Abstract
Neural stem cells (NSCs) in vitro are able to generate clonal structures, “neurospheres,” that exhibit intra-clonal neural cell-lineage diversity; i.e., they contain, in addition to NSCs, neuronal and glial progenitors in different states of differentiation. The present study focuses on a subset of neurospheres derived from fresh clinical specimens of human brain by using an in vitro system that relies on particular growth factors, serum, and anchorage withdrawal. Thirty individual and exemplary cDNA libraries from these neurosphere clones were clustered and rearranged within a panel after characterization of differentially expressed transcripts. The molecular phenotypes that were obtained indicate that clonogenic NSCs in our in vitro system are heterogeneous, with subsets reflecting distinct neural developmental commitments. This approach is useful for the sorting and expansion of NSCs and facilitates the discovery of genes involved in cell proliferation, communication, fate control, and differentiation.
Keywords: stem cell, cDNA panel, microarray, iterative algorithm, temporal profiling
Long-term culture systems allow the continuous propagation of potentially heterogeneous populations of neural stem and progenitor cells in monolayers on substrate-coated plates (1, 2) or as suspended, clonal aggregates of cells known as neurospheres (3, 4). An in vitro system has been developed that exploits serum and anchorage withdrawal, and a medium supplemented with methyl cellulose and pleiotropic growth factors [epidermal growth factor (EGF), basic fibroblast growth factor (FGF2), and insulin] to generate clonal neurospheres from rodent and human brain (5–7). The progeny generated in vitro from a parental clone-forming cell during neurosphere formation demonstrate intra-clonal heterogeneity in the expression of neural lineage-specific markers. Such clonogenic cells can thus be considered to be neural stem cells (NSCs).
Current in vitro models of neurogenesis suggest that neurogenesis is driven by multipotent NSCs that can self-renew and progressively generate more developmentally restricted but still multipotent progenitors and more developmentally committed progeny.
In this study, we asked the question whether a subset of parental cells that are clonogenic in our in vitro system and give rise to neurons and glia could be retrospectively subdivided into distinct subsets of cells with different developmental commitment based on distinct molecular phenotypes of their clonal progeny. This approach has revealed temporal and sequential order in the expression of genes involved in NSC growth and differentiation, and a resulting model can be used to elucidate the molecular phenotype of any clonogenic stem/progenitor cell population; e.g., those involved in developmental and persistent neurogenesis (“neuropoiesis,” ref. 8).
Materials and Methods
Neurosphere Generation and Creation of cDNA Panels.
Human tissue from the forebrain (e.g., subependymal zone and hippocampus, see ref. 6), ranging in age from 13 weeks after conception (a sample from a spontaneous, still-birth) to 57 years of age, was dissociated according to established protocols (5–7) and subjected to the molecular phenotyping method described below. The model presented here was developed on cross comparisons of neurospheres generated from the same brain dissociation (so as not to introduce potential genetic variability). The dissociated cells gave rise to individual neurosphere clones that were used for total RNA extraction and for first-strand cDNA synthesis using SMART cDNA synthesis technology (CLONTECH). The first-strand cDNA pool then was subjected to long distance amplification by using the Advantage 2 PCR Enzyme System (CLONTECH) as recommended by the manufacturer.
Subtraction Procedure, Microarray Screening, and Relative PCR.
Reciprocal subtraction of two exemplary cDNA libraries presented here, no. 22 and no. 30, was performed according to the PCR Select kit (CLONTECH), each step of which was optimized. cDNA differential products were used for random labeling by using a High Prime DNA Labeling kit (Roche Molecular Biochemicals) with some modification of incubation time (20 h) and by enhancing the labeling reaction with an additional 5 units of Klenow enzyme after 10 h. Gene microarray screening was performed by using MICROMAX microarrays according to the manufacturer's instructions (NEN). Relative PCR reactions were made as described (9). Detail descriptions of methods and corresponding technical concerns can be found in the accompanying Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Sequence Analyses.
Analyses of DNA sequences were performed on-line by using the blast algorithm. SwissProt and National Center for Biotechnology Information (NCBI) databases were searched for protein homology, motifs, and potential ORFs. Sequences also were compared with those present in the Stem Cell Database (SCDb, located at http://stemcell.princeton.edu/). The gene expression data were analyzed by using the PubGene database (www.PubGene.org). Expressed Gene Anatomy Database (EGAD) classification of genes was made at the web site www.allgenes.org.
Results
Clonal Neurospheres Exhibit Inter-Clonal Variability in Gene Expression.
It was hypothesized that a population of clonal neurospheres is heterogeneous, and this heterogeneity reflects differences in the developmental potential of parental clone-forming cells. It was also assumed that the relative size of a neurosphere is correlated with the proliferative, differentiation, and developmental potential of a parental clone-forming cell.
Data from individual clonal neurospheres presented here were generated from a single-cell suspension of fetal human forebrain tissue by using the aforementioned in vitro system (5, 6). A representative sample of 30 neurospheres was taken for further analysis and classified according to their size as follows: 7 small neurospheres containing ≈50–100 cells (clones nos. 1–7), 8 neurospheres of medium size containing >100–400 cells (clones nos. 8–15), and 15 large neurospheres containing >400 cells (clones nos. 16–30). The panel of individual cDNA libraries from 30 selected clones was generated and normalized. The quality of normalization was controlled by relative PCR with primers for 18S rRNA. Four libraries (nos. 2, 3, 4, and 6) could not be normalized and were excluded from further experiments.
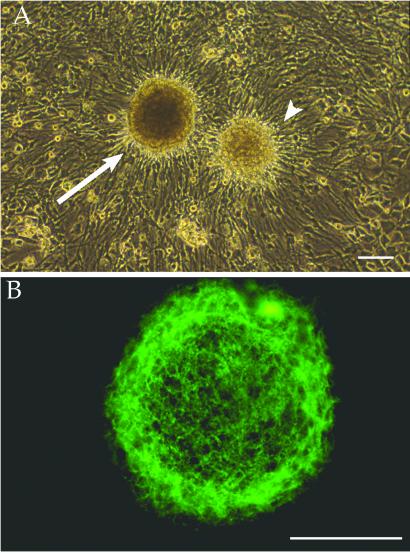
To analyze the inter-clonal heterogeneity exemplified by the two neurospheres shown in Fig. 1A, the panel was screened for the expression of eight markers that are associated with neurogenesis. They include developmentally regulated proteins such as PAX 6 and tenascin-C (hexabrachion, “hxb”; and see neurosphere immunolabeling for this marker in Fig. 1B), the nestin intermediate filament protein (a marker for precursor cells), and the neuronal lineage–specific markers neuron-specific enolase (NSE), or γ-enolase 2 (ENO2), neurofilament protein (NF-M), and microtubule-associated protein-2 (MAP2). Glial fibrillary acidic protein (GFAP), and myelin proteolipid protein (PLP/DM20) were used as astrocyte and oligodendroglial markers, respectively.
Fig 1.
Phase microscopic (A) and immunofluorescence (B) labeling of representative neurosphere clones. (A) Two neurosphere clones (arrow and arrowhead), generated from NSCs that were plated at the same time from the same brain dissociation and next to each other, show neurosphere heterogeneity. (B) A single neurosphere immunostained for tenascin-C (confirming expression of one of the genes screened for in our cDNA libraries and panel) shows dense expression of this developmentally regulated extracellular matrix protein. (Bars = 100 μm.)
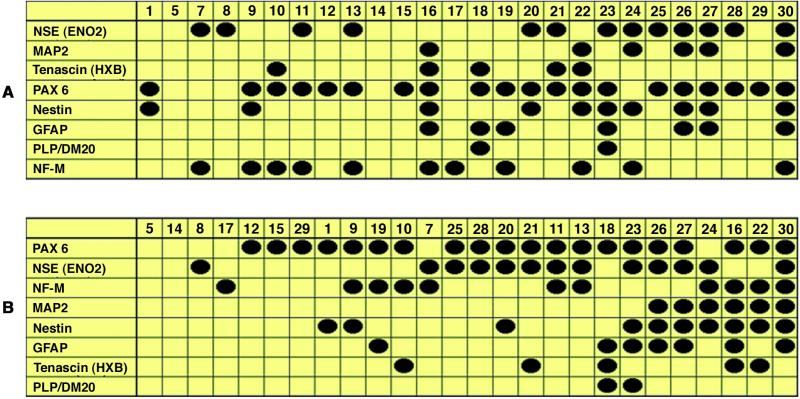
The results from a screening of 26 normalized cDNA libraries are presented in the table shown in Fig. 2A. They demonstrate an inter-clonal heterogeneity of clonal progeny derived from different single parental clone-forming cells. Each individual library expresses a unique combination of eight selected transcripts, which are considered to be “discriminating markers.” These results confirm the hypothesis related to inter-clonal heterogeneity of a neurosphere population, and consequently, the developmental heterogeneity of parental cells capable of forming clones in this culture system.
Fig 2.
Results from screening of 30 human neurosphere cDNA libraries for a representative set of cell phenotype and developmental genes. (A) Nonarranged cDNA panel. (B) cDNA panel rearranged after application of the CLUSTER procedure.
Next, we assumed that there is a limited number of neural clone-forming cell types, and each type of clone-forming cell is capable of forming a distinct cluster of clones with similar or very close combinations of expressed genes by analogy with a developmental hierarchy of hematopoietic cells (10–12). With this assumption, the panel of cDNA libraries was rearranged by using the CLUSTER procedure (STATISTICA 6, HalloGram Publishing, Aurora, CO). The result of this rearrangement is presented in Fig. 2B. The order of rows (marker transcripts) was rearranged according to the frequency of occurrence of marker transcripts in the individual libraries. The order of columns (the numerical numbers of clones) also was rearranged according to the developmental “maturation” status of a parental clone-forming cell. As a result, the clones putatively originating from the most primitive cells (nos. 5, 8, 14, 17) were placed on the left side of the table shown in Fig. 2B, whereas the more “mature” clones (nos. 16, 22, 30) were located on the right side of the table.
This rearrangement does not afford discrimination of clones into separate distinct clusters, suggesting that a set of eight transcripts chosen as discriminating markers is not sufficient for this purpose. Therefore, to achieve a more precise clustering, we attempted to find additional marker transcripts with higher informative values by performing a direct and reverse subtraction of individual cDNA libraries.
A Reciprocal Subtraction of the Individual Libraries from Clonal Neurospheres Reveals Discriminating Markers.
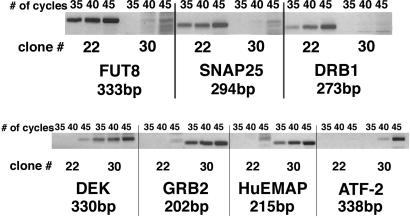
Two cDNA libraries were selected for the subtraction procedure. The products from a direct and a reverse subtraction of cDNA libraries, no. 22 and no. 30, were screened two times on MICROMAX glass microarrays (NEN) enriched with full-size brain-derived genes (80%) and with genes derived from other tissues (20%). The poorly replicated transcripts and those with low-signal intensities were rejected. Thirty transcripts from each library with the highest signal-to-noise ratio were selected from each microarray. Differential expression of each transcript was verified by relative PCR with the corresponding primers. The results of microarray screening confirmed the differential expression of NSE (clone no. 30) and tenascin-C (clone no. 22) transcripts. The differential expression of 42% of selected transcripts was confirmed by relative PCR (Fig. 3). The verified, differentially expressed genes in the subtractive library no. 22/30 is presented in Table 1, which is published as supporting information on the PNAS web site.
Fig 3.
Representative results of 22/30 libraries screened and confirmed by relative PCR. FUT8, SNAP25, DRB1 are differentially expressed in clone no. 22; DEK, GRB2, HuEMAP, and ATF2 are differentially expressed in clone no. 30.
Assuming that our subtractive libraries could contain transcripts not represented in the MICROMAX microarray, we performed a dot-blot screening of subtractive library no. 22. Ninety-six randomly selected transcripts were isolated from a direct cDNA subtracted pool (no. 22) and blotted on nylon membranes. Six transcripts were hybridized primarily to specific probe no. 22. To test these genes for tissue-specific expression, a Multiple Tissue cDNA panel (CLONTECH) was screened. The results of this screening are presented in Table 2 and its accompanying legend, which are published as supporting information on the PNAS web site. These transcripts with unknown functions show no or only partial homology to known gene products in the screened database. Therefore, to generate more structural information for six transcripts (Table 2), the 5′- and 3′-RACE by a modified SMART RACE technology (CLONTECH) was performed.
Analysis of Genes Expressed in the Subtractive Libraries no. 22/30.
Analysis began with a comparison of microarray data with gene expression patterns in a heterogeneous population of mouse brainstem/progenitor cells that was recently reported (13). This comparison demonstrated a potential 36.7% overlap.
Next, we compared transcripts reported in this study with the hematopoietic SCDb (14) for possible homologies. Some of the transcripts expressed in neurospheres no. 22 and no. 30, such as HuEMAP, stathmin, L12, and ERK2, have homologous transcripts in the SCDb. Some of the transcripts that were identified by microarray hybridizations, including DRB1, RASA1, VAMP2 (synaptobrevin 2), Notch 3, UBE2I, PLP, and PTPRZ1 (protein tyrosine phosphatase, receptor-type Z polypeptide1), encoded proteins belonging to the protein families with functions similar to those represented in SCDb. For example, the differentially expressed Notch 3 transcript belongs to the same family as the Notch 1 transcript, which is represented in SCDb. The differential transcript of α-1,6-fucosyltransferase (FUT8) belongs to the same family as α-3-fucosyltransferase (FUT4 or CD15), a well known marker of hematopoietic cells and radial glia (15) which might behave as NSCs (7, 16, 17). The transcript C10, differentially expressed in clone no. 30, reveals a strong nucleotide homology, from 910 to 1,209 bp, to mouse tyrosine-protein kinase Janus kinase 3 (JAK3) from the SCDb. Enhanced erythropoiesis in developing embryonic bodies is accompanied by the overexpression of calcyclin-binding protein (18). This protein is expressed differentially in clone no. 30.
These data support previous findings suggestive of overlap in the developmental programs for neuropoiesis and hematopoiesis (19). In addition, they demonstrate the efficacy of the approach used in this study for discovering differentially expressed, neurogenic gene products.
The results of this study show that differentially expressed transcripts involved in cell signaling and communication were among the most abundant (48.3%, Table 1). The participants of the Ras p21-mitogen-activated protein kinases (MAPK) pathway such as RASA1, Grb2, SOS1/SOS2, ATF2, extracellular signal-regulated kinase 2 (ERK2) are differentially expressed in clone no. 30. Among other important regulators, RAF, the MAPKKK that regulates the ERK activation, is expressed in both clones. The RAF kinase inhibitor protein was differentially expressed in clone no. 30. Moreover, comparing these data with the EGAD allows us to predict that the function encoded by two transcripts with unknown functions, F4 and C10, also could be associated with cell signaling and communication. Again, these results suggest that the present approach might facilitate functional analyses of known gene products and the discovery of genes with previously unknown function involved in distinct stages of neurogenesis.
Extended Reclustering of the Panel of cDNA Libraries with Additional Discriminating Markers.
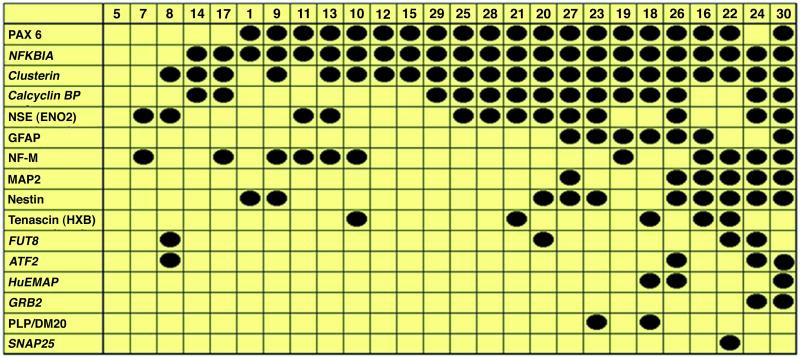
The transcripts that were revealed after subtractive hybridization were used as markers for the next series of clustering and rearrangements of the cDNA panel (Fig. 4). Two transcripts, nuclear factor of κ light chain gene enhancer in B cells inhibitor, α (NFKBIA), one of three major inhibitors of the NF-κB/Rel family of transcription factors (20), and clusterin, heterodimeric apoptosis-associated glycoprotein (21), were present in the cDNA panel with some exclusions (clones nos. 5, 7, and 8 for NFKBIA, and clones nos. 5, 7, 1, and 11 for clusterin). Other transcripts also were present within a limited number of clones. All of these transcripts, except for calcyclin-binding protein (CacyBP), were differentially expressed either in clone no. 22 or no. 30 (Fig. 4). These transcripts included the human echinoderm microtubule-associated protein (HuEMAP) that links alterations in microtubule organization during the cell cycle-to-signal transduction events (22), synaptosome-associated protein (SNAP25), and Grb2 that is responsible for the signaling of a receptor protein tyrosine kinase (R-PTK) to the Ras protein. These transcripts are expressed only on the right side of the cDNA panel where clones from putatively more mature or higher differentiated parental cells are placed. Two other transcripts, FUT8 and ATF2 were also expressed in clones located on the right side of the cDNA panel (except clone no. 8).
Fig 4.
cDNA panel rearrangement using PCR screening with new transcripts. Transcripts used for second screening (NFKBIA, clusterin, calcyclinBP, FUT8, ATF2, HuEMAP, GRB2, SNAP25) are shown in italics.
The rearrangement helped to distinguish at least four distinct clusters of clonal neurospheres. Each cluster includes clones derived from parental cells that were developmentally “close” to each other.
Cluster no. 1 includes five clones of small- and medium-size (clone nos. 5, 7, 8, 14, 17). None of these clones expressed PAX6, GFAP, MAP2, nestin, tenascin-C, HuEMAP, Grb2, PLP/DM20, or SNAP25. The main characteristic of this cluster was the lack of PAX6 expression.
Cluster no. 2 contained five clones (nos. 1, 9, 11, 13, 10) of small and medium sizes that had the following molecular phenotype: PAX6+, NFKBIA+, CacyBP−, GFAP−, NF-M+ (except no. 1), MAP2−, ATF2−, HuEMAP−, and Grb2−, PLP/DM20−, and SNAP25−. Almost all clones within this cluster expressed the neuronal markers NSE (nos. 11 and 13) or NF-M (nos. 9, 11, 13, 10), or nestin (nos. 1 and 9).
Cluster no. 3 included seven clones (nos. 12, 15, 29, 25, 28, 21, 20) of medium and large size. They have the following molecular phenotypes: PAX6+, NFKBIA+, clusterin+, GFAP−, NF-M−, MAP2−, ATF2−, HuEMAP−, Grb2−, PLP/DM20−, and SNAP25−. A predominant feature of this cluster was a high homogeneity of molecular phenotype.
Cluster no. 4, which included nine large clones (nos. 27, 23, 19, 18, 26, 16, 22, 24, 30), was characterized by the expression of NFKBIA+, clusterin+ and (CacyBP–SNAP)+ in different combinations. In addition, most of the clones in this cluster were nestin+ and expressed either MAP2 or GFAP or both. All HuEMAP+ clones expressed GFAP. Finally, the expression of DM20, an isoform of the oligodendrocyte marker PLP that is a structural component of myelin and plays an important role in the early differentiation of oligodendrocytes (23) was confirmed in two GFAP+ spheres (nos. 23 and 18).
Re-clustering of clonal cDNA libraries by molecular phenotype allows us to evaluate the informative value of markers and to classify them into three arbitrary categories:
Restricted. This category includes tenascin-C, FUT8, ATF2, HuEMAP, Grb2, PLP/DM20, and SNAP25 that were expressed in <20% of clones.
Semirestricted. This category includes CacyBP, NSE, GFAP, NF-M, MAP2, and nestin which were expressed in 20–55% of neurosphere clones.
Nonrestricted. This category includes markers such as PAX6, NFKBIA, and clusterin that were expressed in >55% of clones.
Nonrestricted markers are important for the discrimination of major classes of clusters. For example, all clusters can be subdivided into two classes: immature PAX6+ clusters and mature PAX6− clusters. Semirestricted markers such as GFAP, NSE, NF-M, MAP2, and nestin are associated with distinct cellular morphotypes.
All differentially expressed markers that were revealed after subtraction of the two neighboring clones (no. 22 and no. 30) fell into the category “restricted markers.” The majority of clones that expressed the restricted markers such as FUT8, ATF2, HuEMAP, Grb2, SNAP25, and PLP/DM20 were located in the right side of the cDNA panel (Fig. 4), in proximity to clones nos. 22 and 30. Restricted markers thus appear to be a hallmark of heterogeneity within the clones that originated from parental cells having a similar developmental potential.
Discussion
The routine selection of clonal neurospheres generated by NSCs in this culture system allowed us, in previous studies (5, 6, 24), to demonstrate an inter-clonal heterogeneity in the expression of neural lineage-specific markers. These studies led to the hypothesis that a clonal neurosphere, by itself, might be considered as a model of neurogenesis; molecular phenotyping of individual clonal progeny might thus reflect the developmental potential of parental clone-forming cells.
In this report, we use molecular phenotyping of individual clonal progeny to address the issue of whether a set of clone-forming NSCs can be subdivided into additional subsets of cells that exhibit distinct developmental commitment. The results presented here demonstrate that a population of cells from fresh clinical specimens of human brain, capable of forming clones in our in vitro system, is heterogeneous, and includes cells with distinct molecular phenotype in their progeny that is suggestive of variable developmental commitment of the parental clonogenic cells.
Molecular phenotyping of individual neurosphere clones, following several repetitive rounds of subtractive hybridization between two clonal cDNA libraries that was also coupled to microarray screening and the rearrangement of clusters using previously unused transcripts, comprise an iterative procedure or algorithm (see Fig. 6, which is published as supporting information on the PNAS web site). This approach can be used to discriminate functional clusters of clone-forming NSCs with similar developmental potential, as well as to uncover and retrieve a large number of gene products that are involved in genetic and epigenetic programs for neural development. Thus, a clonal neurosphere in our culture system can represent a functional in vitro model of neurogenesis.
Previous culture studies have demonstrated that a population of CNS stem/progenitor cells can be heterogeneous and can include a limited number of cells ranging from immature multipotential stem cells to more restricted progenitors. For example, the forebrain germinal zone and an adult remnant, the subependymal zone, contain at least two distinct populations of stem cells that are FGF2- and EGF-responsive (25), and they appear sequentially during CNS development (26). The population of human CNS clonogenic CD133+ stem cells also reveals at least two subsets of cells: 8GIhi and 8GI−/lo (27). In addition, developmental potential of neural cells also can be modified by the regulatory action of growth conditions and thus contribute to the heterogeneity of NSCs. Cell responsiveness to growth conditions and modification of cell-cycle progression are important for the modulation of epigenetic responses of cells to instructive cues presented from both extracellular and intracellular sources. Stem cells tend to have longer cell cycles than progenitor cells (28), and the selective killing of more rapidly dividing cells in vivo has been used to provide evidence for their existence in the adult CNS (25). FGF-responsive stem cells divide asymmetrically to reproduce themselves, and produce EGF-responsive stem/progenitor cells. The EGF-responsive population increases in size by asymmetric divisions of FGF-responsive cells and by symmetric divisions of EGF-responsive stem cells (25). Thus, clonal brain cell populations are not phenotypically uniform. At least two subpopulations of cells, depending on whether they were derived by means of symmetric or asymmetric division, can be distinguished during the process of clone formation.
At present, however, the proliferative kinetics of stem and progenitor cells in vitro and sequential regulatory epigenetic cascades remain to be determined. Pleiotropic growth factors such as FGF2, EGF, and insulin, which are present in our system, can affect survival, cell cycle, and differentiation potential; this can contribute to clones of different size. In the presence of EGF, the majority of clones were large in diameter, whereas in the presence of FGF2, clones were small. Culturing cells with EGF and FGF2 produce both types of clones (29). Given the fact that large clones of neuronal-restricted progenitor cells have been observed in cultures of spinal cord (30), we propose a clone-forming model that takes into account the proliferative potential of stem and more restricted progenitor cells (Fig. 5B). There is evidence that non-neural cells, e.g., keratinocytes, with characteristics of more restricted progenitor cells produce small clones (31); restricted progenitors from adult mouse brain lack self-renewal potential and give rise to clones of small size (32). This situation is depicted in Fig. 5C.
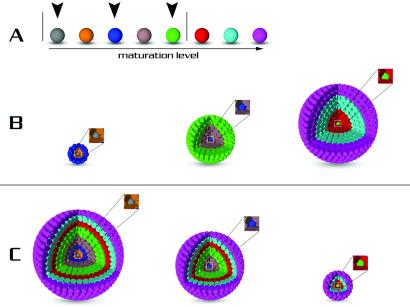
Fig 5.
Hypothetical model for the relationship between neurosphere size and maturation level of clone-forming cells. (A) A row of eight (arbitrary number) cells with varying developmental potential is arranged and color-coded according to presumed maturation level. Five cells within the upright lines can give rise to neurospheres. In this group, the gray cell represents the most immature and the green cell is the most mature clonogenic cell. Three clone-forming cells are depicted (arrowheads); in B and C, alternative architectures of neurospheres are arranged according to the clonogenic potential of the parental cell. (B) In this scenario, the stem cell is less proliferatively active than the progenitor cell. In this model, the most mature (green) cell gives rise to the largest size clone, and the smallest neurosphere contains descendants (e.g., orange and blue cells) from the most immature clonogenic (gray) cell. (C) In this case, the stem cell is more proliferatively active than the progenitor cell. This represents a situation in which the smallest sphere arises from the green neurosphere-forming cell and the largest clone from the more immature gray cell. Both of these simplified models underscore a notion that molecular phenotypic heterogeneity of neurospheres might reflect differences in the developmental potential of distinct stem/progenitor cell groups that co-exist in the brain.
These findings suggest that the size of a clone might reflect the responsiveness to growth factors and the proliferation/differentiation status of the parental clone-forming stem/progenitor cell. Regionally distinct CNS stem cells also may contribute to heterogeneity (33), with multipotent cells present in even mature areas of the CNS including the subependymal zone, septum, striatum, cortex, spinal cord, and optic nerve (7, 34–37), constituting a variety of clonogenic cells that possess distinct developmental and environmental histories (8).
The data presented here demonstrate that 26 clonal neurospheres, which were initially arranged into a panel according to their size (from small to large), display different combinations of transcripts which contribute to their distinct molecular phenotype (Fig. 2). Moreover, these clones were classified into four clusters, suggesting that at least four types of clone-forming stem/progenitor cells are present within this representative sample of clonogenic NSCs in our system (Fig. 4). Modulation of expression of different transcripts within each cluster, e.g., as a result of minor variation in growth conditions, retrospectively reflects distinct developmental potential of the clone-forming cells.
All clones included in cluster no. 1 (small and medium size) are negative for the developmentally regulated transcription factor PAX6. The expression of the PAX6 gene was confined to cells that are not terminally differentiated. We consider PAX6 as a marker of progenitor cells. Consequently, the expression of PAX6 within the panel of clones may reflect the level of maturity of a parental cell: no expression of PAX6 in clones produced by immature cells, high expression in clones formed by progenitor cells, and then again, no expression in clones produced by differentiated cells. This assumption is supported by the results that clone no. 24, which resides in the right side of the panel, does not express PAX6.
In addition to the absence of PAX6 expression, all clones included in cluster no. 1 were negative for nestin. These results are consistent with an earlier study showing that the smallest clones can be immuno-negative for nestin (5). Recently it was shown that a subpopulation of nestin-negative, clone-forming cells exists in the embryonic brain (38). We postulate that nestin expression might sequentially follow the expression of another intermediate filament protein. It is known that an initial expression of keratin K8 and K18 is followed by nestin expression in undifferentiated mammalian neuroblasts (39). We have found that K8 and K18 are expressed together or separately, starting from cluster no. 1 (data not shown).
Clones belonging to cluster no. 1 are also negative for the astroglial marker GFAP, and neuronal markers MAP2 and HuEMAP. At the same time, they were positive for the neuronal markers NSE and NF-M. These data are consistent with the observation that in the developing brain, neuronogenesis precedes gliogenesis (40). The results suggest that nestin−, PAX6− clones might be derived from the less mature cells, and that cluster no. 1 integrates those clones that are generated by the most immature and more restricted stem/progenitor cells.
It is interesting that all GFAP+ clones belong to cluster no. 4, and they are all PAX6+. PAX6 expression has been shown to be upstream from the tenascin-C gene, and PAX6 expression is required for this and other “boundary” extracellular matrix molecule expression (41). Only those clones that are positive for PAX6 (Fig. 2) also expressed tenascin-C. In cluster no. 4, these clones also express the oligodendrocyte marker PLP/DM20 that correlates with GFAP expression. In addition, a transcript, E11, which is expressed in clone nos. 22 and 30 (cluster no. 4), contains a sequence motif specific for crystallin genes, a target of PAX6 (42–45). Thus, according to molecular phenotypic characteristics, we conclude that clones included in cluster no. 4 are derived from the most restricted “mature” progenitor cells. The examples of cluster nos. 1 and 4 demonstrate the feasibility of the method for classifying clonogenic cells and their progeny based on distinct molecular phenotype.
Application of our iterative algorithm suggests that repetitive rounds of subtraction and screening, through hybridization on microarrays with differentially expressed transcripts, affords a more precise grouping of clonal cDNA libraries into functional clusters than their arrangement according to neurosphere size. The reciprocal subtraction between two closely situated clones within a panel helps to reveal transcripts that are specific for a single difference between the two clones. Each round of the algorithm brings one closer and closer to a functional ordering of clones that presumably reflects degree of developmental commitment of parental clone-forming cells.
An additional rationale for this approach includes the assumption that a varying continuum of molecular phenotype of the progeny of individual clone-forming cells reflects, retrospectively, the developmental status of parental cells. These are the cells that presumably give rise to heterogeneous populations of neurons and glia during the building of the CNS. Thus, panels of clones can be analyzed and used in dynamic models of neurogenic gene expression. Molecular phenotyping of individual clonal progeny provide a wealth of information for clustering clones, as well as for predicting the existence of clusters based on successive panel rearrangements. For example, knowing that a molecular phenotype such as 5F3(CD133)+, 5E12+, CD34−, CD45−, CD24−/lo is associated with a neural stem cell (27), one can focus on this distinct continuum for additional, extensive analyses. We believe that the approach described here, using neural as well as potentially non-neural stem/progenitor cell clones as microsystems for analyses of developmentally diverse cDNA libraries, can be useful for the discovery of genes specific for defined stages of neurodevelopmental programs.
Supplementary Material
Acknowledgments
We thank Marina Juravleva for technical assistance, Thomas Reiniger for assistance with the generation of Fig. 5, and Dr. Bjorn Scheffler for help generating the manuscript. This work was supported by National Institutes of Health/National Institute of Neurological Disorders and Stroke Grant NS37556, the McKnight Brain Institute, and the University of Florida Shands Cancer Center.
Abbreviations
EGF, epidermal growth factor
FGF2, fibroblast growth factor
NSC, neural stem cell
NSE, neuron-specific enolase
NF-M, neurofilament protein
MAP-2, microtubule-associated protein-2
GFAP, glial fibrillary acidic protein
PLP/DM20, myelin proteolipid protein
SCDb, Stem Cell Database
References
- 1.McKay R. (1997) Science 276, 66-71. [DOI] [PubMed] [Google Scholar]
- 2.Ray J., Peterson, D. A., Schinstine, M. & Gage, F. H. (1993) Proc. Natl. Acad. Sci. USA 90, 3602-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reynolds B. A. & Weiss, S. (1992) Science 255, 1707-1710. [DOI] [PubMed] [Google Scholar]
- 4.Weiss S., Dunne, C., Hewson, J., Wohl, C., Wheatley, M., Peterson, A. C. & Reynolds, B. A. (1996) J. Neurosci. 16, 7599-7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kukekov V. G., Laywell, E. D., Thomas, L. B. & Steindler, D. A. (1997) Glia 21, 399-407. [DOI] [PubMed] [Google Scholar]
- 6.Kukekov V. G., Laywell, E. D., Suslov, O., Davies, K., Scheffler, B., Thomas, L. B., O'Brien, T. F., Kusakabe, M. & Steindler, D. A. (1999) Exp. Neurol. 156, 333-344. [DOI] [PubMed] [Google Scholar]
- 7.Laywell E. D., Rakic, P., Kukekov, V. G., Holland, E. C. & Steindler, D. A. (2000) Proc. Natl. Acad. Sci. USA 97, 13883-13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steindler D. A. & Pincus, D. (2002) Lancet 359, 1047-1054. [DOI] [PubMed] [Google Scholar]
- 9.Suslov O. N., Kukekov, V. G., Laywell, E. D., Scheffler, B. & Steindler, D. A. (2000) J. Neurosci. Methods 96, 57-61. [DOI] [PubMed] [Google Scholar]
- 10.Lemischka I. (2001) Rev. Clin. Exp. Hematol. 51, 15-25. [DOI] [PubMed] [Google Scholar]
- 11.Oh I. H., Lau, A. & Eaves, C. J. (2000) Blood 96, 4160-4168. [PubMed] [Google Scholar]
- 12.Singh N., Phillips, R. A., Iscove, N. N. & Egan, S. E. (2000) Exp. Hematol. 28, 527-534. [DOI] [PubMed] [Google Scholar]
- 13.Geschwind D. H., Ou, J., Easterday, M. C., Dougherty, J. D., Jackson, R. L., Chen, Z., Antoine, H., Terskikh, A., Weissman, I. L., Nelson, S. F. & Kornblum, H. I. (2001) Neuron 29, 325-339. [DOI] [PubMed] [Google Scholar]
- 14.Phillips R. L., Ernst, R. E., Brunk, B., Ivanova, N., Mahan, M. A., Deanehan, J. K., Moore, K. A., Overton, G. C. & Lemischka, I. R. (2000) Science 288, 1635-1640. [DOI] [PubMed] [Google Scholar]
- 15.Gocht A. (1992) Acta Anat. 145, 434-441. [DOI] [PubMed] [Google Scholar]
- 16.Hartfuss E., Galli, R., Heins, N. & Gotz, M. (2001) Dev. Biol. 229, 15-30. [DOI] [PubMed] [Google Scholar]
- 17.Heins N., Malatesta, P., Cecconi, F., Nakafuku, M., Tucker, K. L., Hack, M. A., Chapouton, P., Barde, Y. A. & Gotz, M. (2002) Nat. Neurosci. 5, 308-315. [DOI] [PubMed] [Google Scholar]
- 18.Xia Z. B., Dai, M. S., Magoulas, C., Broxmeyer, H. E. & Lu, L. (2000) J. Hematother. Stem Cell Res. 9, 651-658. [DOI] [PubMed] [Google Scholar]
- 19.Terskikh A. V., Easterday, M. C., Li, L., Hood, L., Kornblum, H. I., Geschwind, D. H. & Weissman, I. L. (2001) Proc. Natl. Acad. Sci. USA 98, 7934-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Memet S., Laouini, D., Epinat, J. C., Whiteside, S. T., Goudeau, B., Philpott, D., Kayal, S., Sansonetti, P. J., Berche, P., Kanellopoulos, J. & Israel, A. (1999) J. Immunol. 163, 5994-6005. [PubMed] [Google Scholar]
- 21.Park I. S., Che, Y. Z., Bendayan, M., Kang, S. W. & Min, B. H. (1999) J. Endocrinol. 162, 57-65. [DOI] [PubMed] [Google Scholar]
- 22.Li Q. & Suprenant, K. A. (1994) J. Biol. Chem. 269, 31777-31784. [PubMed] [Google Scholar]
- 23.Yang X. & Skoff, R. P. (1997) J. Neurosci. 17, 2056-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ignatova T. N., Kukekov, V. G., Laywell, E. D., Suslov, O. N., Vrionis, F. D. & Steindler, D. A. (2002) Glia 39, 191-204. [DOI] [PubMed] [Google Scholar]
- 25.Martens D. J., Tropepe, V. & van Der Kooy, D. (2000) J. Neurosci. 20, 1085-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciccolini F. (2001) Mol. Cell. Neurosci. 17, 895-907. [DOI] [PubMed] [Google Scholar]
- 27.Uchida N., Buck, D. W., He, D., Reitsma, M. J., Masek, M., Phan, T. V., Tsukamoto, A. S., Gage, F. H. & Weissman, I. L. (2000) Proc. Natl. Acad. Sci. USA 97, 14720-14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison S. J., Shah, N. M. & Anderson, D. J. (1997) Cell 88, 287-298. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S. C., Lipsitz, D. & Duncan, I. D. (1998) J. Neurosci. Res. 54, 181-190. [DOI] [PubMed] [Google Scholar]
- 30.Mayer-Proschel M., Kalyani, A. J., Mujtaba, T. & Rao, M. S. (1997) Neuron 19, 773-785. [DOI] [PubMed] [Google Scholar]
- 31.Zhu A. J., Haase, I. & Watt, F. M. (1999) Proc. Natl. Acad. Sci. USA 96, 6728-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiasson B. J., Tropepe, V., Morshead, C. M. & van der Kooy, D. (1999) J. Neurosci. 19, 4462-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Temple S. (2001) Nature 414, 112-117. [DOI] [PubMed] [Google Scholar]
- 34.Gage F. H., Coates, P. W., Palmer, T. D., Kuhn, H. G., Fisher, L. J., Suhonen, J. O., Peterson, D. A., Suhr, S. T. & Ray, J. (1995) Proc. Natl. Acad. Sci. USA 92, 11879-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer T. D., Ray, J. & Gage, F. H. (1995) Mol. Cell. Neurosci. 6, 474-486. [DOI] [PubMed] [Google Scholar]
- 36.Palmer T. D., Markakis, E. A., Willhoite, A. R., Safar, F. & Gage, F. H. (1999) J. Neurosci. 19, 8487-8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shihabuddin L. S., Ray, J. & Gage, F. H. (1997) Exp. Neurol. 148, 577-586. [DOI] [PubMed] [Google Scholar]
- 38.Kawaguchi A., Miyata, T., Sawamoto, K., Takashita, N., Murayama, A., Akamatsu, W., Ogawa, M., Okabe, M., Tano, Y., Goldman, S. A. & Okano, H. (2001) Mol. Cell. Neurosci. 17, 259-273. [DOI] [PubMed] [Google Scholar]
- 39.Leake D., Asch, W. S., Canger, A. K. & Schechter, N. (1999) Differentiation (Berlin) 65, 181-189. [DOI] [PubMed] [Google Scholar]
- 40.Qian X., Shen, Q., Goderie, S. K., He, W., Capela, A., Davis, A. A. & Temple, S. (2000) Neuron 28, 69-80. [DOI] [PubMed] [Google Scholar]
- 41.Stoykova A., Gotz, M., Gruss, P. & Price, J. (1997) Development (Cambridge, U.K.) 124, 3765-3777. [DOI] [PubMed] [Google Scholar]
- 42.Cvekl A., Kashanchi, F., Sax, C. M., Brady, J. N. & Piatigorsky, J. (1995) Mol. Cell. Biol. 15, 653-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cvekl A., Sax, C. M., Li, X., McDermott, J. B. & Piatigorsky, J. (1995) Proc. Natl. Acad. Sci. USA 92, 4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meech R., Kallunki, P., Edelman, G. M. & Jones, F. S. (1999) Proc. Natl. Acad. Sci. USA 96, 2420-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson J., Cvekl, A. & Wistow, G. (1995) Proc. Natl. Acad. Sci. USA 92, 4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.