Abstract
Genes encoding a homolog of Escherichia coli OxyR (oxyR) and an alkyl hydroperoxide reductase system (ahpC and ahpD) have been isolated from Streptomyces coelicolor A3(2). The ahpC and ahpD genes constitute an operon transcribed divergently from the oxyR gene. Expression of both ahpCD and oxyR genes was maximal at early exponential phase and decreased rapidly as cells entered mid-exponential phase. Overproduction of OxyR in Streptomyces lividans conferred resistance against cumene hydroperoxide and H2O2. The oxyR mutant produced fewer ahpCD and oxyR transcripts than the wild type, suggesting that OxyR acts as a positive regulator for their expression. Both oxyR and ahpCD transcripts increased more than fivefold within 10 min of H2O2 treatment and decreased to the normal level in 50 min, with kinetics similar to those of the CatR-mediated induction of the catalase A gene (catA) by H2O2. The oxyR mutant failed to induce oxyR and ahpCD genes in response to H2O2, indicating that OxyR is the modulator for the H2O2-dependent induction of these genes. Purified OxyR protein bound specifically to the intergenic region between ahpC and oxyR, suggesting its direct role in regulating these genes. These results demonstrate that in S. coelicolor OxyR mediates H2O2 induction of its own gene and genes for alkyl hydroperoxide reductase system, but not the catalase gene (catA), unlike in Escherichia coli and Salmonella enterica serovar Typhimurium.
OxyR is an H2O2-sensing transcriptional regulator inducing more than 10 genes in response to H2O2 in Escherichia coli and Salmonella enterica serovar Typhimurium (15, 43). It is activated by a disulfide bond formation between two cysteine residues and induces the expression of oxyS (which encodes a small, nontranslated regulatory RNA), katG (which encodes hydrogen peroxidase I), ahpC (which encodes alkyl hydroperoxide reductase), gorA (which encodes glutathione reductase), dps (which encodes DNA binding protein), and grxA (which encodes glutaredoxin 1). Glutaredoxin 1 deactivates OxyR by reducing the disulfide bond, forming an autoregulatory feedback loop (49). Irrespective of its redox state, OxyR also acts as a repressor of its own expression like other LysR family of transcriptional regulators do. Several oxyR genes have been identified in other organisms, such as Haemophilus influenzae (35), various Mycobacterium species (20, 22, 36), Xanthomonas species (34), and even the anaerobic bacterium Bacteroides fragilis (40).
In Mycobacterium and Xanthomonas species, the oxyR gene is tightly linked to the genes for alkyl hydroperoxide reductase system. In E. coli and S. enterica serovar Typhimurium, the alkyl hydroperoxide reductase system is composed of two components, AhpC (22 kDa) and AhpF (54 kDa) (30). The reduced form of AhpC converts alkyl hydroperoxides to the corresponding alcohols with concomitant oxidation of the two sulfhydryls to a disulfide bond between two subunits. It has recently been reported that AhpC is the main defense system against endogenously generated hydrogen peroxide (18, 19). The oxidized AhpC contains two intersubunit disulfide bonds per dimer (39). AhpF, which shows homology to the thioredoxin reductase family, reduces the oxidized AhpC by transferring reducing equivalents from NAD(P)H to the disulfide of AhpC. The reduction of AhpC is mediated by two cysteine disulfide centers in AhpF (7). Likewise, in Bacillus subtilis (2, 5) and Xanthomonas species (34), AhpF is involved in the reduction of AhpC. In Xanthomonas species, the ahpF, oxyR, and orfX genes are arranged in an operon, and the ahpC gene is located upstream of ahpF as a monocistronic transcription unit. On the other hand, in Mycobacterium species, the ahpC gene is divergently transcribed from the oxyR gene, whereas the ahpF homologue has not been identified. Instead, the ahpD gene, encoding a protein with a thioredoxin fold, is located downstream of ahpC (48). It has recently been reported that AhpD in complex with Lpd (dihydrolipoamide dehydrogenase) and SucB (dihydrolipoamide succinyltransferase) reduces AhpC in an NADH-dependent manner (4).
AhpC homologues, named thiol-specific antioxidant or thioredoxin peroxidase (TPx), are also distributed among eukaryotic organisms (8, 10). In Saccharomyces cerevisiae, two AhpC homologous proteins, Tsa1p and Ahp1p, have been identified (9, 11, 32). Both proteins form intermolecular disulfide bonds, which can be specifically reduced by thioredoxin. Therefore, thioredoxin and the thioredoxin reductase system have the function of AhpF in this organism. In spite of the similarity in structure and activation mechanism between Tsa1p and Ahp1p, their activity is known to be specific for H2O2 and organic peroxide, respectively, in budding yeast.
Streptomyces is a gram-positive soil bacterium with high GC content which undergoes a complex cycle of morphological and physiological differentiation during growth. In previous studies, adaptive response to H2O2 was observed in Streptomyces coelicolor (33). Two-dimensional protein gel analysis revealed that S. coelicolor induced synthesis of more than 100 proteins when exposed to H2O2, and the catA gene encoding catalase A was identified as one of the H2O2-inducible genes (13, 17). The production of catalase A, which is the major vegetative catalase, is regulated by a peroxide-sensing repressor, CatR (26). Another peroxide-sensing transcriptional regulator found in S. coelicolor, RsrA, is an antisigma factor for σR, which directs the expression of thioredoxin genes (31, 37, 38). In this study, we present our finding of a third peroxide sensor in this organism, OxyR, and its role in regulating the expression of alkyl hydroperoxide reductase and its own gene product.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. coelicolor M145 and Streptomyces lividans TK24 cells were grown as described previously (29). E. coli DH5α and BL21(DE3)pLysS were used for cloning and overexpression, respectively. E. coli ET12567 was used to prepare unmethylated DNA to transform S. coelicolor. XL1-Blue MRA was used as a host for the λEMBL3 genomic library of S. coelicolor.
Cloning of ahpC, ahpD, and oxyR genes.
An internal ahpC gene fragment of 407 bp was generated from S. coelicolor by PCR using primers ACN (5′TTCTTCTGGCC[C/G]AAGGACTTCAC3′) and ACC (5′TTCAGAGT[C/G]GGGTCGCCGTT[C/G]C3′) designed from the conserved regions among known bacterial AhpC proteins. The PCR product was used as a probe to screen the λEMBL3 genomic library of S. coelicolor M145. The common 3.8-kb PstI fragment in positive clones was sequenced and found to contain ahpC, ahpD, and oxyR genes (Fig. 1A). The nucleotide sequence information was deposited in GenBank under accession number AF186371.
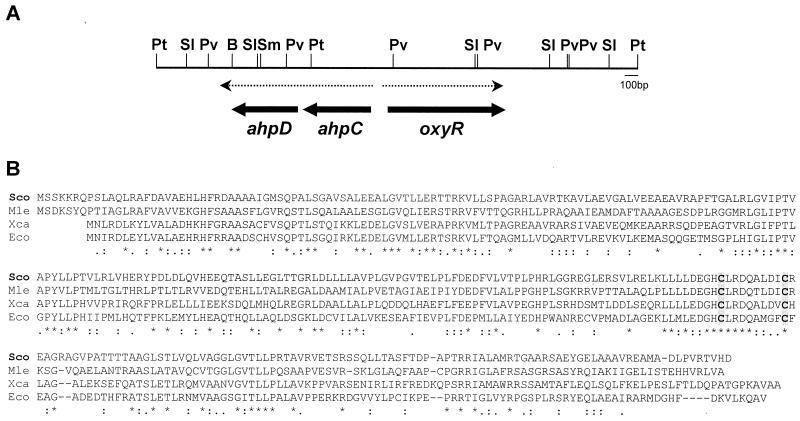
FIG. 1.
(A) Restriction map and organization of the ahpC, ahpD, and oxyR genes. A restriction map of the 3.8-kb PstI fragment containing the ahpCD and oxyR genes is presented. Thick arrows indicate the positions and directions of the three genes. The two divergent transcripts are shown as dashed arrows. Abbreviations: B, BamHI; Pt, PstI; Pv, PvuII; SI, SalI; Sm, SmaI. (B) Comparison of the predicted amino acid sequence of OxyR with its homologues. The amino acid sequence of OxyR from S. coelicolor (Sco) (AF186371) was aligned with those from M. leprae (Mle) (Al035300), E. coli (Eco) (J04553), and X. campestris (Xca) (U94336). The position of two cysteine residues involved in disulfide bond formation and activation of OxyR in E. coli are shaded and presented in boldface type.
RNA isolation.
RNA was isolated from M145 cells grown in YEME (29). Cells were resuspended in modified Kirby mixture (1% sodium-triisopropyl naphthalene sulfonate, 6% sodium 4-amino salicylate, 6% phenol equilibrated with 10 mM Tris-HCl buffer [pH 8.3]) and disrupted by sonication with a microtip (Sonics and Materials Inc.) at 25% of the maximum amplitude (600 W, 20 kHz).
Northern blot analysis.
Northern blot analysis of the ahpC transcript was performed according to standard procedures (41). Fifty micrograms of RNA was electrophoresed on a 1.2% agarose gel containing formamide. The probe used for detecting ahpC transcript was a 597-bp ahpC gene fragment, produced by PCR using primers ACON (5′TTGGAGAGCATATGCTCACTGTCG3′) and ACOC (5′CGGACTTCAGGGATCCGAGGGAC3′), and was labeled with [α-32P]dATP.
S1 nuclease protection analysis.
To generate the probe for S1 mapping the 5′ end of the ahpCD transcript, a 261-bp fragment was amplified by PCR using primers ACS1 (5′GGCGGTCAGGTCGAACTCGGGG3′) and OXYS1 (5′CACGGAAGTGCAGGTGCTCGG3′) from pJH101 containing a 3.7-kb SmaI fragment in pUC18 (Fig. 2D). The amplified fragment was end labeled and digested with PvuII, and the 218-bp probe was uniquely labeled at the ACS1 5′ end 45 nucleotides (nt) downstream from the start codon and was eluted from the agarose gel. For the oxyR probe, an 802-bp fragment was generated by PCR using primers OXYS1 and ACOC from pJH101. Since the 5′ end of the OXYS1 primer is located upstream of the NarI site used to disrupt the oxyR gene, this probe can detect transcripts generated from the oxyR promoter in oxyR mutants as well. The amplified fragment was end labeled and digested with PstI to prepare a 691-bp probe uniquely labeled at the OXYS1 5′ end, which is 79 nt downstream from the start codon. S1 mapping analysis was carried out with 5 to 50 μg of RNA as described previously (42). The hybridization products were analyzed on sequencing gels with the sequencing ladder generated from the primers ACS1 or OXYS1 and pJH101 as a template. To S1 map the catA gene, a 0.6-kb SalI/BglII fragment uniquely labeled at the BglII site was used as a probe as previously described (26).
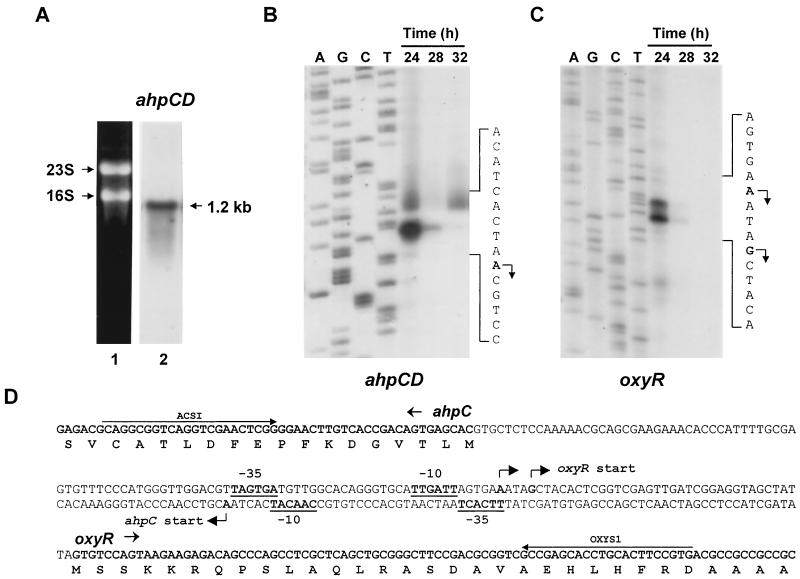
FIG. 2.
Transcription of the ahpCD and oxyR genes. (A) Northern blot analysis of the ahpC-ahpD transcript. RNA was isolated from S. coelicolor M145 cells grown in YEME for 24 h. Fifty micrograms of RNA was loaded on 1.2% agarose gel containing formamide, and Northern blot analysis was carried out using a 0.6-kb ahpC DNA probe as described in Materials and Methods (lane 2). Lane 1 shows 23S and 16S rRNA bands stained with ethidium bromide. (B and C) High-resolution S1 mapping of the 5′ ends of ahpCD (B) and oxyR (C) mRNA. RNAs prepared from M145 cells grown in YEME at 30°C for 24, 28, and 32 h, were subjected to S1 mapping analysis as described in Materials and Methods. The protected fragments were analyzed on sequencing gels with sequencing ladders generated from the same primers and templates used for the preparation of the probes. The transcription start sites are shown in boldface type and designated by arrows on the sense sequence of each transcript. (D) Nucleotide sequence of the intergenic region of oxyR and ahpCD. The oxyR sense strand sequence is presented, except in the center line, where both strands are presented to show divergent promoter elements. Transcription start sites of the ahpCD (ahpCp) and oxyR (oxyRp) are indicated by bent arrows. The putative −10 and −35 elements of ahpCD and oxyR promoters are in boldface type and underlined. Primers used to generate S1 probes (ACS1 and OXYS1) are indicated by arrows.
Disruption of the oxyR gene in S. coelicolor.
A NarI/SalI fragment (0.4 kb) of the oxyR gene cloned in pUC18 (pJH110) was excised as a HindIII fragment using polylinker sites and cloned into pKC1139 (3), which contains a temperature-sensitive replication origin, resulting in pJH405. The pJH405 plasmid DNA was prepared from E. coli ET12567 and then introduced into S. coelicolor M145 protoplasts. The transformants were selected on plates containing apramycin (50 μg/ml) at 30°C. Spores of the transformants were plated on NA medium (29) containing apramycin and incubated at 37°C for 2 days to select plasmid-integrated clones. Disruption of the oxyR gene was confirmed by Southern hybridization.
Overproduction and partial purification of OxyR.
Mutagenic primers OXYON (5′AGGTAGCTACATATGTCCAGTAAG3′; the NdeI site is underlined) and OXYOB (5′GGGTGGTCGCCGGATCCCTCA3′; the BamHI site is underlined) were used to amplify the oxyR coding region by PCR. The 972-bp PCR product digested with NdeI and BamHI was cloned into pET21c (Novagen) to generate pJH4. E. coli BL21(DE3)pLysS cells harboring pJH4 were grown in 200 ml of Luria-Bertani medium to an A600 of 0.5 and treated with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. Following harvest, cells were resuspended in lysis buffer (20 mM Tris-HCl [pH 7.9], 0.15 M NaCl, 5 mM EDTA, 0.1 mM dithiothreitol [DTT], 10 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 10% glycerol) and disrupted by sonication. The lysate was centrifuged at 16,000 × g for 10 min, and the pellet was washed with lysis buffer containing 2% sodium deoxycholate and recentrifuged. The washed pellet was dissolved in lysis buffer containing 8 M urea, and the insoluble residue was removed by centrifugation at 16,000 × g for 10 min. The dissolved protein was dialyzed twice for 8 h against 10 volumes of lysis buffer at 4°C. The dialyzed extract was centrifuged at 16,000 × g for 20 min to remove any precipitated material, and the clarified solution was loaded onto 10 ml of heparin-Sepharose (CL 6B column). Proteins were eluted using a gradient of 0.2 to 1.0 M NaCl in TGED buffer (10 mM Tris-HCl [pH 7.9], 0.1 mM EDTA, 0.1 mM DTT, 10% glycerol). OxyR was eluted with NaCl at a concentration between 0.4 and 0.6 M NaCl. The fractions enriched in OxyR were pooled and dialyzed against the storage buffer (10 mM Tris-HCl [pH 7.9], 0.1 mM EDTA, 10 mM MgCl2, 0.1 M KCl, 50% glycerol).
Gel mobility shift assay.
The DNA fragment spanning the ahpC-oxyR intergenic region was generated by PCR using primers ACS1 (5′ end at position −182 relative to the oxyR start codon) and OXYS1 (5′ end at position +79). The promoter fragment of the furA-catC operon spanning from nt −99 to +92 relative to the furA start site was also generated by PCR as described previously (27). The PCR product was end labeled with [γ-32P]ATP and T4 polynucleotide kinase. A 0.6-kb SalI/BglII fragment containing the catA promoter up to nt −364 was end labeled with [α-32P]dATP and Klenow enzymes according to the method of Hahn et al. (26). The unlabeled isotope was removed by centrifugation through a Sephadex G-50 spin column. The labeled probe was incubated with 200 ng of purified OxyR in 20 μl of binding buffer [25 mM Tris-HCl (pH 7.8), 0.5 mM EDTA, 6 mM MgCl2, 50 mM KCl, 0.5 mM DTT, 100 μg of poly(dI-dC) per ml, 5% glycerol] at 30°C for 10 min. The DNA-protein mixture was electrophoresed on a 4% native polyacrylamide gel in 20 mM Tris-borate buffer. The gels were dried and analyzed by autoradiography.
Expression of the ahpC, ahpD, and oxyR genes in S. lividans.
The recombinant plasmid pJHOxyR was constructed by cloning a 2.6-kb PstI fragment containing the oxyR gene into the PstI site of pIJ702, a Streptomyces multicopy plasmid vector. To generate pJHAhpCD containing the complete ahpC and ahpD genes, a 1.9-kb PstI/PvuII fragment was cloned into pUC18 and then into the EcoRI/HindIII sites of pIJ718, a pIJ702-derivative containing modified polycloning sites (donated by Y.-H. Cho). To generate pJHAhpC carrying the complete ahpC gene, a 0.8-kb PvuII fragment was cloned into pUC18 and then into pIJ718 using the EcoRI and HindIII sites in the polylinker region. Preparation of protoplast and transformation were done as described elsewhere (29). The transformants were selected and maintained in the presence of thiostrepton (50 μg/ml).
Western blot analysis.
Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the gel was soaked in transfer buffer (25 mM Tris, 192 mM glycine, 20% [vol/vol] methanol) for 10 min and then transferred to nitrocellulose membrane (BA79; Schleicher & Schuell) at 60 V for 60 min using Trans-Blot Cell (Bio-Rad). The membrane was blocked for 1 h in Tris-buffered saline containing 0.1% Triton X-100 (TBST) supplemented with 0.5% bovine serum albumin. The membrane was then incubated with a 1:10,000 dilution of polyclonal mouse antibodies raised against AhpC and AhpD for 1 h and then washed twice with TBST for 10 min each. The reacting signal was detected by goat anti-mouse immunoglobulin G conjugated with horseradish peroxidase using a Western ECL detection system (Amersham Biosciences, Ltd.).
RESULTS
Cloning and sequence analysis of the oxyR and ahpCD genes in S. coelicolor.
A gene fragment containing an ahpC gene homologue was isolated from the λEMBL3 genomic library of S. coelicolor A3(2) M145. The nucleotide sequence analysis of the common 3.8-kb PstI fragment from positive clones revealed that this region contains three open reading frames (ORFs), one showing high homology to known ahpC genes and another showing high homology to known oxyR genes (Fig. 1A).
The ahpC gene encodes a protein of 184 amino acids with a calculated molecular mass of 20,679 Da. This protein is highly homologous to other known bacterial alkyl hydroperoxide reductases (AhpC; about 60% identity with those from Mycobacterium spp.) and its eukaryotic homologues known as thiol-specific antioxidants or thioredoxin peroxidases. Two active-site cysteine residues are conserved among these AhpC homologues. The ahpD gene is located 7 nt downstream of the ahpC gene, encoding a protein of 178 amino acids (19,000 Da) with significant homology (about 55 to 80% identity) to ahpD genes also located downstream of the ahpC gene in Mycobacterium tuberculosis, Mycobacterium bovis, and Streptomyces viridosporus.
The oxyR gene is located 138 nt upstream from the ahpC gene in a divergent orientation. It encodes a protein of 313 amino acids (33,096 Da) showing homology to other known OxyR proteins from E. coli (16), Mycobacterium leprae, H. influenza (35), Xanthomonas campestris, and S. viridosporus (Fig. 1B). Two cysteine residues (C199 and C208) known to be involved in the disulfide bond formation and activation of OxyR in E. coli are also conserved in the S. coelicolor OxyR protein (C206 and C215).
The organization of the ahpC, ahpD, and oxyR genes in S. coelicolor is the same as those found in S. viridosporus and several Mycobacterium species such as M. leprae, M. tuberculosis, and M. bovis (36, 48). In M. tuberculosis, oxyR is naturally inactivated by multiple mutations (20). In M. leprae, translation of the region downstream of ahpC gene revealed the presence of an AhpD homologue. However, this ORF contains a frameshift and hence is interrupted by stop codons. It is not certain whether this truncation is due to a sequencing error.
Analysis of transcripts from ahpC, ahpD, and oxyR.
Since the start codon of the ahpD coding region is located just 7 nt downstream of the ahpC stop codon, the possibility of their cotranscription was examined by Northern hybridization analysis. The size of the mRNA hybridized with the randomly labeled ahpC gene fragment was about 1.2 kb (Fig. 2A), indicating that the ahpC and ahpD genes are cotranscribed from a single promoter.
To determine the transcription start site, S1 nuclease mapping analysis was done. The RNA samples were prepared from M145 cells grown for 24, 28, and 32 h in YEME. The examined time span corresponded to the growth from the early to the mid-exponential phases. When hybridized with the 168-bp S1 probe, end labeled at the 5′ end of primer ACS1, a single species of protected band was detected, suggesting that the transcription starts from the A residue 63 nt upstream of the ahpC start codon (Fig. 2B). The level of ahpCD transcript was high at the early exponential phase and decreased rapidly when cells entered the mid-exponential phase. Putative −35 (TTCACT) and −10 (CAACAT) promoter elements resembling EσhrdB-type consensus sequences (TTGaCA-N17-18-TAgaaT, lowercase letters indicate less conserved nucleotides) were identified upstream of the transcription start site (Fig. 2D).
The transcription start site of the oxyR gene was also determined using the same RNA samples (Fig. 2C). Two species of protected bands were detected with the 691-bp S1 probe end labeled at the 5′ end of primer OXYS1 (Fig. 2D). The transcription start sites of the oxyR gene were mapped to the A and G residues located at 40 and 36 nt upstream of the oxyR start codon, respectively (Fig. 2D). The expression pattern of oxyR mRNA during growth was similar to that of ahpCD mRNA. Putative −35 (TAGTGA) and −10 (TTGATT) promoter elements resembling EσhrdB-type consensus sequences were identified upstream of the transcription start site (Fig. 2D).
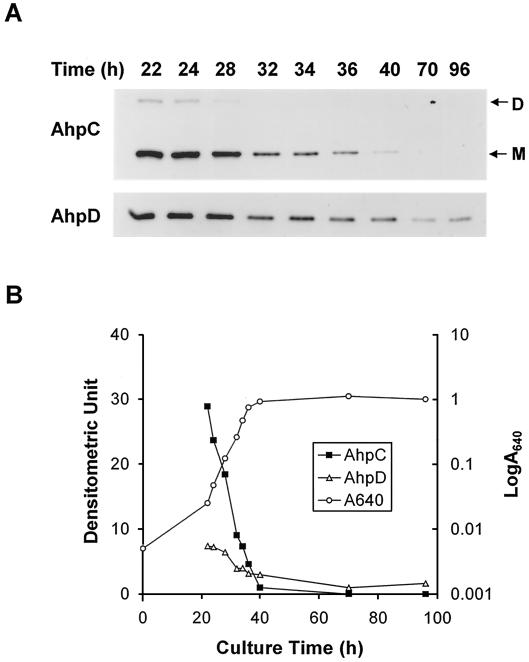
AhpC and AhpD proteins were overproduced in E. coli using the pET21c overexpression vector. The apparent molecular masses determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis were a little higher (AhpC, 25 kDa; AhpD, 21 kDa) than those predicted form the nucleotide sequences. The overexpressed proteins were used to raise mouse antibodies. The change in the level of AhpC and AhpD proteins was then monitored during growth by immunoblot analysis (Fig. 3). The decrease in the level of AhpC and AhpD proteins during growth reflected their mRNA levels. The change in the level of AhpC protein was more dramatic than that in the AhpD protein levels, most likely reflecting their difference in stability.
FIG. 3.
Growth-dependent expression of AhpC and AhpD proteins. M145 cells were grown in YEME. At various time points aliquots were taken for measurement of optical density at 640 nm and lysed to prepare cell extracts. (A) The AhpC and AhpD proteins were detected by Western blot analysis as described in Materials and Methods. The positions of the monomer (M) and dimer (D) of AhpC are shown. (B) The growth curve and the relative band intensity of the densitometric tracing data in panel A are presented. The densitometric value was set to 1 for AhpC at 40 h (closed square) and for AhpD at 70 h (open triangle).
Effect of OxyR and AhpCD overproduction.
To investigate the role of AhpC, AhpD, and OxyR in defense against oxidative stress, we introduced these genes into S. lividans TK24 on multicopy plasmid pIJ702 and investigated the effect of their overproduction on the synthesis of various antiperoxide enzymes and resistance against oxidants.
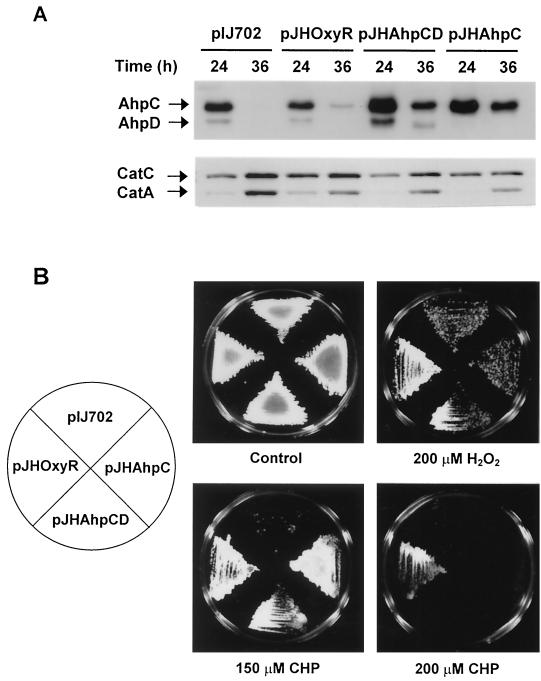
The level of antiperoxide enzymes in cells grown on NA plate for 24 and 36 h, corresponding to the substrate mycelium and aerial mycelium stage, was examined by immunoblotting (Fig. 4A). Control cells harboring pIJ702 vector only exhibited growth phase-dependent expression of AhpC and AhpD on NA plate as observed in liquid culture of S. coelicolor. Overproduction of OxyR led to prolonged synthesis of AhpC and reduced production of CatA as observed at 36 h. When AhpC and AhpD (pJHAhpCD) or AhpC (pJHAhpC) alone were overproduced, catalase A and catalase C expression decreased compared with that observed in the control cells. These results imply the existence of a mechanism balancing the expression of AhpC and catalases. Synthesis of other antioxidant enzymes such as superoxide dismutases (SodN and SodF) and glucose-6-phosphate dehydrogenase were not affected by the elevation of OxyR or AhpC levels (data not shown).
FIG. 4.
Overproduction of AhpC, AhpD, and OxyR and their contribution to resistance against oxidants. (A) Overproduction of AhpC and AhpD proteins. S. lividans cells containing oxyR (pJHOxyR), ahpCD (pJHAhpCD), or ahpC (pJHAhpC) genes on multicopy plasmid pIJ702 were grown on NA plates containing thiostrepton (50 μg/ml) until they formed substrate mycelium (24 h) or aerial mycelium (36 h). The amount of CatA, CatC, AhpC, and AhpD proteins in cell extracts was determined by Western blot analysis. (B) Effect of overproduction on resistance against hydrogen peroxide and cumene hydroperoxide. About 106 spores of cells harboring plasmids were transferred to plates of NA medium containing 200 μM hydrogen peroxide, 150 to 200 μM cumene hydroperoxide (CHP), or no oxidants (control) with thiostrepton (50 μg/ml). Cells were incubated at 30°C for 3 days.
We then examined the effect of these genes on resistance against oxidative stress by plating spores on NA plates containing various oxidants (Fig. 4B). Overproduction of AhpC alone was enough to confer resistance to cumene hydroperoxide but not to H2O2. Cells containing pJHOxyR were more resistant to cumene hydroperoxide and to H2O2 than the cells containing pJHAhpC or pJHAhpCD. These results demonstrates that (i) AhpC can function in removing the toxic effect of organic peroxide as predicted and (ii) OxyR induces the defense system against H2O2 and organic peroxide, involving further antioxidant proteins in addition to AhpCD and catalases.
Positive regulation of ahpCD and oxyR genes by OxyR.
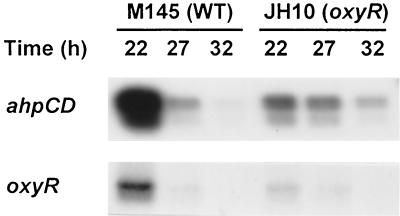
To examine the role of OxyR in transcriptional regulation, oxyR gene was disrupted by integration of a pKC1139-derived recombinant plasmid containing an internal fragment of oxyR gene. The growth rate of the oxyR disruptant (JH10) was significantly reduced compared to the wild type, similar to the behavior of E. coli oxyR mutant (24). Furthermore, JH10 exhibited the bald phenotype with a failure in the formation of aerial mycelium on R2YE plates (29). It differentiated normally on SFM or minimal medium (29). When the level of ahpCD and oxyR transcripts in wild type (M145) and oxyR mutant (JH10) was examined by S1 mapping, we found that both transcripts were markedly decreased in the mutant, implying that OxyR acts as a positive regulator of both ahpCD and oxyR gene expression (Fig. 5). For oxyR mutant, the oxyR-specific probe detects nonfunctional RNA containing only the N-terminal part of oxyR gene and some vector sequence. Hence, there is a possibility that some probable change in RNA stability may interfere with accurate measurement. Expression of other genes such as catC (which codes for catalase-peroxidase), catA (which codes for catalase A), sodN (which codes for Ni-superoxide dismutase [SOD]), sodF (which codes for Fe-SOD), and zwf (which codes glucose-6-phosphate dehydrogenase) were not affected by the loss of oxyR (data not shown).
FIG. 5.
Effect of oxyR mutation on the production of ahpCD and oxyR transcripts. M145 (WT) and JH10 (oxyR mutant) cells were grown in YEME for 22, 27, and 32 h. The amounts of ahpCD and oxyR transcripts were determined by S1 mapping as described in the text.
H2O2 induction of ahpCD and oxyR genes mediated by OxyR.
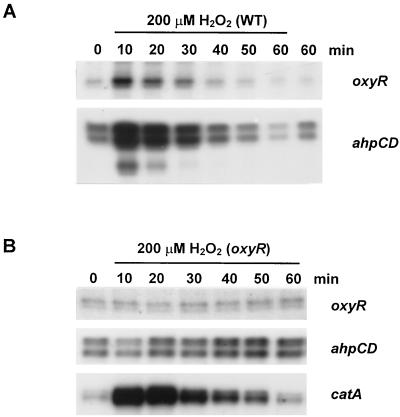
H2O2 inducibility of ahpCD and oxyR genes in S. coelicolor was then investigated. Wild-type S. coelicolor cells were grown to early exponential phase and treated with 200 μM H2O2 for different lengths of time up to 1 h. The levels of ahpCD and oxyR transcripts were analyzed by S1 mapping (Fig. 6A). The level of both oxyR and ahpCD transcripts increased more than fivefold within 10 min of H2O2 treatment, returning to the prestimulus level in 50 min. The level of AhpC and AhpD proteins increased about twofold within 30 min of H2O2 induction as monitored by immunoblot analysis (data not shown).
FIG. 6.
OxyR-dependent induction of the ahpCD and oxyR genes by H2O2. Induction of ahpCD and oxyR by H2O2 in wild-type (A) and oxyR mutant (B) cells. S. coelicolor M145 and JH10 cells were grown in YEME to the early exponential phase and treated with 200 μM H2O2. Samples were taken at 10-min intervals over 1 h, and S1 mapping analysis was carried out for oxyR and ahpCD transcripts.
In the oxyR mutant (JH10), neither ahpCD nor oxyR was induced by H2O2 treatment, demonstrating that H2O2 induction of these genes is mediated by OxyR (Fig. 6B). The gene for catalase A (catA), known to be induced by H2O2 via inactivation of repressor CatR (26), was induced normally in oxyR mutant. These results clearly demonstrate that the two separate peroxide-sensing regulators independently control the expression of two different antiperoxide enzymes in S. coelicolor.
Binding of OxyR protein to the oxyR-ahpCD intergenic region.
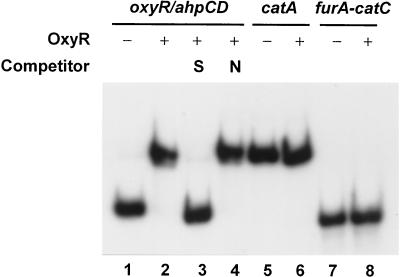
Participation of OxyR as a direct regulator of ahpCD and oxyR gene expression was further examined by DNA binding analysis. OxyR protein was purified from E. coli following overproduction using the pET system. The binding activity to the promoter region of ahpCD-oxyR, as well as the catA and furA-catC genes, was examined by gel mobility shift assay. As presented in Fig. 7, the purified OxyR bound specifically only to the ahpCD-oxyR intergenic region, but not to catA or furA-catC promoters. The result suggests that OxyR most likely regulates ahpCD and its own gene transcription via direct interaction with the binding site located in the intergenic promoter region.
FIG. 7.
Binding of OxyR to ahpCD-oxyR intergenic region. A gel mobility shift assay of OxyR binding to different promoter fragments was performed. Two hundred nanograms of purified OxyR (6 pmol) was incubated with 32P-labeled promoter fragments of oxyR-ahpCD, catA, or furA-catC as described in Materials and Methods. OxyR binding was detected by electrophoresis on 4% polyacrylamide gel and autoradiography. Lanes 1, 5, and 7 contain only the radiolabeled probes. Probes were incubated with OxyR without any competitors except poly(dI-dC) (lanes 2, 6, and 8) or with a 200-fold molar excess of unlabeled probe DNA (S; lane 3) or nonspecific DNA fragments (N; lane 4).
DISCUSSION
In the present study, the ahpCD and oxyR genes were isolated from S. coelicolor, and OxyR was proposed as a positive regulator of these genes. In S. coelicolor, the oxyR gene is transcribed divergently from the ahpCD operon as observed in S. viridosporus and mycobacterial species examined, such as M. leprae, Mycobacterium avium, M. tuberculosis, M. bovis, and Mycobacterium marinum (20, 21, 36). It is an unexpected observation that S. coelicolor OxyR acts a transcriptional activator of its own gene. Most members of the LysR family of transcriptional regulators, including E. coli OxyR, autoregulate their own expression as negative regulators. In E. coli, oxyR expression decreases rather than increases during the first 10 min of H2O2 treatment (44). In S. coelicolor, by contrast, both ahpCD and oxyR are induced by H2O2 in an OxyR-dependent manner.
In E. coli and B. subtilis, the ahpC gene consists of an operon with ahpF encoding NAD(P)H-dependent AhpC reductase (2, 5, 46). However, the ahpD genes found in S. coelicolor and Mycobacterium spp. reveal little homology to the ahpF gene. The AhpD protein is about 19 kDa, much smaller than AhpF (52 kDa), and does not contain FAD or NAD(P)H binding domains conserved in AhpF proteins. However, the conservation of cysteine residues in the C-X-X-C motif among AhpD proteins from S. coelicolor and Mycobacterium spp. suggests their function as thioredoxin-like proteins involved in reducing AhpC (28). Since the overproduction of AhpC alone could render resistance to cumene hydroperoxide, it can be postulated that a catalytic amount of AhpD might be required. This idea is consistent with a recent finding in M. tuberculosis that AhpD acts as a thioredoxin-like molecule and reduces AhpC using electrons transferred from NADH through dihydrolipoamide dehydrogenase and dihydrolipoamide succinyltransferase (4).
The pattern of growth phase-dependent expression of the oxyR and ahpCD seems not uniform among organisms. In E. coli, oxyR mRNA showed biphasic expression with a peak at early exponential phase followed by a decline at the stationary phase. The induction of oxyR expression at exponential phase is dependent on the cyclic AMP receptor protein (CRP) regulator (25). We observed similar peak expression of oxyR (as well as ahpC) during early exponential growth of S. coelicolor. This growth phase-dependent expression of oxyR is modulated by OxyR itself, in contrast to the corresponding action of CRP in E. coli. This difference can be justified from the fact that the catabolite repression in S. coelicolor does not proceed through the glucose-phosphotransferase system and hence cAMP-CRP complex as occurs in E. coli (1). If the activity of OxyR is regulated by H2O2 produced by endogenous aerobic respiration as postulated for E. coli (24, 25), the positive regulation of the oxyR gene by OxyR may allow rapid amplification of response via positive feedback, ensuring a rapid production of alkyl hydroperoxide reductase and other oxyR regulon components in response to a subtle increase in H2O2 due to aerobic respiration.
The regulation of ahpC expression in S. coelicolor is still different from that of another gram-positive bacterium, B. subtilis. In B. subtilis, for which the oxyR homologue has not been identified, the ahpCF expression increases at the stationary phase. In this organism, a repressor Fur homologue, PerR, which is similar to CatR in S. coelicolor, is responsible for H2O2 induction and metal-dependent stationary-phase induction of genes like katA (which codes for catalase), mrgA (which codes for nonspecific DNA binding protein), and hemAXCDBL (which code for the heme biosynthesis operon) as well as ahpCF (6, 12).
Regulation of genes for the peroxide-removing system in S. coelicolor is achieved by at least four separate regulators. Alkyl hydroperoxide reductase (AhpC) is maximally produced during early exponential phase and is induced by exogenous H2O2, all under the control of OxyR (this study). Catalase A (CatA), a major monofunctional catalase, is produced maximally at the late exponential phase, maintaining its level throughout stationary phase, and is induced by exogenous H2O2 under the control by a Fur-type repressor, CatR (26). Catalase B, another monofunctional catalase, is produced only after stationary phase and upon differentiation of S. coelicolor under the control of a stationary phase-specific sigma factor σB (14). Catalase C, a catalase-peroxidase, is produced transiently during late exponential phase and is suggested to be regulated by another Fur-type repressor, FurA, in a metal-dependent manner (27). Therefore, in S. coelicolor, antioxidant genes are regulated by a wider variety of regulators than those observed in other organisms examined so far.
It can be postulated that in aerobically growing S. coelicolor, endogenous production of H2O2 from aerobic respiration during early phase of growth triggers rapid induction of the antioxidant system, including alkyl hydroperoxide reductase, to protect membrane lipids and genetic material, via increasing the synthesis and the activity of OxyR. In later growth phases when OxyR is no longer produced, the catA gene may be derepressed, via inactivation of repressor CatR by H2O2, to maintain the level of H2O2 under a certain limit. Therefore, the peroxide-sensitive regulators OxyR and CatR may divide their labor depending on the growth phase. Through the action of OxyR, CatR, and σB, S. coelicolor can be equipped with antiperoxide systems in all growth phases, whereas corresponding functions are exerted by PerR and SigB in B. subtilis and OxyR and σS in E. coli. A third peroxide-sensitive regulator in S. coelicolor, the anti-sigma factor RsrA, also plays a role in responding against a peroxide stress, by inducing thioredoxin and other thiol-reducing systems (31, 38), which may play some role in reducing AhpC. In B. subtilis and X. campestris, OhrR, a new regulator of the MarR family of transcription repressors, regulates the organic hydroperoxide resistance (ohr) gene in response to organic hydroperoxides (23, 45). Inspection of the S. coelicolor genome reveals three ohr homologs, and one of them neighbors a divergent ORF with moderate amino acid sequence homology to OhrR. This implies that there may exist yet another regulator of OhrR type involved in peroxide stress response in S. coelicolor.
In S. coelicolor OxyR did not regulate the production of other antioxidant enzymes such as Ni-containing SOD, Fe-containing SOD, or glucose-6-phosphate dehydrogenase. Nevertheless, cells overproducing OxyR were more resistant to H2O2 and cumene hydroperoxide than cells overproducing AhpC and AhpD, implying the presence of other components of the OxyR regulon having antioxidant function. Moreover, the oxyR disruptant showed a conditional bald phenotype, suggesting the role of oxyR in morphological differentiation as well (Hahn et al., unpublished data). An apparently related observation in E. coli has demonstrated the involvement of OxyR in the control of some surface properties, including colony morphology and auto-aggregation (47). Further studies on the genes regulated by OxyR as well as their regulation mechanism are expected to reveal the interesting function of OxyR in this organism.
Acknowledgments
This work was supported by a research grant (2000-2-20200-001-1) from the Korea Science and Engineering Foundation to J.-H.R. So-Young Oh was a recipient of BK-21 fellowship for graduate students.
REFERENCES
- 1.Angell, S., C. G. Lewis, M. J. Buttner, and M. J. Bibb. 1994. Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Mol. Gen. Genet. 244:135-143. [DOI] [PubMed] [Google Scholar]
- 2.Antelmann, H., S. Engelmann, R. Schmid, and M. Hecker. 1996. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J. Bacteriol. 178:6571-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Bryk, R., C. D. Lima, H. Erdjument-Bromage, P. Tempst, and C. Nathan. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295:1073-1077. [DOI] [PubMed] [Google Scholar]
- 5.Bsat, N., L. Chen, and J. D. Helmann. 1996. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J. Bacteriol. 178:6579-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 7.Calzi, M. L., and L. B. Poole. 1997. Requirement for the two AhpF cysteine disulfide centers in catalysis of peroxide reduction by alkyl hydroperoxide reductase. Biochemistry 36:13357-13364. [DOI] [PubMed] [Google Scholar]
- 8.Chae, H. Z., I.-H. Kim, K. Kim, and S. G. Rhee. 1993. Cloning, sequencing, and mutation of thiol-specific antioxidant gene of Saccharomyces cerevisiae. J. Biol. Chem. 268:16815-16821. [PubMed]
- 9.Chae, H. Z., S. J. Chung, and S. G. Rhee. 1994. Thioredoxin-dependent peroxide reductase from Yeast. J. Biol. Chem. 269:27670-27678. [PubMed] [Google Scholar]
- 10.Chae, H. Z., K. Robison, L. B. Poole, G. Church, G. Storz, and S. G. Rhee. 1994. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: Alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. USA 91:7017-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chae, H. Z., T. B. Uhm, and S. G. Rhee. 1994. Dimerization of thiol-specific antioxidant and the essential role of cysteine 47. Proc. Natl. Acad. Sci. USA 91:7022-7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, L., L. Keramati, and J. D. Helmann. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 92:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho, Y.-H., and J.-H. Roe. 1997. Isolation and expression of the catA gene encoding the major vegetative catalase in Streptomyces coelicolor Müller. J. Bacteriol. 179:4049-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho, Y.-H., E.-J. Lee, B.-E. Ahn, and J.-H. Roe. 2001. SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol. Microbiol. 42:204-214. [DOI] [PubMed] [Google Scholar]
- 15.Christman, M. F., R. W. Morgan, F. S. Jacobson, and B. N. Ames. 1985. Positive control of a regulon for defences against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753-762. [DOI] [PubMed] [Google Scholar]
- 16.Christman, M. F., G. Storz, and B. N. Ames. 1989. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl. Acad. Sci. USA 86:3484-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung, H.-J., and J.-H. Roe. 1993. Profile analysis of proteins related with hydrogen peroxide response in Streptomyces coelicolor (Müller). Kor. J. Microbiol. 31:166-174. [Google Scholar]
- 18.Costa Seaver, L., and J. A. Imlay. 2001. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J. Bacteriol. 183:7182-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa Seaver, L., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deretic, V., W. Philipp, S. Dhandayuthapani, M. H. Mudd, R. Curcic, T. Garbe, B. Heym, L. E. Via, and S. T. Cole. 1995. Mycobacterium tuberculosis is a natural mutant with an inactivated oxidative-stress regulatory gene: implications for sensitivity to isoniazid. Mol. Microbiol. 17:889-990. [DOI] [PubMed] [Google Scholar]
- 21.Dhandayuthapani, S., Y. Zhang, M. H. Mudd, and V. Deretic. 1996. Oxidative stress response and its role in sensitivity to isoniazid in mycobacteria: characterization and inducibility of ahpC by peroxides in Mycobacterium smegmatis and lack of expression in M. aurum and M. tuberculosis. J. Bacteriol. 178:3641-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhandayuthapani, S., M. H. Mudd, and V. Deretic. 1997. Interaction of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J. Bacteriol. 179:2401-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuangthong, M., S. Atichartpongkul, S. Mongkolsuk, and J. D. Helmann. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.González-Flecha, B., and B. Demple. 1997. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J. Bacteriol. 179:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.González-Flecha, B., and B. Demple. 1997. Transcriptional regulation of the Escherichia coli oxyR gene as a function of cell growth. J. Bacteriol. 179:6181-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn, J.-S., S.-Y. Oh, K. F. Chater, Y.-H. Cho, and J.-H. Roe. 2000. H2O2-sensitive fur-like repressor CatR regulating the major catalase gene in Streptomyces coelicolor. J. Biol. Chem. 275:38254-38260. [DOI] [PubMed] [Google Scholar]
- 27.Hahn, J.-S., S.-Y. Oh, and J.-H. Roe. 2000. Regulation of the furA and catC operon, encoding a ferric uptake regulator homologue and catalase-peroxidase, respectively, in Streptomyces coelicolor A3(2). J. Bacteriol. 182:3767-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmgren, A. 1985. Thioredoxin. Annu. Rev. Biochem. 54:237-271. [DOI] [PubMed] [Google Scholar]
- 29.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. John Innes Foundation, Norwich, England.
- 30.Jacobson, F. S., R. W. Morgan, M. F. Christman, and B. N. Ames. 1989. An alkylhydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. J. Biol. Chem. 264:1488-1496. [PubMed] [Google Scholar]
- 31.Kang, J.-G., M. S. B. Paget, Y.-J. Seok, M.-Y. Hahn, J.-B. Bae, J.-S. Hahn, C. Kleanthous, M. J. Buttner, and J.-H. Roe. 1999. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 18:4292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, J., D. Spector, C. Godon, J. Labarre, and M. B. Toledano. 1999. A new antioxidant with alkyl hydroperoxide defense properties in yeast. J. Biol. Chem. 274:4537-4544. [DOI] [PubMed] [Google Scholar]
- 33.Lee, J.-S., Y.-C. Hah, and J.-H. Roe. 1993. The induction of oxidative enzymes in Streptomyces coelicolor upon hydrogen peroxide treatment. J. Gen. Microbiol. 139:1013-1018. [Google Scholar]
- 34.Loprasert, S., S. Antichartpongkun, W. Whangsuk, and S. Mongkolsuk. 1997. Isolation and analysis of the Xanthomonas alkyl hydroperoxide reductase gene and the peroxide sensor regulator genes ahpC and ahpF-oxyR-orfX. J. Bacteriol. 179:3944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maciver, I., and E. J. Hansen. 1996. Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype. Infect. Immun. 64:4618-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagán-Ramos, E., J. Song, M. McFalone, M. H. Mudd, and V. Deretic. 1998. Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J. Bacteriol. 180:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paget, M. S., J.-G. Kang, J.-H. Roe, and M. J. Buttner. 1998. SigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). EMBO J. 17:5776-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paget, M. S., V. Molle, G. Cohen, Y. Aharonowitz, and M. J. Buttner. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the σR regulon. Mol. Microbiol. 42:1007-1020. [DOI] [PubMed] [Google Scholar]
- 39.Poole, L. B. 1996. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 2 cystine disulfides involved in catalysis of peroxide reduction. Biochemistry 35:65-75. [DOI] [PubMed] [Google Scholar]
- 40.Rocha, E. R., G. Owens, Jr., and C. J. Smith. 2000. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 182:5059-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Smith, C. P. 1991. Methods for mapping transcribed DNA sequences, p. 237-252. In T. A. Brown (ed.), Essential molecular biology: a practical approach. Oxford University Press, New York, N.Y.
- 43.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 44.Storz, G., L. A. Tartaglia, and B. N. Ames. 1990. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248:189-194. [DOI] [PubMed] [Google Scholar]
- 45.Sukchawalit, R., S. Loprasert, S. Atichartpongkul, and S. Mongkolsuk. 2001. Complex regulation of the organic hydroperoxide resistance gene (ohr) from Xanthomonas involves OhrR, a novel organic peroxide-inducible negative regulator, and posttranscriptional modifications. J. Bacteriol. 183:4405-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tartaglia, L. A., G. Storz, M. H. Brodsky, A. Lai, and B. N. Ames. 1990. Alkyl hydroperoxide reductase from Salmonella typhimurium. Sequence and homology to thioredoxin reductase and other flavoprotein disulfide oxidoreductases. J. Biol. Chem. 265:10535-10540. [PubMed] [Google Scholar]
- 47.Warne, S. R., J. M. Varley, G. J. Boulnois, and M. G. Norton. 1990. Identification and characterization of a gene that controls colony morphology and auto-aggregation in Escherichia coli K12. J. Gen. Microbiol. 136:455-462. [DOI] [PubMed] [Google Scholar]
- 48.Wilson, T. M., and D. M. Collins. 1996. ahpC, a gene involved in isoniazid resistance of the Mycobacterium tuberculosis complex. Mol. Microbiol. 19:1025-1034. [DOI] [PubMed] [Google Scholar]
- 49.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of OxyR transcription factor by reversible disulfide bond formation. Science 278:1718-1721. [DOI] [PubMed] [Google Scholar]