Abstract
The kinesin superfamily proteins (KIFs) play essential roles in receptor transportation along the microtubules. KIF17 transports the N-methyl-d-aspartate receptor NR2B subunit in vitro, but its role in vivo is unknown. To clarify this role, we generated transgenic mice overexpressing KIF17 tagged with GFP. The KIF17 transgenic mice exhibited enhanced learning and memory in a series of behavioral tasks, up-regulated NR2B expression with the potential involvement of a transcriptional factor, the cAMP-dependent response element-binding protein, and increased phosphorylation of the cAMP-dependent response element-binding protein. Our results suggest that the motor protein KIF17 contributes to neuronal events required for learning and memory by trafficking fundamental N-methyl-d-aspartate-type glutamate receptors.
In a neuron, most of the proteins are synthesized in the cell body and need to be transported to pre/postsynaptic sites of utilization. Kinesin superfamily proteins (KIFs) are responsible for many of the major microtubule- and ATP-dependent transport pathways in neuronal cells (1, 2). Among these KIFs (3–5), KIF17 (6), a homodimeric microtubule (plus end-directed) motor protein, binds to a cargo molecule NR2B through the scaffolding Mint 1 (mLin10) complex (7, 8) in vitro, raising the possibility that it supports neuronal functions mediated by NR2B. Functional N-methyl-d-aspartate (NMDA) receptors contain heteromeric combinations of the NR1 subunit and one or more of NR2A-D subunits (9), among which the NR2B subunit predominates in forebrain structures (10).
The biology of learning, particularly the molecular mechanism, without a doubt has been vigorously studied recently (11). Glutamate receptor subunits being trafficked from cell bodies to synapses may play an important role in learning and memory (12–15). Overexpression of NR2B in a mouse model results in enhancement of learning and memory (16, 17). Here we examine the in vivo role of KIF17 by investigating what happens if we overexpress the motor of NR2B, KIF17, in mice. We generated transgenic mice in which KIF17 is overexpressed mainly in the postnatal forebrain by using the CAMK II promoter (17–19) and examined whether the NMDA receptor-dependent behavioral patterns of mice are altered by overexpression of KIF17.
Materials and Methods
Generation of GFPKIF17 Transgenic Mice.
A mouse fusion cDNA of GFPKIF17 (3.8 kb) (6) was subcloned into the NotI site of the 8.5-kb upstream region of the CaMKIIα gene with the pCMVβ vector (a gift from S. Okabe) (20). For microinjection, the inserted DNA fragment was released by digestion with SalI, purified by using a QIAEX gel extraction kit (Qiagen, Chatsworth, CA), and passed through a spin-X column (Costar). Microinjection of the purified DNA into BDF1 (CLEA, Osaka) mouse-derived pronuclei of fertilized oocytes and transplantation of these oocytes to the oviducts of pseudopregnant ICR outbred foster mothers were performed as previously described (19, 21–24). Hybrids of BDF were used as hosts of the transgene and were backcrossed with WT BDF mice. The genetic status of the mice was further confirmed by PCR of genomic DNA from mice tails (3–4 wk) by using GFP primers as previously described (19). Lines 1 and 4 were used primarily in this study. Basically, identical results were obtained for these two lines; therefore, data from line 1 are presented except where noted. Control WT mice were chosen from the same littermates. For the behavioral experiments, the same groups of male mice were used (10–12 wk).
Histological Analysis and Electron Microscopy.
Mice (10–12 wk) were anesthetized and fixed in FEA solution (5% formalin/70% ethanol/5% acetic acid). The brain tissues were then removed, washed, dehydrated in a graded series of ethanol, and embedded in Paraplast (Oxford Labware, St. Louis). The blocks were cut by using a rotatory microtome (HM355; Rotary Microtome Zeiss) and sectioned serially at 12-μm thickness by using the same microtome. They were mounted on glass slides, deparafinized, and stained with hematoxylin/eosin or Nissl solution. For electron microscopy, mice were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). Brains were dissected and sectioned by using a microslicer. Matching areas of each tissue were chosen, fixed overnight, processed by the conventional method, and observed under a transmission electron microscope (JEOL-2000 EX or 2010 H). Synaptic densities for each genotype were observed from prints. For each genotype, at least three mice were examined. For observation of the GFP expression pattern, coronal sections of whole brains were dissected as previously described (19). For forebrain subregional observation, brains of mice were dissected and fixed in 2% paraformaldehyde in 0.1 M PBS (pH 7.4) for 2 h at room temperature. Cryostat coronal sections of the cerebrum (20–40 μm) were prepared by using a cryomicrotome (CM3000, Leica, Deerfield, IL), mounted on amino–propylethyldiethoxysilane-coated microscope slides, and air dried; observations were carried out by using the charge-coupled device camera system (TS100 Nihon Kogaku, Tokyo). For observing GFPKIF17 movement, the fresh forebrain region was dissected and sectioned by using a microslicer. Sections were immediately mounted on slides and observed under a Zeiss LSM510 confocal laser-scanning microscope within 5 min.
Immunoprecipitation and Immunoblotting.
Brains were dissected at 4°C, and both cerebral cortices and hippocampi were dissected on ice and placed separately in ice-cold RIPA buffer (50 mM Tris/1% Triton X-100/0.1% SDS/150 mM NaCl, pH 8.0) containing the protease inhibitor cocktail tablets (Roche Applied Science, Mannheim, Germany). For each genotype, 10 mice were examined, data of which are shown in Fig. 5A. The tissue was homogenized and centrifuged at 13,000 × g for 30 min at 4°C. The resultant pellets were resuspended in RIPA buffer, and the protein concentration was determined by using the BCA Protein Assay kit (Pierce) according to the manufacturer's protocol. Aliquots of the suspension were obtained and stored at −70°C.
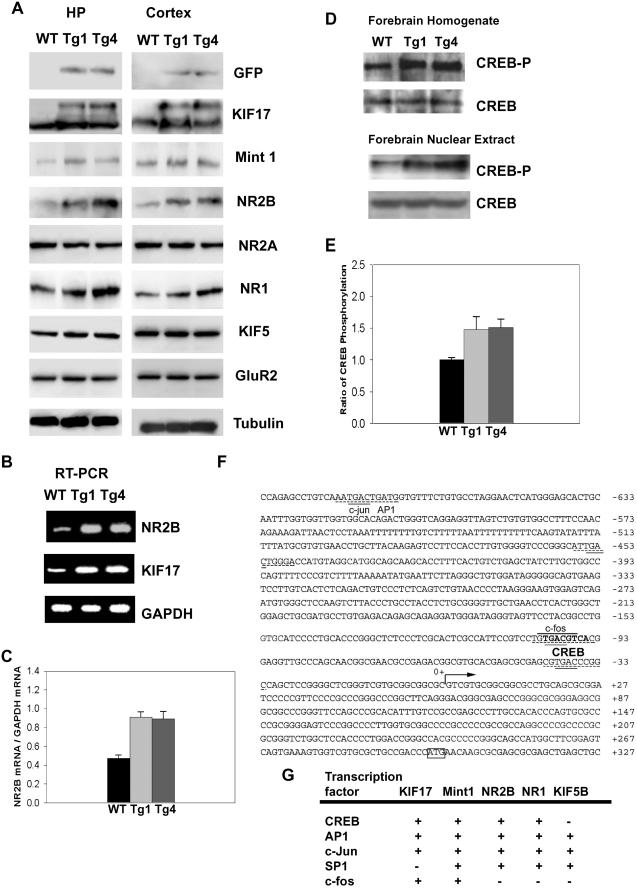
Fig 5.
Biochemical and genomic studies of GFPKIF17 mice. (A) Immunoblot analysis of KIF17 protein and NR2B-associated proteins in total forebrain extracts from hippocampus (HP) and cortex (CTX) of 10-wk-old mice of both transgenic lines (Tg-1 and -4) and WT (Left, hippocampus; Right, cortex panels). (B) Expression of NR2B mRNA in GFPKIF17 transgenic mice as determined by RT-PCR analysis. (C) Ratio of NR2B mRNA to GAPDH in GFPKIF17 and WT mice. (D) Immunoblot analysis of the expression level of CREB in forebrain homogenate and nucleus extract of WT and transgenic mice using an antibody specific for unphosphorylated CREB and CREB that is phosphorylated at Ser-133. (E) Ratio of phosphorylated CREB in GFPKIF17 and WT mice forebrain extract. (F) Characterization and nucleotide sequence of the 5′ end of the mouse KIF17 upstream of the 1.0-kb promoter region. (G) Summary of transcription factors present in KIF17 and associated proteins.
For immunoprecipitation, 50 μg of forebrain homogenates was incubated with the anti-GFP antibody (CLONTECH) at 4°C overnight. Protein-A-coupled agarose beads (Pharmacia) were added to the extracts, and the mixture was incubated at 4°C overnight. After centrifugation at 3,000 × g for 5 min, the beads were then washed three times in TBST [Tris-buffered saline (pH 7.4) and 0.05% Tween-20]. The washed beads were then resuspended in 2× sample loading buffer, and boiled for 5 min. The proteins were resolved by SDS/PAGE, and separated proteins were then transferred onto poly(vinylidene difluoride) membranes, following the same procedure as that for immunoblotting. After blocking the reaction (5% nonfat milk/BSA) in TBST, the blots were incubated overnight at 4°C in blocking buffer with one of the primary antibodies that recognizes KIF17 (6), Mint 1 (mLin10) (Transduction Laboratories, Lexington, KY), NR1 (Chemicon), NR2A (Molecular Probes), NR2B (Transduction Laboratories), GluR2 (PharMingen), KIF5b/kinesin (Sigma), tubulin (Sigma), cAMP-response element-binding protein (CREB) (NEB), or CREB-P (NEB). For CREB-P stimulation and immunoblotting, eight mice (10–12 wk old) of each genotype, from Tg1, Tg4, and their WT littermates, were used and processed as described (25) and according to the protocol (Activemotif). We further collected the nuclear extracts as described in ref. 26 for the CREB/CREB phosphorylation immunoblotting. After incubation with a secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit/mouse antibody, Amersham Pharmacia), the band signals were developed by using an enhanced chemiluminescence (ECL) procedure (Amersham Pharmacia). The membranes were then incubated in ECL solution, and the bands were visualized by developing the blots in ECL Hyperfilm (Amersham Pharmacia). For detection of CREB phosphorylation, the same membranes were reblotted by the ECL Stripping kit (Chemicon). Optical densities of images were measured by using software (scion image; Scion, Frederick, MD), and the values of means SD were obtained from three independent experiments.
RT-PCR.
mRNA was extracted from the forebrain of six mice (10–12 wk old) of each genotype, Tg1, Tg4, and their WT littermates (Miltenyi Biotec, Auburn, CA). mRNA (1 μg) used for the reverse transcription of first-strand complementary DNA was synthesized by using the SuperScript first-strand synthesis system for the RT-PCR kit (GIBCO/BRL). PCR was conducted by using 25 cycles at 96°C for 30 sec, 55°C for 90 sec, and 72°C for 60 sec in a GeneAmp PCR system 9700 Thermal cycler (Perkin–Elmer). The primers used were as follows: for the NR2B, (forward) 5′-AAA GAT CTG CAA ATC CTA CTT CTT CAG GC-3′ (reverse) 5′-AAG GAT CCT CAG ACA TCA GAC TCA ATA CT-3′; for the KIF17, (forward) 5′-TGG GTG CTG CTC AAC GTC TAT GAC TCT ATC-3′, (reverse) 5′-GGA GAA GGG GAT GTC AAG GGA CTC TAG-G3′; and for GAPDH, (forward) 5′-CCT GCA CCA CCA ACT GCT TAG-C3′, (reverse) 5′-GCC AGT GAG CTT CCC GTT CAG C-3′.
Behavioral Tests.
Adult transgenic and WT male mice (10- to 12-wk-old littermates) were used in all behavioral tests in a blind manner.
Open Field Test.
The anxiety and general locomotor activity of the mice were evaluated as described (25). Mice were put inside an open field area and allowed to explore 10 min. Mouse activities in the open field were quantitated by a computer-operated Digiscan optical animal system (target/3, Neuroscience). Running velocity, total distance, and time spent on center and margin area were recorded. Data were analyzed by ANOVA.
Delay Matching Place (DMP) Task.
The test was conducted as previously described (27), with minor modifications. The mice were first pretrained to navigate their way to a visible platform for 2 days, with four trials per day. The mice were then repeatedly trained to navigate their way to a hidden platform at a fixed location until they met a rigorous criterion of successfully locating the hidden platform in three consecutive trials with an average escape latency of less than 30 sec, or until completing a maximum of 16 trials. Each mouse performed up to eight trials per day with intertrial intervals of 10–40 min. If a mouse met the criterion in less than five trials, it was continually trained to complete five trials, so that a complete set of latency data for all mice could be obtained in the first five trials. After the training was completed, starting the next day, the mice were trained to navigate their way to a new hidden platform location in the same manner as the training to the first location, except that the maximal number of trials performed was reduced to eight. This protocol was repeated two more times until a total of four platform locations were learned. All water maze experiments, including this DMP task and the Morris water maze, were videotaped and later digitized and analyzed with the software (target/2, Neuroscience).
Water Maze Task.
The water maze procedures were essentially the same as previously described (17, 28). The training protocol consisted of six sessions (four trials per session per day). The navigation of the mice was recorded by a video camera, and the escape latency to the platform was recorded. In addition, we carried out two transfer tests. The first was carried out at the end of the third session and the second at the end of the last session. During the transfer test, the platform was removed and the mice were allowed to swim in the pool for 60 sec. The time spent in each quadrant was recorded. One-way ANOVA was used to determine the effect of genotype on spatial preference.
Results
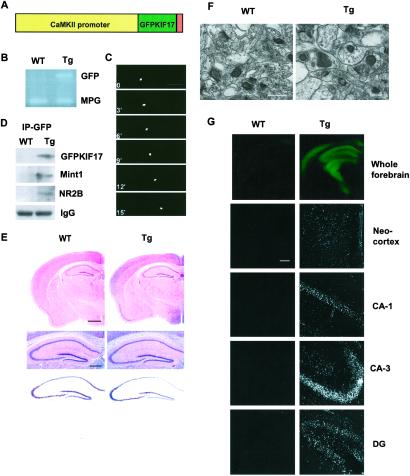
We generated transgenic mice in which overexpression of KIF17 is restricted to the postnatal forebrain by using the CAMK IIα promoter (Fig. 1A). The genetic status of the mice was confirmed by PCR; mice were genotyped at 3–4 wk by using GFP-specific primers and MPG as the internal control gene (Fig. 1B). We performed laser-scanning microscopy with real-time imaging, which showed that GFPKIF17 moves in neuronal dendrites in brain slices at the same velocity (0.7 μm/s) as observed in cultured hippocampal neurons transfected with GFPKIF17 (L. Guillaud, M.S., and N.H., unpublished results), supporting the finding that GFPKIF17 is biologically functional (Fig. 1C). Immunoprecipitation with GFP, which is consistent with results previously reported (6), showed that in the transgenic mice, GFP coimmunoprecipitated with GFPKIF17, Mint 1 (mLin10), and NR2B (Fig. 1D). Of the six lines produced, we report here the results from two independent lines (Tg1 and Tg4) that we have systematically analyzed. They showed similar patterns of GFPKIF17 expression and nearly identical behavioral patterns. Therefore, we used line Tg1 for this study and mainly described the results for it here. Light microscopy and electron microscopy showed no gross structural abnormalities in the transgenic mice (Fig. 1 E and F). In both sets of mice, we found no major difference between mutant and WT mice in hippocampal CA1 synapse morphology (Fig. 1F). We further investigated histologically the GFPKIF17 distribution of the transgene using coronal sections and directly observed that the transgene was highly enriched in the forebrain region (Fig. 1G). The GFPKIF17 transgenic mice showed normal growth and body weights and mated normally. These results indicate that GFPKIF17 is biologically functional, and an increased expression level of KIF17 or GFP is not, by itself, neurotoxic and does not affect the basic architecture of the brain.
Fig 1.
Construction and histological characterization of GFPKIF17 transgenic mice. (A) The construct for production of GFPKIF17 transgenic mice. (B) PCR screening with GFP primers, using endogenous MPG as internal control. (C) Visualization of motility of GFPKIF17. (Bar = 20 μm.) (D) Immunoprecipitation of the forebrain fraction from mouse using anti-GFP antibody; the same immunoblot was used for antibodies against KIF17, Mint 1(mLin10), NR2B, and IgG. (E) Normal brain morphology in transgenic mice compared with WT, hematoxylin/eosin staining (Top), higher magnification of the hematoxylin/eosin staining (Middle), and Nissl staining (Bottom) of the hippocampus. (Bars = 500 μm.) (F) The ultrastructure of axospinous synapses in CA1 hippocampus shown in each case. (Bar = 10 μm.) (G) GFPKIF17 localization in forebrain region of mice. Neocortex and hippocampal subregions (CA1, CA3, DG). (Bar = 100 μm.)
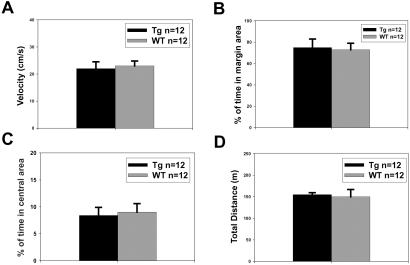
We next investigated the behavioral phenotypes of GFPKIF17 mice by using a series of behavioral tests in a blind manner. GFPKIF17 mice did not exhibit seizures or convulsions. We found that GFPKIF17 mice and WT littermates were indistinguishable in the open-field test. There were no significant differences in their running velocity (P > 0.5, ANOVA) (Fig. 2A) and percentage of time spent on margin region from the wall (P > 0.5, ANOVA) (Fig. 2B), percentage of time spent in the central area (P > 0.05, ANOVA) (Fig. 2C), and total distance traveled (P > 0.05, ANOVA) (Fig. 2D) between GFPKIF17 and WT mice. The result indicated that GFPKIF17 mice exhibit normal exploratory activity and anxiety-related responses with their WT littermate.
Fig 2.
Normal performance of GFPKIF17 transgenic mice in open field test. The results show that two groups of mice explore the activity chamber similarly. There were no significant group differences (Tg-1, n = 12; WT, n = 12) in activity indices such as (A) running velocity (P > 0.5 ANOVA) and (B) percentage of time spent on margin region from the wall (P > 0.5 ANOVA), (C) percentage of time spent in central area (P > 0.05 ANOVA), and (D) total distance traveled (P > 0.05 ANOVA) during the 10-min test.
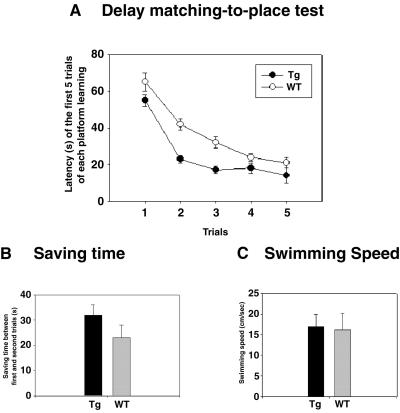
To investigate working/episodic-like memory, we then assessed performance in the DMP task (27, 29), which measures the ability of mice to encode ongoing events rapidly and also involves NMDA receptor. A total of four different hidden platform locations were learned sequentially. The reduction in escape latency in the second trial compared with that in the first within a single session reflects the mouse's ability to acquire memory of the platform location based on a single exposure against interference by memories of other platform locations acquired in previous sessions. We found that GFPKIF17 mice could rapidly remember the new platform location and reduced their escape latency at a faster rate than WT mice [F (1, 22) = 5.655, P < 0.05, one-way ANOVA] (Fig. 3A). The reduction in the escape latency in the second trial compared with that in the first indicated a significant difference between the genotypes (Fig. 3B). In addition, swimming velocities were not significantly different (Fig. 3C) (P > 0.6), showing that GFPKIF17 mice acquired memory of the platform location faster than their WT littermates (Movies 1–4, which are published as supporting information on the PNAS web site, www.pnas.org).
Fig 3.
Enhanced performance of working memory task by GFPKIF17 transgenic mice. (A) Escape latencies in the first five trials of new platform training, averaged from the last two training sessions (Tg-1, n = 12; WT, n = 12) [F (1, 22) = 5.655, P < 0.05 ANOVA]. (B) The time saved (reduction in latencies) between the first and second trials of each session averaged from the last two training sessions (P < 0.05 ANOVA). (C) Swimming velocities averaged from the first two trials of the last two training sessions. No significant difference was observed between the mutant and WT mice.
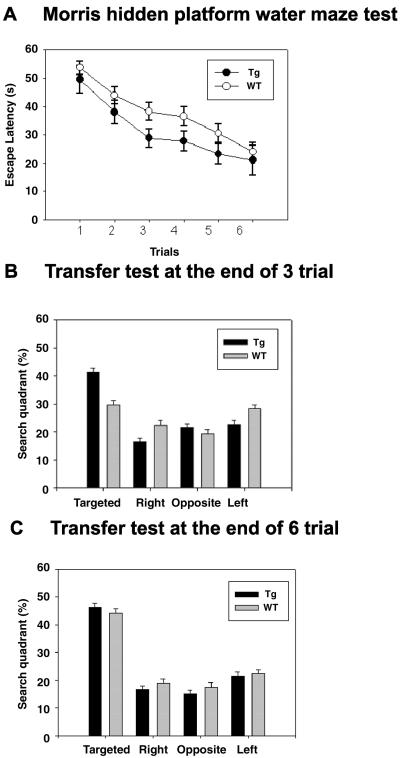
We next investigated performance in the hidden-platform version of the Morris water maze task, because the task depends on NMDA receptors (28, 30). Adult littermate WT (n = 12) and transgenic (n = 12) mice swam in the pool at comparable velocities. After the training session, the latency to mount on the platform (Fig. 4A) and the path length decreased in both WT and transgenic mice, but the values were significantly shorter in GFPKIF17 mice than in WT mice throughout the session [F (1, 22) = 8.945, P < 0.01, one-way ANOVA]. In a transfer test on day 3 (P < 0.05, Dunnett's test), GFPKIF17 mice exhibited an obvious preference for the targeted quadrant in which the platform was previously located compared with control (Fig. 4B). After further training, on day 6, WT mice showed the same magnitude of preference as GFPKIF17 mice (Fig. 4C). These results suggest that GFPKIF17 transgenic mice learned more quickly than WT littermates in these water maze tasks (Movies 5 and 6, which are published as supporting information on the PNAS web site).
Fig 4.
Enhanced performance of spatial learning and memory task by GFPKIF17 transgenic mice. (A) Escape latency in Morris water maze task (Tg-1, n = 12; wild-type, n = 12, mean ± SEM) [F (1, 22) = 8.945, P < 0.01 ANOVA]. The GFPKIF17 mice showed significantly shorter escape latencies than WT mice on the sessions. (B) Location preference in the transfer test conducted at the end of the third training session. GFPKIF17 transgenic mice spent more time in the target quadrant than other quadrants, whereas WT mice did not show any preference for the target quadrant at this stage (P < 0.05, Dunnett's test). (C) Location preference in the second transfer test carried out at the end of the sixth training session. Both transgenic and WT mice exhibited a strong preference for the target quadrant where the hidden platform was previously located. There were no significant differences between the genotypes.
To determine what underlies the genetic enhancement of learning and memory in GFPKIF17 mice, we first assessed the protein expression pattern in the mice forebrain, because overexpression of the NR2B protein in the mice forebrain enhanced memory (17). From immunoblotting results, we found that GFPKIF17 levels were ≈30% of the total KIF17, and the NR2B protein in the cortex and hippocampus of transgenic mice was up-regulated 1.5-fold as in WT mice (Table 1, which is published as supporting information on the PNAS web site). There was also a slight increase in the NR1 protein expression level but a slight decrease in the NR2A expression level in these regions, indicating that the ratio of NR2B to NR2A in the receptor complex may have increased (Table 1). KIF5B, GluR2, or tubulin as control (Fig. 5A), and the relative ratios of different proteins expressed in mice were estimated from the immunoblots of three independent experiments (Table 1).
We next measured the mouse forebrain NR2B mRNA level by RT-PCR and detected a significant difference between WT and GFPKIF17 mice. To our surprise, NR2B mRNA levels in the adult GFPKIF17 Tg mouse forebrain were nearly 2-fold more than those in their littermates, based on results from four independent experiments, indicating that NR2B mRNA levels, as well as KIF17 mRNA levels, might be regulated by a pretranscriptional process in GFPKIF17 Tg mice (Fig. 5 B and C).
As evidence from a variety of species, genetic manipulations of CREB-associated protein transcriptional regulation in Aplysia, Drosophila, mouse, and rat suggest their crucial roles in long-term memory (31). Because CREB was found in the NR2B subunit promoter region (32), we further determined whether the expression levels of CREB and phosphorylated CREB are affected in our mice. The expression level of CREB was not altered by KIF17 expression in either group. We measured directly the expression level of phosphorylated CREB (25) using an antibody specific for CREB that is phosphorylated at Ser-133. We observed the up-regulation of phosphorylated CREB in the adult GFPKIF17 mice forebrain homogenate and even further by nuclear extract preparation in three independent experiments (Fig. 5 D and E).
Does the phosphorylation of CREB also explain the up-regulation of intrinsic KIF17 in GFPKIF17 mice (Table 1) (Fig. 3B)? To answer this question, we used GenBank and Celera mouse genome databases, mining the KIF17 transcription promoter region. Similar to the NR2B promoter (32), KIF17 contains a CREB consensus sequence, TGACGTCA, which is found within the 1-kb promoter region (Fig. 5F), whereas other KIFs expressed in neurons, such as KIF5B, do not have this sequence in the region upstream of the 1.0-kb promoter (Fig. 5G). On the basis of these results, we hypothesize that the transcription factor CREB may also participate in the regulation of KIF17.
Discussion
Motor protein kinesins appeared as short leg-like structures on vesicles in electron microscope images (33–36). On the basis of the transport of various kinds of cargoes and variety of structural candidates of motor proteins in vivo, we and other researchers isolated and characterized multiple kinesin genes and proteins (3–5, 37, 38). In neurons, sorting and delivery of organelles depend on KIFs (1). Among these, KIF17, a homodimeric motor protein, through its COOH domain interacted with the PDZ domain of Mint 1 (mLin10) scaffolding protein trafficking NMDA receptor subunits complex in vitro (6).
We establish a gain-in-function mouse model of motor protein (KIFs). Because the GFPKIF17 mice are viable, we are able to examine them in a variety of behavioral paradigms. This approach allows us to identify candidate neuronal populations or specific brain regions in which KIF17 function appears critical. To our surprise, we found that the molecular motor through transportation plays a fundamental role in the higher brain function in vivo.
Working Memory Is Improved with Overexpression of KIF17.
Working memory is an immediate and rapidly decaying memory thought to be anatomically sustained by a prefrontal cortex–hippocampus network (39, 40). Previous studies of human patients showed that the hippocampus is important for both episodic (event) and semantic (fact) declarative memories (41, 42) The rodent hippocampus is critically involved in the formation of spatial working memory (27, 43) and more recently has been suggested to be involved in underlying the formation of episodic-like memory (27, 44, 45). In this study, enhancement of working/episodic-like memory was revealed by the DMP task in the GFPKIF17 mice. The difference of saving time between genotypes in the DMP task strongly suggests that the mutant mice have enhanced ability to learn the new location of the platform quickly just by one trial, at the same time suppressing the interfering memory of the previous platform location (27).
Spatial Learning and Memory Are Facilitated by Overexpression of KIF17.
To investigate whether memory was enhanced in other behavioral tasks, we used the Morris water maze task, a hippocampal-dependent task that challenges spatial learning and memory (28). During this task, animals learn the location of a hidden platform in a circular pool using distal cues. During acquisition, mice from all groups showed a decrease in escape latency (Fig. 4A). Strikingly, however, GFPKIF17 mice displayed significantly lower escape latencies and shorter path length than control mice (Fig. 4A). Because basal behaviors, locomotor activity, and swimming velocity were similar between the genotypes, the improved performance in spatial learning and working memory tasks in GFPKIF17 mice cannot be due to improvements in sensorimotor function or from changes in motivational/emotional processes.
How Does Forebrain KIF17 Overexpression Enhance Learning and Memory?
We examined the relationship of NR2B, Mint 1(mLin10) and KIF17 in mice and found that they colocalize by immunoprecipitation (Fig. 1D). We then assessed the mouse forebrain protein expression pattern, because overexpression of the NR2B protein in the mouse forebrain enhanced memory (17). From immunoblotting results, we found that KIF17 and the NR2B protein in the cortex and hippocampus of transgenic mice were up-regulated compared with WT mice (Fig. 5A). We next measured the mouse forebrain NR2B and KIF17 mRNA levels by RT-PCR and detected a significant enhancement between WT and GFPKIF17 mice (Fig. 5B), which might be regulated by a pretranscriptional process in GFPKIF17 Tg mice. Similar findings are consistently and independently obtained in hippocampal culture studies of KIF17 and NR2B dynamics (L. Guillaud, M.S., and N.H., unpublished results).
As demonstrated by an extensive, if not daunting, body of literature, CREB is activated as a response to a vast array of physiological stimuli (46). The CREB pathway, from upstream activating proteins to transcription factor, has been implicated in learning and memory (11, 31, 47–53). Activation of CREB is thought to be important in the formation of long-term memory, which leads to the phosphorylation of CREB (31, 54, 55). Our study suggests that the formation of long-term memory through the activation of CREB could be attributed at least to some extent to the activation and induction of KIF17 gene expression, which could result in enhanced transport of NR2B in dendrites and further activation of NR2B gene expression. Regulation of transport of vesicles containing functional molecules, such as receptors in cells, is not simple. It could involve multiple mechanisms such as synthesis of motor proteins, control of motor protein activity (56), and regulation of motor interaction with cargoes (57). Our data may raise the possibility that nuclear transcriptional signaling also regulates the transport of specific cargoes in cell.
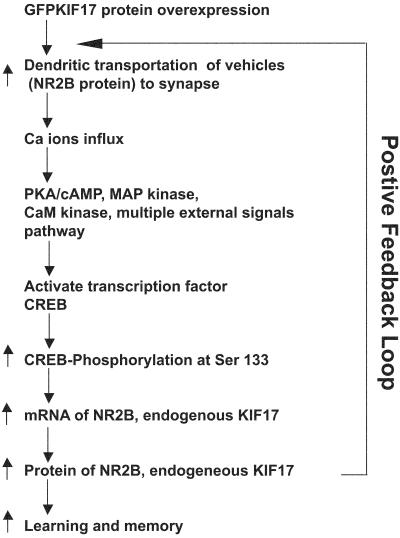
We propose that CREB phosphorylation leads to NR2B and KIF17 protein up-regulation based on our biochemical data. Because this proposal is still in the speculative stage, the direct linking between cAMP response-element-binding protein phosphorylation, NR2B, and KIF17 up-regulation remains to be addressed in future studies. In Fig. 6, we propose a potential mechanism of how KIF17 overexpression leads to the improvement of learning and memory. The enhancement of KIF17 synthesis leads to an increase in the amount of NMDA receptor subunits (NR2B) transported in dendrites, which may result in enhancement of synaptic activity. The resulting activation of the Ca2+ influx and signaling back to the nucleus somehow increase the level of phosphorylated CREB, which could further activate transcription of KIF17 mRNA directly. This process could also enhance transcription of NR2B mRNA. The entire positive feedback loop of KIF17 trafficking may underlie the improvement of learning and memory in vivo (Fig. 6). We propose that the function of KIF17 should be further investigated by complementary electrophysiological analyses in the future. Finally, our findings shed new light on the function of the motor proteins, KIFs, which might lead to new insights and the development of treatment for learning deficits in neuronal encoding, storage, and retrieval in the future.
Fig 6.
Model for KIF17 function. Schematic of the involvement of motor protein KIF17 in the transport of NMDA receptors and enhancement of learning and memory. The relationship between transport of NR2B by KIF17, synaptic events, involvement of CREB, and transcriptional regulation of NR2B and of KIF17 by phosphorylated CREB.
Supplementary Material
Acknowledgments
We thank T. Takahashi, K. Kobayashi, F. Mori-Kawakami, Y. P. Tang, G. Collingridge, T. Hensch, R. G. Morris, and R. Sprengel for discussion and advice; S. Okabe (Tokyo Medical and Dental University, Tokyo) and M. Mayford (The Scripps Research Institute) for supply of the CaMK II promoter; and H. Miki, L. Guillaud, M. Fukuda, M. Sugaya, N. Onouchi, our animal working staff, and other members of the Hirokawa laboratory for stimulating discussions and valuable advice throughout the study. R.W. is the recipient of an Overseas Outstanding Young Scholar Award from the Li Po Chun Foundation (Hong Kong, 2000). This work is supported by a fellowship from the Japanese Society for Promotion for Science Research and Grants to Young Scientists [to R.W. (DC-1, 2001) and J.T. (Postdoc, 2002)] and by a Special Grant-in-Aid from the Center of Excellence, Japanese Ministry of Education, Culture, Sports, Science and Technology (to N.H.).
Abbreviations
DMP, delay matching place
NMDA, N-methyl-d-aspartate
KIF, kinesin superfamily protein
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hirokawa N. (1998) Science 279, 519-526. [DOI] [PubMed] [Google Scholar]
- 2.Hirokawa N., Noda, Y. & Okada, Y. (1998) Curr. Opin. Cell Biol. 10, 60-73. [DOI] [PubMed] [Google Scholar]
- 3.Aizawa H., Sekine, Y., Takemura, R., Zhang, Z., Nangaku, M. & Hirokawa, N. (1992) J. Cell Biol. 119, 1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakagawa T., Tanaka, Y., Matsuoka, E., Kondo, S., Okada, Y., Noda, Y., Kanai, Y. & Hirokawa, N. (1997) Proc. Natl. Acad. Sci. USA 94, 9654-9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miki H., Setou, M., Kaneshiro, K. & Hirokawa, N. (2001) Proc. Natl. Acad. Sci. USA 98, 7004-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Setou M., Nakagawa, T., Seog, D. H. & Hirokawa, N. (2000) Science 288, 1796-1802. [DOI] [PubMed] [Google Scholar]
- 7.Butz S., Okamoto, M. & Sudhof, T. C. (1998) Cell 94, 773-782. [DOI] [PubMed] [Google Scholar]
- 8.Okamoto M. & Sudhof, T. C. (1997) J. Biol. Chem. 272, 31459-31464. [DOI] [PubMed] [Google Scholar]
- 9.Nakanishi S. (1992) Science 258, 597-603. [DOI] [PubMed] [Google Scholar]
- 10.Monyer H., Sprengel, R., Schoepfer, R., Herb, A., Higuchi, M., Lomeli, H., Burnashev, N., Sakmann, B. & Seeburg, P. H. (1992) Science 256, 1217-1221. [DOI] [PubMed] [Google Scholar]
- 11.Kandel E. R. (2001) Science 294, 1030-1038. [DOI] [PubMed] [Google Scholar]
- 12.Bliss T. V. & Collingridge, G. L. (1993) Nature 361, 31-39. [DOI] [PubMed] [Google Scholar]
- 13.Collingridge G. L. & Bliss, T. V. (1995) Trends Neurosci. 18, 54-56. [PubMed] [Google Scholar]
- 14.Shi S., Hayashi, Y., Esteban, J. A. & Malinow, R. (2001) Cell 105, 331-343. [DOI] [PubMed] [Google Scholar]
- 15.Wong R. W., Setou, M. & Hirokawa, N. (2001) Trends Cell Biol. 11, 320. [DOI] [PubMed] [Google Scholar]
- 16.Tang Y. P., Wang, H., Feng, R., Kyin, M. & Tsien, J. Z. (2001) Neuropharmacology 41, 779-790. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y. P., Shimizu, E., Dube, G. R., Rampon, C., Kerchner, G. A., Zhuo, M., Liu, G. & Tsien, J. Z. (1999) Nature 401, 63-69. [DOI] [PubMed] [Google Scholar]
- 18.Mayford M., Bach, M. E., Huang, Y. Y., Wang, L., Hawkins, R. D. & Kandel, E. R. (1996) Science 274, 1678-1683. [DOI] [PubMed] [Google Scholar]
- 19.Krestel H. E., Mayford, M., Seeburg, P. H. & Sprengel, R. (2001) Nucleic Acids Res. 29, E39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okabe S., Collin, C., Auerbach, J. M., Meiri, N., Bengzon, J., Kennedy, M. B., Segal, M. & McKay, R. D. (1998) J. Neurosci. 18, 4177-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong R. W., Sham, M. H., Lau, Y. L. & Chan, S. Y. (2000) Mol. Biotechnol. 15, 155-159. [DOI] [PubMed] [Google Scholar]
- 22.Wong R. W., Kwan, R. W., Mak, P. H., Mak, K. K., Sham, M. H. & Chan, S. Y. (2000) J. Biol. Chem. 275, 18297-18301. [DOI] [PubMed] [Google Scholar]
- 23.Wong R. W. & Chan, S. Y. (2000) Mol. Biotechnol. 15, 65-67. [DOI] [PubMed] [Google Scholar]
- 24.Chan S. Y. & Wong, R. W. (2000) J. Biol. Chem. 275, 38693-38698. [DOI] [PubMed] [Google Scholar]
- 25.Kang H., Sun, L. D., Atkins, C. M., Soderling, T. R., Wilson, M. A. & Tonegawa, S. (2001) Cell 106, 771-783. [DOI] [PubMed] [Google Scholar]
- 26.Dignam J. D., Lebovitz, R. M. & Roeder, R. G. (1983) Nucleic Acids Res. 11, 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng H., Chattarji, S., Barbarosie, M., Rondi-Reig, L., Philpot, B. D., Miyakawa, T., Bear, M. F. & Tonegawa, S. (2001) Cell 107, 617-629. [DOI] [PubMed] [Google Scholar]
- 28.Morris R. G., Garrud, P., Rawlins, J. N. & O'Keefe, J. (1982) Nature 297, 681-683. [DOI] [PubMed] [Google Scholar]
- 29.Chen G., Chen, K. S., Knox, J., Inglis, J., Bernard, A., Martin, S. J., Justice, A., McConlogue, L., Games, D., Freedman, S. B. & Morris, R. G. (2000) Nature 408, 975-979. [DOI] [PubMed] [Google Scholar]
- 30.Tsien J. Z., Huerta, P. T. & Tonegawa, S. (1996) Cell 87, 1327-1338. [DOI] [PubMed] [Google Scholar]
- 31.Mayford M. & Kandel, E. R. (1999) Trends Genet. 15, 463-470. [DOI] [PubMed] [Google Scholar]
- 32.Klein M., Pieri, I., Uhlmann, F., Pfizenmaier, K. & Eisel, U. (1998) Gene 208, 259-269. [DOI] [PubMed] [Google Scholar]
- 33.Hirokawa N. (1982) J. Cell Biol. 94, 129-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirokawa N., Pfister, K. K., Yorifuji, H., Wagner, M. C., Brady, S. T. & Bloom, G. S. (1989) Cell 56, 867-878. [DOI] [PubMed] [Google Scholar]
- 35.Hirokawa N. (1996) Trends Cell Biol. 6, 135-141. [DOI] [PubMed] [Google Scholar]
- 36.Wells W. A. (2002) J. Cell Biol. 158, 609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vale R. D., Reese, T. S. & Sheetz, M. P. (1985) Cell 42, 39-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brady S. T. (1985) Nature 317, 73-75. [DOI] [PubMed] [Google Scholar]
- 39.Olton D. S. & Feustle, W. A. (1981) Exp. Brain Res. 41, 380-389. [DOI] [PubMed] [Google Scholar]
- 40.Jarrard L. E. (1993) Behav. Neural. Biol. 60, 9-26. [DOI] [PubMed] [Google Scholar]
- 41.Viskontas I. V., McAndrews, M. P. & Moscovitch, M. (2000) J. Neurosci. 20, 5853-5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Squire L. R. & Zola, S. M. (1998) Hippocampus 8, 205-211. [DOI] [PubMed] [Google Scholar]
- 43.Olton D. S. & Papas, B. C. (1979) Neuropsychologia 17, 669-682. [DOI] [PubMed] [Google Scholar]
- 44.Morris R. G. & Frey, U. (1997) Philos. Trans. R. Soc. London B Biol. Sci. 352, 1489-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris R. G. (2001) Philos. Trans. R. Soc. London B 356, 1453-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lonze B. E., Riccio, A., Cohen, S. & Ginty, D. D. (2002) Neuron 34, 371-385. [DOI] [PubMed] [Google Scholar]
- 47.Yin J. C. & Tully, T. (1996) Curr. Opin. Neurobiol. 6, 264-268. [DOI] [PubMed] [Google Scholar]
- 48.Silva A. J., Kogan, J. H., Frankland, P. W. & Kida, S. (1998) Annu. Rev. Neurosci. 21, 127-148. [DOI] [PubMed] [Google Scholar]
- 49.Abel T. & Kandel, E. (1998) Brain Res. Brain Res. Rev. 26, 360-378. [DOI] [PubMed] [Google Scholar]
- 50.Lamprecht R. (1999) Cell Mol. Life Sci. 55, 554-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kida S., Josselyn, S. A., de Ortiz, S. P., Kogan, J. H., Chevere, I., Masushige, S. & Silva, A. J. (2002) Nat. Neurosci. 5, 348-355. [DOI] [PubMed] [Google Scholar]
- 52.Pittenger C., Huang, Y. Y., Paletzki, R. F., Bourtchouladze, R., Scanlin, H., Vronskaya, S. & Kandel, E. R. (2002) Neuron 34, 447-462. [DOI] [PubMed] [Google Scholar]
- 53.Barco A., Alarcon, J. M. & Kandel, E. R. (2002) Cell 108, 689-703. [DOI] [PubMed] [Google Scholar]
- 54.Impey S. & Goodman, R. H. (2001) Sci. STKE 2001, PE1. [DOI] [PubMed] [Google Scholar]
- 55.Mayr B. & Montminy, M. (2001) Nat. Rev. Mol. Cell. Biol. 2, 599-609. [DOI] [PubMed] [Google Scholar]
- 56.Reilein A. R., Rogers, S. L., Tuma, M. C. & Gelfand, V. I. (2001) Int. Rev. Cytol. 204, 179-238. [DOI] [PubMed] [Google Scholar]
- 57.Terada S. & Hirokawa, N. (2000) Curr. Opin. Neurobiol. 10, 566-573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.