
To confirm this hypothesis, we tested complex disruption in a RanGTPase hydrolysis assay by using heterologous NES from mouse minute virus NS2 and PKI proteins. In vitro, both PKF050-638 and LMB disrupt CRM1-NS2-RanGTP and CRM1-PKI-RanGTP complex formation, whereas the enantiomer PKF050-637 does not. Inhibition of PKI complex by PKF050-638 is better than the inhibition of the NS2-RanGTP, probably because the NS2 peptide is binding more strongly to CRM1-RanGTP than PKI (21). In addition, the concentrations of drug needed to block NS2 or PKI are expected to be higher than the concentrations needed to inhibit the Rev-NES because of their stronger binding to CRM1 (18). The fact that this compound also interferes with the NS2 and PKI binding to CRM1 suggests that it is a general inhibitor of CRM1-mediated nuclear export.
To identify the site of interaction of PKF050-638 with CRM1, we tested its effect on the growth of S. cerevisiae carrying a threonine at position 539 of its CRM1 (this strain is resistant to LMB) and on an S. cerevisiae mutant carrying a cysteine at position 539 of its CRM1 (susceptible to the toxic effect of LMB). PKF050-638 showed a similar activity as LMB, although the S. cerevisiae strain was, to some extent, also inhibited. The enantiomer showed no activity in this assay. These data indicate that the specific and selective target of the new drug is CRM1. Our experiments indicate a direct interaction with CRM1 in the region containing Cys-539. Therefore, amino acid 539 is important for the effects of both LMB and PKF050-638 on CRM1.
In all experiments described here, the enantiomer PKF050-637 was much less active than PKF050-638. The fact that both structures have the same chemical properties but different conformation suggests that the conformation of PKF050-638 is optimal to fit in the NES-binding pocket of CRM1. The spatial conformation of the methylester group seems to be crucial for activity.
We showed that PKF050-638 acts similarly to LMB. The active concentrations are higher but still workable, and the selectivity window is sufficient for most experiments. Considering that PKF050-638 interferes with the NES-binding site of CRM1, a general nucleocytoplasmic export factor for proteins, it is not surprising that the compound exhibits cellular toxicity. This toxicity precludes its activity as an antiviral drug in clinical practice. Despite its toxicity, this new inhibitor is a useful tool in exploring CRM1-mediated nuclear export pathways. Because its enantiomer is inactive, there must be some structural preference for the NES-binding site of CRM1 which could be explored. Currently, only LMB is available for such research. Because its production is in Streptomyces cultures, the quality of the LMB lots is variable. PKF050-638 is a chemically synthesized compound; it should not suffer from these drawbacks. An additional attractive feature is that the effect of PKF050-638 is more readily reversible than the effect of LMB. PKF050-638 could, therefore, be used to study CRM1 interactions, export mechanisms, and import rates of proteins by using the CRM1-dependent export pathway. In addition, because of its reversible effect, PKF050-638 gives us the ability to study the export rates of these proteins.
The question arises whether the Rev-CRM1 interaction is a good target for anti-HIV therapy. The Rev-NES-binding site on CRM1 is also used by cellular leucine-rich NESs that make the interference on CRM1 very unspecific, with, as a result, the expected toxicity.
Currently reported Rev inhibitors mainly target the Rev-RRE interaction (31, 32). Interference with this interaction is expected to be more specific with less cellular toxicity than interaction with CRM1 because it is a strictly viral target. This approach, however, allows for viral escape through drug resistance. Low molecular weight inhibitors of Rev-RRE interaction have been identified, but they have limited success in inhibiting the viral replication. It can been argued that even partial inhibition of Rev function may dramatically affect the pathogenic potential of HIV. Rev-independent HIV or simian immunodeficiency virus molecular clones have been constructed and have been shown to be attenuated in animals. Rev function is an important determinant of HIV pathogenic potential, and partial Rev inhibition may contribute to virus control in vivo. Therefore, Rev should become a high priority candidate target for antiviral strategies. Attempts to screen for anti-Rev compounds have not been very successful, but the knowledge of the molecular interactions involved in Rev function may facilitate future strategies. PKF050-635 in a chemically synthesized small molecule Rev inhibitor, acting through blocking CRM1-mediated nuclear export, and is important, therefore, both in terms of Rev inhibition and in terms of unraveling the CRM1-mediated export complex.
Acknowledgments
We thank Christophe Pannecouque for helpful discussions. These investigations were supported in part by the Geconcerteerde Onderzoeksacties (GOA 00/12) van de Vlaamse Gemeenschap and the Vlaams Fonds voor Wetenschappelijk Onderzoek (FWO: G.0140.98). D.D. acknowledges a grant from the Flemish Institute supporting Scientific-Technological Research in Industry (IWT), and the Belgian American Educational Foundation (BAEF).
Abbreviations
RRE, Rev responsive element
NES, nuclear export signal
BFP, blue fluorescent protein
PKI, protein kinase inhibitor
LMB, leptomycin B
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kalland K. H., Szilvay, A. M., Brokstad, K. A., Saetrevik, W. & Haukenes, G. (1994) Mol. Cell. Biol. 14, 7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer B. E. & Malim, M. H. (1994) Genes Dev. 8, 1538-1547. [DOI] [PubMed] [Google Scholar]
- 3.Stauber R. H., Afonina, E., Gulnik, S., Erickson, J. & Pavlakis, G. N. (1998) Virology 251, 38-48. [DOI] [PubMed] [Google Scholar]
- 4.Szilvay A. M., Brokstad, K. A., Boe, S. O., Haukenes, G. & Kalland, K. H. (1997) Virology 235, 73-81. [DOI] [PubMed] [Google Scholar]
- 5.Fornerod M., Ohno, M., Yoshida, M. & Mattaj, I. W. (1997) Cell 90, 1051-1060. [DOI] [PubMed] [Google Scholar]
- 6.Fukuda M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M. & Nishida, E. (1997) Nature 390, 308-311. [DOI] [PubMed] [Google Scholar]
- 7.Stade K., Ford, C. S., Guthrie, C. & Weis, K. (1997) Cell 90, 1041-1050. [DOI] [PubMed] [Google Scholar]
- 8.Gorlich D., Dabrowski, M., Bischoff, F. R., Kutay, U., Bork, P., Hartmann, E., Prehn, S. & Izaurralde, E. (1997) J. Cell Biol. 138, 65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fornerod M., van Deursen, J., van Baal, S., Reynolds, A., Davis, D., Murti, K. G., Fransen, J. & Grosveld, G. (1997) EMBO J. 16, 807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattaj I. W. & Englmeier, L. (1998) Annu. Rev. Biochem. 67, 265-306. [DOI] [PubMed] [Google Scholar]
- 11.Dahlberg J. E. & Lund, E. (1998) Curr. Opin. Cell Biol. 10, 400-408. [DOI] [PubMed] [Google Scholar]
- 12.Kutay U., Bischoff, F. R., Kostka, S., Kraft, R. & Gorlich, D. (1997) Cell 90, 1061-1071. [DOI] [PubMed] [Google Scholar]
- 13.Kehlenbach R. H., Dickmanns, A., Kehlenbach, A., Guan, T. & Gerace, L. (1999) J. Cell Biol. 145, 645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorlich D. (1998) EMBO J. 17, 2721-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribbeck K., Lipowsky, G., Kent, H. M., Stewart, M. & Gorlich, D. (1998) EMBO J. 17, 6587-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith A., Brownawell, A. & Macara, I. G. (1998) Curr. Biol. 8, 1403-1406. [DOI] [PubMed] [Google Scholar]
- 17.Askjaer P., Jensen, T. H., Nilsson, J., Englmeier, L. & Kjems, J. (1998) J. Biol. Chem. 273, 33414-33422. [DOI] [PubMed] [Google Scholar]
- 18.Askjaer P., Bachi, A., Wilm, M., Bischoff, F. R., Weeks, D. L., Ogniewski, V., Ohno, M., Niehrs, C., Kjems, J., Mattaj, I. W. & Fornerod, M. (1999) Mol. Cell. Biol. 19, 6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolff B., Sanglier, J. J. & Wang, Y. (1997) Chem. Biol. 4, 139-147. [DOI] [PubMed] [Google Scholar]
- 20.Kudo N., Matsumori, N., Taoka, H., Fujiwara, D., Schreiner, E. P., Wolff, B., Yoshida, M. & Horinouchi, S. (1999) Proc. Natl. Acad. Sci. USA 96, 9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishi K., Yoshida, M., Fujiwara, D., Nishikawa, M., Horinouchi, S. & Beppu, T. (1994) J. Biol. Chem. 269, 6320-6324. [PubMed] [Google Scholar]
- 22.Stauber R. H., Horie, K., Carney, P., Hudson, E. A., Tarasova, N. I., Gaitanaris, G. A. & Pavlakis, G. N. (1998) BioTechniques 24, 462-471. [DOI] [PubMed] [Google Scholar]
- 23.D'Agostino D. M., Ciminale, V., Pavlakis, G. N. & Chieco-Bianchi, L. (1995) AIDS Res. Hum. Retroviruses 11, 1063-1071. [DOI] [PubMed] [Google Scholar]
- 24.Stauber R. H. & Pavlakis, G. N. (1998) Virology 252, 126-136. [DOI] [PubMed] [Google Scholar]
- 25.Neville M. & Rosbash, M. (1999) EMBO J. 18, 3746-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kudo N., Khochbin, S., Nishi, K., Kitano, K., Yanagida, M., Yoshida, M. & Horinouchi, S. (1997) J. Biol. Chem. 272, 29742-29751. [DOI] [PubMed] [Google Scholar]
- 27.Zolotukhin A. S. & Felber, B. K. (1999) J. Virol. 73, 120-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bischoff F. R. & Ponstingl, H. (1995) Methods Enzymol. 257, 135-144. [DOI] [PubMed] [Google Scholar]
- 29.Hamamoto T., Gunji, S., Tsuji, H. & Beppu, T. (1983) J. Antibiot. 36, 639-645. [DOI] [PubMed] [Google Scholar]
- 30.Ossareh-Nazari B., Bachelerie, F. & Dargemont, C. (1997) Science 278, 141-144. [DOI] [PubMed] [Google Scholar]
- 31.Zapp M. L., Young, D. W., Kumar, A., Singh, R., Boykin, D. W., Wilson, W. D. & Green, M. R. (1997) Bioorg. Med. Chem. 5, 1149-1155. [DOI] [PubMed] [Google Scholar]
- 32.Ratmeyer L., Zapp, M. L., Green, M. R., Vinayak, R., Kumar, A., Boykin, D. W. & Wilson, W. D. (1996) Biochemistry 35, 13689-13696. [DOI] [PubMed] [Google Scholar]