Abstract
RIMs are presynaptic active zone proteins that regulate neurotransmitter release. We describe two related genes that encode proteins with identical C-terminal sequences that bind to the conserved PDZ domain of RIMs via an unusual PDZ-binding motif. These proteins were previously reported separately as ELKS, Rab6-interacting protein 2, and CAST, leading us to refer to them by the acronym ERC. Alternative splicing of the C terminus of ERC1 generates a longer ERC1a variant that does not bind to RIMs and a shorter ERC1b variant that binds to RIMs, whereas the C terminus of ERC2 is synthesized only in a single RIM-binding variant. ERC1a is expressed ubiquitously as a cytosolic protein outside of brain; ERC1b is detectable only in brain, where it is both a cytosolic protein and an insoluble active zone component; and ERC2 is brain-specific but exclusively localized to active zones. Only brain-specific ERCs bind to RIMs, but both ubiquitous and brain-specific ERCs bind to Rab6, a GTP-binding protein involved in membrane traffic at the Golgi complex. ERC1a and ERC1b/2 likely perform similar functions at distinct localizations, indicating unexpected connections between nonneuronal membrane traffic at the Golgi complex executed via Rab6 and neuronal membrane traffic at the active zone executed via RIMs.
RIM1α and -2α are multidomain adaptor proteins that were discovered as putative effectors for Rab3, a synaptic vesicle protein that binds GTP and regulates neurotransmitter release (1–4). RIMs are composed of an N-terminal Zn2+-finger domain, a central PDZ domain, and C-terminal C2A and C2B domains (3, 4). The binding of RIMs to Rab3 and the localization of RIM1α to presynaptic active zones suggested a function in neurotransmitter release, which was confirmed by genetic experiments in Caenorhabditis elegans and mice (5–7). Deletion of RIM1α in mice caused distinct phenotypes in different types of synapses. In excitatory synapses capable of N-methyl-D-aspartate (NMDA)-dependent long-term potentiation (e.g., Schaffer collateral/commissural fiber synapses in the CA1 region of the hippocampus) and in inhibitory synapses, deletion of RIM1α decreased the neurotransmitter release probability in response to Ca2+ influx, leading to a decline in synaptic transmission and changes in short-term synaptic plasticity (6). In excitatory synapses that lack NMDA-dependent long-term potentiations but experience protein kinase A-dependent long-term potentiation (e.g., mossy fiber synapses in the CA3 region of the hippocampus), deletion of RIM1α had no detectable effect on acute neurotransmitter release but prevented protein kinase A-dependent potentiation (7). Although these results confirmed an important role for RIM1α and, by extension, RIM2α, in presynaptic function, the spectrum of mutant phenotypes suggests that this role is incompletely understood.
It is likely that RIMs regulate release by interacting with other proteins. Several binding partners were identified. The N-terminal Zn2+-finger domain of RIM1α directly binds to Rab3 [which led to the initial discovery of RIMs (3, 4)] and to the N terminus of Munc13–1, another active zone protein that functions in neurotransmitter release (6, 8, 9). A proline-rich sequence in the linker between the C2A and C2B domain interacts with the SH3 domain of brain-specific adaptor proteins called RIM-BPs (4) that have been implicated in Ca2+-channel regulation (10). The C-terminal C2B domain binds to α-liprins (6), putative adaptor proteins of the active zone that were shown in invertebrates to be essential for regulating active zone size (11–13), and to synaptotagmin 1, which in turn regulates release (6, 14). RIMs thus behave like a molecular scaffold that tethers multiple synaptic proteins. However, no binding proteins for the central PDZ domain of RIMs, their evolutionarily most conserved domain, were identified.
In the present study, we characterize a family of active zone proteins whose binding to RIMs is regulated by tissue-specific alternative splicing. These proteins, here tentatively referred to by the acronym ERC, based on independent previous namings as ELKS (15), Rab6-interacting protein 2 (16), and CAST (17), are components of active zones in neurons but appear to be involved in general intracellular membrane traffic in all cells, indicating an unexpected connection between the active zone and nonspecialized cellular trafficking pathways.
Methods
Plasmids.
Yeast two-hybrid bait vectors: pLexNRim1-PDZ (residues 492–772 in the SmaI site); pLexNRim2-PDZ (residues 548–781 in the SmaI site). ERC1b prey clones: pPreyPDZ-16 (residues 578–947), pPreyPDZ-41 (579–947), pPreyPDZ-94 (606–947), pPreyPDZ-54 (780–947), pPreyPDZ-24 (807–947), pPreyPDZ-37 (911–947), pPreyPDZ-94–3 (606–944), pPreyPDZ-9 (939–947); pPreyERC-1 to pPreyERC-12 = point mutants of pPreyPDZ-9 (Table 2). Mammalian expression vectors (in pCMV5) pCMV5 ERC1b and pCMV5ERC2: full-length rat ERC1b/2, respectively (in EcoRI/BamHI sites). Bacterial expression vectors: pMalC2-ERC-16 (residues 578–947 of ERC1b in SalI site); pGexERC1(residue 117–425); pGex-Rim1-PDZ (residues 492–772 in the SmaI site); pGexRab6 (full-length Rab6a in BamHI/NcoI sites).
Table 2.
Sequence specificity of the interaction of the RIM PDZ domains with the C-terminal 9 residues of ERC1b
| Sequence | Position of mutant | RIM1 PDZ domain | RIM2 PDZ domain |
|---|---|---|---|
| ERC1b | NA | ++ | ++ |
| QDEEEGIWA* | WT | ++ | + |
| QDEEEGIWD* | 0 | − | − |
| QDEEEGIWL* | 0 | + | − |
| QDEEEGIDA* | −1 | − | − |
| QDEEEGIAA* | −1 | − | − |
| QDEEEGDWA* | −2 | − | − |
| QDEEEGAWA* | −2 | − | − |
| QDEEEDIWA* | −3 | − | − |
| QDEEEAIWA* | −3 | − | − |
| QDEEKGIWA* | −4 | ++ | + |
| QDEEAGIWA* | −4 | ++ | + |
| QDEKEGIWA* | −5 | ++ | + |
| QDEAEGIWA* | −5 | ++ | + |
Interactions were measured by yeast two-hybrid by using survival on selection plates and β-galactosidase activation as criteria. The RIM PDZ domains were supplied as bait vectors in pLexN, and the C-terminal sequences of ERC1b as prey vectors in pVP16 containing either the C-terminal half of ERC1b or only the nine residues shown. Interactions were scored as strong (++), moderate (+), and absent (−), depending on the relative strength of color development and survival of colonies. Asterisk denotes a stop codon. NA, not applicable.
Yeast two-hybrid screening and interaction assays were carried out with pLexN bait vectors in L40 yeast essentially as described (3, 4) by using β-galactosidase filter assays for confirmation.
Antibodies.
Most antibodies were described (4, 6, 18). Polyclonal sera to ERCs were raised in rabbits to the protein encoded by pMalC2-ERC-16 (serum P224) and to synthetic peptides coupled to keyhole limpet hemocyanin via an N-terminal cysteine residue (19): serum 4790 (ERC1b/2) to peptide CDQDEEEGIWA; serum 4791 (ERC1a) to peptide CDILEQVVNALESS; and serum U5004 (ERC2) to peptide CDIEDDSRMNPEFADRLK. Affinity purifications were performed as described (3).
cDNA cloning was performed by standard procedures (4). Sequences were analyzed by using the National Center for Biotechnology Information program suite and public databases in addition to the Celera sequence databases with standard blast programs (20).
Tissue Fractionations.
Mouse embryos were obtained after timed matings at the indicated ages. Various tissues were harvested from embryonic and adult wild-type and RIM1α knockout mice. Brain subcellular fractionations to isolate synaptic vesicles and synaptic plasma membranes were carried out as described (21). Equivalent protein amounts of each fraction were analyzed by immunoblotting.
Immunocytochemistry.
Double immunofluorescence labeling of cultured rat hippocampal neurons (12 days in vitro) was carried out essentially as described (22) by using affinity-purified ERC1b antibodies (p224) and monoclonal antibodies to RIM1α or synapsin, with secondary antibodies coupled with Alexa Fluor 488 and 546 (Molecular Probes). Immunoelectron microscopy of brain sections was performed by using a preembedding 1.4-nm immunogold procedure with signal amplification with the HQ silver enhancement kit (Nanoprobes, Yaphank, NY) essentially as described (23).
Miscellaneous Procedures.
SDS/PAGE and immunoblotting analyses were executed by using standard procedures (24, 25). GST fusion protein affinity chromatography was performed essentially as described (4) by using total brain extracts from frozen rat brains (obtained from Pel-Freeze Biologicals) or solubilized proteins from transfected COS cells.
Results
Identification of Proteins Binding to the RIM PDZ Domain.
We screened a rat-brain cDNA library by yeast two-hybrid selection with a bait encoding the RIM1α PDZ domain (3). Of 46 prey clones sequenced, 16 independent clones encoded overlapping sequences from the C terminus of a single protein referred to as ERC1b (see below). To test the validity of the yeast two-hybrid interaction, we raised an antibody to a recombinant ERC1b fusion protein and affinity-purified the antibody on the immobilized antigen. Immunoblots of rat-brain proteins revealed that this antibody reacted with a single ≈125-kDa protein, which was largely but not completely insoluble (Fig. 1A). We then used pulldown experiments to determine whether rat-brain ERC1b binds to the RIM1α PDZ domain, as suggested by the yeast two-hybrid interaction. Efficient capture of brain ERC1b by a GST fusion protein containing the RIM1α PDZ domain, but not by a control GST protein, was observed (Fig. 1B). The reverse pulldown could not be performed with brain homogenates, because RIMs are completely insoluble (3, 4). We therefore expressed a fragment of RIM1α, including the PDZ domain by transfection in COS cells. Pulldowns of the recombinant RIM PDZ domain protein with maltose-binding protein (MBP) fusion protein of one of the ERC1b prey clones (pPreyPDZ-16), but not with MBP alone, confirmed the yeast two-hybrid interaction (Fig. 1C).
Fig 1.
Binding of ERC1b to RIM1 PDZ-domain. (A) Immunoblot analysis of rat brain proteins with affinity-purified ERC1b antibodies to the prey clone pPreyPDZ-16 that was isolated by yeast two-hybrid screening with the RIM1α PDZ domain. Total homogenates, supernatant, and pellet obtained after high-speed centrifugation were tested. (B) Pulldown of soluble brain ERC1b with a GST-RIM1α PDZ domain fusion protein but not with GST alone. (C) Pulldown of recombinant RIM PDZ domain expressed in transfected COS cells with an ERC1b maltose-binding protein fusion protein.
Sequence Analyses of ERC Proteins.
Databank searches with the sequence of the yeast two-hybrid prey clones revealed that it was closely related to two proteins encoded by the random human cDNAs KIAA0378 and KIAA1081 (26). These two proteins were separately described previously as ELKS (15), Rab6-interacting protein 2 (Rab6IP2) (16), and CAST (17), leading us to refer to the two proteins by the acronyms ERC1 and ERC2. ELKS (which corresponds to ERC2) is an anonymous gene fused to RET tyrosine kinase in thyroid carcinomas with a chromosomal translocation (15). As revealed by its name, Rab6-interacting protein 2 (which corresponds to ERC1) was identified in yeast two-hybrid screens as a second interacting protein for the GTPase Rab6 that primarily functions in the Golgi complex (16, 27). CAST (which corresponds to ERC2) was described as an active zone protein that binds to RIM1α (17).
We cloned full-length rat cDNAs for ERC1/2 and compared their sequences to those of human and mouse ERCs compiled from EST and genome sequences (Fig. 2A and data not shown). ERC1/2 sequences are conserved in human, mouse, and rat (99.6% and 98.8% identity for ERC1/2, respectively, in all three species) and are similar to each other (71% identity in the nonalternatively spliced human sequences). As noted previously (15–17), the majority of the ERC sequences are occupied by coiled-coil regions with no specific homology to other proteins. A single ERC homolog was detected in C. elegans (F42A6.9; GenBank accession no. NM_067928) that resembles ERCs weakly throughout the protein but exhibits patches of strong similarity, most notably at the C terminus, which is identical to that of vertebrate ERC1b/2.
Fig 2.
Structure of ERC1a, -b, and -2. (A) Alignment of the human sequences. Identical residues are highlighted, and similar residues are boxed. Alternatively spliced regions identified as variable sequences in rat, mouse, or human cDNA or EST sequences are marked by “ = ” on the top of the sequences. (B) Structure of the 3′ end of the human ERC1 gene to illustrate mechanism of alternative splicing that creates ERC1a and -b (see supporting information).
Gene Structures and Alternative Splicing of ERCs.
The human and mouse genes encoding ERCs are inordinately large (Table 1). For example, ERC2 contains only 955 residues but is encoded by a 0.75-megabase gene with four introns of >100 kilobases (see Tables 3–6, which are published as supporting information on the PNAS web site, www.pnas.org). Analysis of the ERC gene structures uncovered an unusual organization in that most of the exons are in-frame (see supporting information on the PNAS web site and ref. 28), suggesting that ERCs could be alternatively spliced, which was confirmed by databank searches and cDNA cloning (Fig. 2A). One short homologous sequence (“SLTS” at position 775 in ERC1) is variably inserted in both ERC1 and -2. Two separate interior sequences are alternatively spliced in either ERC1 or -2; these are highly conserved in mouse, rat, and human ERC1 or -2. Genome analyses detected these alternatively spliced regions only in either ERC1 or -2, indicating that they diverge among ERCs. Possibly most importantly, the C terminus of ERC1 but not ERC2 is synthesized in two splice variants that encode a longer sequence (ERC1a) without homologies in GenBank, and a shorter sequence (ERC1b) that is almost identical to ERC2 (Fig. 2A). Analyses of the human and mouse genome indicated that these C termini are encoded by distinct conserved exons that are separated by large introns (Fig. 2B).
Table 1.
Characteristics of ERC genes
| Species
|
ERC1 | ERC2 | ||
|---|---|---|---|---|
| Chromosome | Size, kb | Chromosome | Size, kb | |
| Human | 12p13 | 464.1 | 3p12 | 755.0 |
| Mouse | 6E3 | 252.1 | 14A3 | 671.8 |
Genes were analyzed in the genome sequences deposited in the public databanks of the National Center for Biotechnology Information and in the proprietary Celera databanks. For a precise description of the genes, including exon–intron junctions, see Tables 3 and 4.
The RIM PDZ Domains Recognize a Novel Sequence Motif in ERCs.
In the yeast two-hybrid screens, we isolated ERC1b because it binds to the RIM1α PDZ domain, whereas ERC2 as CAST was independently described as a RIM1α-binding protein (17). Because all ERC1 prey clones isolated in our yeast two-hybrid screens included the ERC1b C terminus that is identically found in ERC2 (Fig. 2A), the C termini of ERC1b/2 may directly bind to the PDZ domain of RIMs. This hypothesis was confirmed in yeast two-hybrid assays. Deleting the last three residues from ERC1b abolished its interaction with the RIM1 PDZ domain, whereas prey clones expressing only the last nine residues of ERC1b capably interacted with the PDZ domains of RIM1α/2 (Table 2 and data not shown).
To determine which C-terminal residues of ERCs are essential for binding to the RIM PDZ domains, we mutagenized each of the last six residues of ERC1b/2 and tested binding to RIM PDZ domains (Table 2). The last four residues (positions P0–P−3) but not the preceding residues (positions P−4 and P−5) were found to be critical for all interactions (Table 2). The sequence requirement for binding to the RIM PDZ domains appears to be more restricted than for other PDZ domains, because even conservative substitutions (e.g., exchanging alanine at P0 for isoleucine) abolished binding (Table 2). In contrast, in classical PDZ domains such as those of PSD-95, similar substitutions are tolerated (reviewed in refs. 29 and 30). Different from RIM binding, which is specific for ERC1b as opposed to ERC1a, but consistent with previous reports (16), both ERC1 splice variants were found to bind to Rab6 (data not shown).
Tissue Distribution of ERCs.
The original ERC1b antibody (Fig. 1) was generated against a conserved region, suggesting that it crossreacts with ERC1a and possibly ERC2. To study the biochemical properties of the various ERC forms, we raised three subtype-specific antibodies. These antibodies were directed to synthetic peptides from the alternatively spliced ERC1a-specific C-terminal sequence, the common C-terminal sequence of ERC1b/2, and a unique ERC2-specific sequence.
The original ERC1b antibody detects ERC proteins in all rat tissues tested, with a slightly smaller size in brain (≈120 kDa) than in peripheral tissues (≈125 kDa; Fig. 3A). In contrast, the ERC1a-specific antibody reacts only with ERCs in peripheral tissues, whereas the ERC1b/2-specific and the ERC2-specific antibodies label ERCs only in brain. Thus the longer alternatively spliced ERC1a is exclusively expressed outside of brain, whereas the shorter ERC1b and the similar ERC2 (Fig. 2) are restricted to brain (Fig. 3A). The ubiquitous ERC1a was observed in development at the earliest time tested (embryonic day 8.5; Fig. 3B), consistent with its widespread presence in adult tissues (Fig. 3A). By contrast, the brain-specific ERC1b/2 became detectable only when other synaptic proteins, such as synaptotagmin 1, were synthesized (Fig. 3B). The differences in expression patterns between ERC1a vs. ERC1b/2 are also reflected in their subcellular localization in that the ubiquitous ERC1a was found to be largely cytosolic (data not shown), whereas the brain ERC1b/2 were largely particulate (see below).
Fig 3.
Tissue distribution and developmental expression of ERCs analyzed by immunoblotting with subtype-specific antibodies. (A) Proteins from the indicated rat tissues were probed with four ERC antibodies and a vasolin-containing protein control antibody. Antibodies used were: ERC1b (top section), raised against the conserved domain of ERC1b (Fig. 2); ERC1a (second section), raised against a synthetic peptide from the ERC1a-specific C terminus; ERC1b/2 (third section), raised against a peptide from the common C terminus of ERC1b and ERC2; and ERC2 (fourth section), raised against an internal ERC2-specific peptide. Numbers on the left indicate positions of molecular mass markers. (B) Expression of ERC1a and -1b/2 in mouse embryos at different stages of gestation examined with subtype-specific antibodies. Protein loads were normalized for samples from whole embryos (embryonic day E8.5–E12.5) or embryo heads (E14.5–E18.5) using the levels of VCP determined by immunoblotting. The low molecular mass band in the E8.5 and E9.5 samples (asterisk) is due to crossreactivity with the synaptotagmin 1 antibody (Syt 1) used as a positive control for a synaptic protein.
Overlapping but Nonidentical Localizations of ERC1b/2 in Brain.
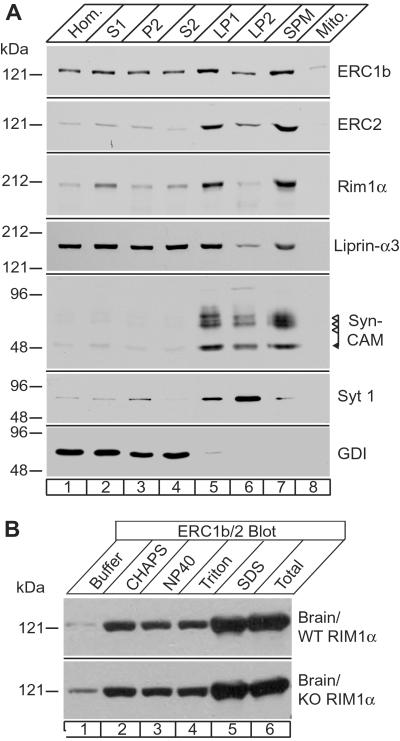
The insolubility and detergent resistance of ERCs in brain resembles the behavior of RIMs to which ERC1b/2 bind. To test whether both ERC isoforms are colocalized with RIMs in presynaptic plasma membranes, we performed subcellular fractionations (Fig. 4A). Immunoblotting of various fractions with ERC1b/2- and ERC2-specific antibodies revealed that both proteins were coenriched in synaptic plasma membranes with RIM1α and with SynCAM, a synaptic cell adhesion molecule (18). However, the ERC1b/2 antibodies detected an appreciable pool of soluble protein, whereas the ERC2-specific antibodies detected only insoluble ERC2 (Fig. 4A). Because the ERC1b/2 antibodies recognize both forms but the ERC2 antibody only ERC2, the soluble ERC pool observed with the former likely corresponds to ERC1b. α-Liprins, active zone proteins that, like ERC1, are also widely distributed in many cell types outside of active zones (11) and also bind to RIMs (6), exhibited a distribution very similar to ERC1b (Fig. 4A). These results suggest that ERC1b and α-liprins are present in a soluble cytosolic and an insoluble active zone form, whereas ERC2 is exclusively localized to active zones, as suggested by Ohtsuka et al. (17).
Fig 4.
Subcellular distribution of ERCs in brain. (A) Rat brain homogenate (Hom.; lane 1) was used to prepare a low-speed supernatant (S1; lane 2), which was separated into crude synaptosomes (P2; lane 3) and synaptosomal supernatant (S2; lane 4). Synaptosomes were lysed and subjected to sequential low- and high-speed centrifugations to yield crude synaptosomal membranes (LP1; lane 5) and synaptic vesicles (LP2). LP1 was used to isolate synaptic plasma membranes (SPM; lane 7) and mitochondria (Mito.; lane 8) by centrifugation on a sucrose step gradient (21). Fractions were analyzed by immunoblotting for the proteins indicated on the right (GDI, GDP-dissociation inhibitor). Numbers on the left indicate positions of molecular mass markers. (B) Brain homogenates from wild-type (WT RIM1) and RIM1α knockout mice (KO RIM1) were treated with buffer alone or with 1% of the indicated detergents, centrifuged at 100,000 × g, and the supernatants were analyzed by immunoblotting with the ERC1b/2 antibody. Numbers on the left indicate positions of molecular mass markers.
That RIMs bind ERCs and that both are active zone components raises the possibility that RIM-binding recruits ERCs to active zones like Munc13–1 (6). To test this hypothesis, we examined whether the abundance and solubility of ERCs are altered when RIM1α, the major RIM isoform in forebrain, is deleted. We homogenized brains from wild-type and RIM1α knockout mice in various detergent buffers and determined the amount of soluble ERCs in the supernatant after high-speed centrifugation (Fig. 4B). In wild-type brains, the majority of ERC1b/2 was insoluble with nondenaturing detergents but was solubilized by SDS. No major change in the abundance or solubility of ERCs was observed in RIM1α knockout mice, indicating that ERCs are not simply immobilized by RIMs at the active zone.
Synaptic Localization of ERC.
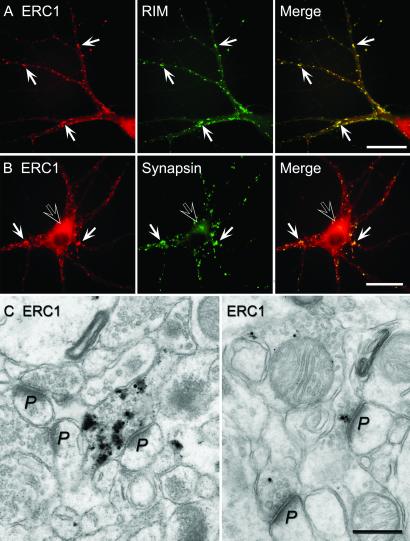
To confirm the dual synaptic and cytoplasmic localization of ERC1b in neurons suggested by the subcellular fractionation (Fig. 4A), we double-labeled cultured hippocampal neurons with affinity-purified ERC1b antibodies and antibodies to RIM1α or to synapsin (Fig. 5 A and B). ERC1b was detected in synapses colocalized with RIM1α and synapsin and in the neuronal cell body distinct from RIM1α and synapsin. To confirm that ERC1b is present in presynaptic nerve terminals, we performed immunoelectron microscopy (Fig. 5C). Strong labeling of nerve terminals was observed throughout the mouse hippocampus. Not all terminals were labeled, probably because of limited antibody penetration, because parallel staining experiments for synaptophysin, a synaptic vesicle protein present in all nerve terminals (31), exhibited a similarly restricted distribution of immunoreactivity (data not shown).
Fig 5.
Immunolocalization of ERC1b in cultured neurons and brain sections. (A and B) Double immunofluorescence labeling of cultured neurons with affinity-purified ERC1b antibodies and monoclonal antibodies to RIM (A) or to synapsin (B). (Bar = 30 μm.) (C) Immunolabeling of rat brain sections from hippocampus by immunoelectron microscopy using pre-embedding labeling with silver enhancement. (Bar = 300 nm.)
Discussion
The presynaptic active zone is an electron-dense biochemically insoluble structure that serves to integrate synaptic vesicle exo- and endocytosis with intracellular signaling in the nerve terminal (reviewed in ref. 32). Although several active zone proteins have been described (e.g., Munc13s, Liprins, Bassoon, and Piccolo in addition to RIMs; reviewed in ref. 32), the molecular composition of active zones and the function of their components remain unclear. RIMs are evolutionarily conserved active zone proteins that are composed of multiple independently folded domains (3, 4). Genetic studies in mice and worms showed that RIMs regulate active zone function (5–7), but their precise action is incompletely understood (33). Important binding partners for the N-terminal Zn2+-finger domain [Rab3 and Munc13–1 (3, 4, 6, 8, 9)] and the C-terminal C2B domain of RIMs [α-liprins and synaptotagmin 1 (6, 14)] were isolated, but the significance of their highly conserved central PDZ domains is unclear. In the present study, we have characterized a family of proteins that bind to these PDZ domains, and that we refer to here by the acronym “ERC” based on previous separate namings of members of this protein family, in the order in which they were described: ELKS, which is involved in a chromosomal translocation (15); Rab6IP2, which interacts with rab6 (16); and CAST, which is localized to the active zone (17).
ERCs are largely composed of a “pioneer” sequence of ≈1,000 residues that is predicted to form coiled coils. This sequence is highly conserved between ERC1/2 but interrupted by short subtype-specific alternatively spliced regions (Fig. 2). In addition, alternative splicing at the C terminus of ERC1 but not -2 creates two variants, referred to as ERC1a/b, that are expressed in a strictly tissue-specific distribution (Fig. 3). ERC2 is a brain-specific protein similar to ERC1b, although its mRNA appears to be ubiquitously expressed (15). ERC1b/2 contain identical C termini that bind to the PDZ domains of RIM1α/2 (Table 2). The binding sequence is highly specific; even conservative substitutions abolish the interaction, suggesting that the RIM PDZ domains do not belong to the traditional three classes of PDZ domains (29, 30). Consistent with the binding of RIMs to ERC1b/2, both ERCs appear to be components of presynaptic active zones (Figs. 4 and 5; ref. 17). However, two independent lines of evidence demonstrate that the localization of ERCs to active zones in brain is not mediated by their interaction with RIMs. First, although ERC1b/2 interact with RIMs via identical C-terminal sequences, ERC1b is partly soluble, whereas ERC2 is not (Fig. 4A). Second, in RIM1α knockouts the abundance and solubility of ERCs are unchanged. In this respect, ERCs are similar to α-liprins but differ from Munc13–1, which is severely impaired in RIM1α knockouts (6).
The three principal forms of ERCs, ERC1a, -b, and -2, exhibit a spectrum of subcellular distributions and expression patterns despite their strong sequence similarity. ERC1a is found only outside of brain as a largely soluble protein, whereas ERC2 is restricted to brain as an insoluble protein, and ERC1b is brain-specific and both partly insoluble and synaptic, and partly soluble and cytoplasmic. These data suggest that ERCs have at least two principal functions, one that is ubiquitously executed in the cytosol and one that is synapse-specific and associated with the active zone. A possible clue to the ubiquitous function of ERCs was derived in the original description of ERC1a/b as a rab6-interacting protein (16), which was confirmed here (data not shown), suggesting a role in Rab-dependent membrane traffic. This is not the first instance of an active zone protein that has an additional role in a ubiquitous process, because α-liprins, which also bind to RIMs but are not regulated by alternative splicing, also do this (6, 12). At least part of the active zone thus appears to be built by reusing existing materials in the cell, α-liprins, and ERCs, which are presumably recruited by active zone-specific adaptor proteins like RIMs. The question arises whether the ubiquitous and active zone-specific functions of ERCs are connected. Such a connection is supported by the assembly of active zone components from precursor vesicles (34, 35) that may be generated in the Golgi complex by a Rab6-dependent process, which in turn could require ERC binding to Rab6. Furthermore, at least in neurons Rab6 may participate in a post-Golgi trafficking function (36), which could involve an interaction with ERCs.
Supplementary Material
Acknowledgments
We thank Dr. A. Ho for providing protein samples from embryonic mice, Drs. B. Goud and S. Monier (Institut Curie, Paris) for providing mouse Rab6-interacting protein/ERC1 cDNA clones, and Ms. I. Kornblum and E. Borowicz (Howard Hughes Medical Institute, Dallas) for excellent technical assistance.
References
- 1.Geppert M., Bolshakov, V. Y., Siegelbaum, S. A., Takei, K., De Camilli, P., Hammer, R. E. & Südhof, T. C. (1994) Nature 369, 493-497. [DOI] [PubMed] [Google Scholar]
- 2.Geppert M., Goda, Y., Stevens, C. F. & Südhof, T. C. (1997) Nature 387, 810-814. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., Okamoto, M., Schmitz, F., Hofman, K. & Südhof, T. C. (1997) Nature 388, 593-598. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Sugita, S. & Südhof, T. C. (2000) J. Biol. Chem. 275, 20033-20044. [DOI] [PubMed] [Google Scholar]
- 5.Koushika S. P., Richmond, J. E., Hadwiger, G., Weimer, R. M., Jorgensen, E. M. & Nonet, M. L. (2001) Nat. Neurosci. 4, 997-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoch S., Castillo, P. E., Jo, T., Mukherjee, K., Geppert, M., Wang, Y., Schmitz, F., Malenka, R. C. & Südhof, T. C. (2002) Nature 415, 321-326. [DOI] [PubMed] [Google Scholar]
- 7.Castillo P. E., Schoch, S., Schmitz, F., Südhof, T. C. & Malenka, R. C. (2002) Nature 415, 327-330. [DOI] [PubMed] [Google Scholar]
- 8.Betz A., Thakur, P., Junge, H. J., Ashery, U., Rhee, J. S., Scheuss, V., Rosenmund, C., Rettig, J. & Brose, N. (2001) Neuron 30, 183-196. [DOI] [PubMed] [Google Scholar]
- 9.Wang X., Hu, B., Zimmermann, B. & Kilimann, M. W. (2001) J. Biol. Chem. 276, 32480-32488. [DOI] [PubMed] [Google Scholar]
- 10.Hibino H., Pironkova, R., Onwumere, O., Vologodskaia, M., Hudspeth, A. J. & Lesage, F. (2002) Neuron 34, 411-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serra-Pages C., Medley, Q. G., Tang, M., Hart, A. & Streuli, M. (1998) J. Biol. Chem. 273, 15611-15620. [DOI] [PubMed] [Google Scholar]
- 12.Zhen M. & Jin, Y. (1999) Nature 401, 371-375. [DOI] [PubMed] [Google Scholar]
- 13.Kaufmann N., DeProto, J., Ranjan, R., Wan, H. & Van Vactor, D. (2002) Neuron 34, 27-38. [DOI] [PubMed] [Google Scholar]
- 14.Coppola T., Magnin-Luthi, S., Perret-Menoud, V., Gattesco, S., Schiavo, G. & Regazzi, R. (2001) J. Biol. Chem. 276, 32756-32762. [DOI] [PubMed] [Google Scholar]
- 15.Nakata T., Kitamura, Y., Shimizu, K., Tanaka, S., Fujimori, M., Yokoyama, S., Ito, K. & Emi, M. (1999) Genes Chromosomes Cancer 25, 97-103. [DOI] [PubMed] [Google Scholar]
- 16.Monier S., Jollivet, F., Janoueix-Lerosey, I., Johannes, L. & Goud, B. (2002) Traffic. 3, 289-297. [DOI] [PubMed] [Google Scholar]
- 17.Ohtsuka T., Takao-Rikitsu, E., Inoue, E., Inoue, M., Takeuchi, M., Matsubara, K., Deguchi-Tawarada, M., Satoh, K., Morimoto, K., Nakanishi, H., et al. (2000) J. Cell Biol. 158, 577-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biederer T., Sara, Y., Mozhayeva, M., Atasoy, D., Liu, X., Kavalali, E. T. & Südhof, T. C. (2002) Science 297, 1525-1531. [DOI] [PubMed] [Google Scholar]
- 19.Johnston P. A., Jahn, R. & Südhof, T. C. (1989) J. Biol. Chem. 264, 1268-1273. [PubMed] [Google Scholar]
- 20.Tabuchi K. & Südhof, T. C. (2002) Genomics 79, 849-859. [DOI] [PubMed] [Google Scholar]
- 21.Butz S., Fernandez-Chacon, R., Schmitz, F., Jahn, R. & Südhof, T. C. (1999) J. Biol. Chem. 274, 18290-18296. [DOI] [PubMed] [Google Scholar]
- 22.Biederer T., Cao, X., Südhof, T. C. & Liu, X. (2002) J. Neurosci. 22, 7340-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugita S., Han, W., Butz, S., Liu, X., Fernandez-Chacon, R., Lao, Y. & Südhof, T. C. (2001) Neuron 30, 459-473. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U. K. (1970) Nature 277, 680-685. [DOI] [PubMed] [Google Scholar]
- 25.Towbin H., Staehelin, T. & Gordon, J. (1979) Proc. Natl. Acad. Sci. USA 76, 4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kikuno R., Nagase, T., Waki, M. & Ohara, O. (2002) Nucleic Acids Res. 30, 66-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janoueix-Lerosey I., Jollivet, F., Camonis, J., Marche, P. N. & Goud, B. (1995) J. Biol. Chem. 270, 14801-14808. [DOI] [PubMed] [Google Scholar]
- 28.Yokota T., Nakata, T., Minami, S., Inazawa, J. & Emi, M. (2000) J. Hum. Genet. 45, 6-11. [DOI] [PubMed] [Google Scholar]
- 29.Harris B. Z. & Lim, W. A. (2001) J. Cell Sci. 114, 3219-3231. [DOI] [PubMed] [Google Scholar]
- 30.Hung A. Y. & Sheng, M. (2002) J. Biol. Chem. 277, 5699-6702. [DOI] [PubMed] [Google Scholar]
- 31.Jahn R., Schiebler, W., Ouimet, C. & Greengard, P. A. (1985) Proc. Natl. Acad. Sci. USA 82, 4137-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garner C. C., Kindler, S. & Gundelfinger, E. D. (2000) Curr. Opin. Neurobiol. 10, 321-327. [DOI] [PubMed] [Google Scholar]
- 33.Lonart G. (2002) Trends Neurosci. 25, 329-332. [DOI] [PubMed] [Google Scholar]
- 34.Ahmari S. E., Buchanan, J. & Smith, S. J. (2000) Nat. Neurosci. 3, 445-451. [DOI] [PubMed] [Google Scholar]
- 35.Zhai R. G., Vardinon-Friedman, H., Cases-Langhoff, C., Becker, B., Gundelfinger, E. D., Ziv, N. E. & Garner, C. C. (2001) Neuron 29, 131-143. [DOI] [PubMed] [Google Scholar]
- 36.Tixier-Vidal A., Barret, A., Picart, R., Mayau, V., Vogt, D., Wiedenmann, B. & Goud, B. (1993) J. Cell Sci. 105, 935-947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.