Abstract
Insulators are elements that shelter genes from the effects of silencers or enhancers. CTCF is the only vertebrate protein that has a recognized role in transcriptional insulation, but how it exerts its effect is unknown. In an attempt to better understand how CTCF functions, we have used an insulation assay in Saccharomyces cerevisiae. We show that CTCF acts as an insulator in yeast, where it can efficiently block the spreading of repressive telomeric chromatin. We identify two domains of the protein that are responsible for this activity: a short and very potent N-terminal domain, as well as the C-terminus of the protein.
INTRODUCTION
Insulators are DNA elements that protect genes from the influence of neighboring regulatory sequences. They can be subdivided into two classes: barrier elements shield genes from the encroachment of repressive chromatin, while enhancer-blockers protect genes from unwanted activation by positively acting elements (1). Insulators have been described in many different eukaryotic organisms, including vertebrates, Drosophila, Schizosaccharomyces pombe and Saccharomyces cerevisiae, but the mechanisms underlying their function are still unclear (2–4). However, several results suggest that these mechanisms might be conserved between species. For instance the 5′ β-globin insulator, initially characterized in chicken, can protect transgenes against position effects in Drosophila (5). Also, the Drosophila proteins BEAF and Su(Hw), known to act at the scs′ and gypsy insulators, respectively, can generate barriers against repressive heterochromatin in yeast (6,7).
CTCF is the only insulator protein known in vertebrates at this time. Its activity was first evidenced at the 5′ β-globin insulator (8). This locus has both barrier and enhancer-blocking properties. CTCF is necessary and sufficient for the enhancer-blocking function, while recent research suggests that another factor is responsible for the barrier activity (9). Further investigations have revealed that the role of CTCF is not limited to the β-globin locus, but is in fact very widespread. Indeed, all vertebrate enhancer-blocking sequences characterized to date seem to involve CTCF (1). Therefore, CTCF has a role of paramount importance in insulation, yet the mechanisms by which it fulfills its function are totally unknown.
The budding yeast S.cerevisiae has been a very valuable model system in the study of transcriptional phenomena. Mechanisms of transcriptional activation and transcriptional repression that were first discovered in yeast are now known to be well conserved in higher eukaryotes (10,11). We reasoned that a yeast system might also be helpful in the investigation of transcriptional insulation. Using an assay we have previously well characterized (12,13), we show that CTCF has insulating properties in yeast. We use this system to map down two insulation domains in the protein. These results will guide future experiments in vertebrates.
MATERIALS AND METHODS
Yeast strains and plasmids
The yeast strain GF97 is derived from W303 and has been described earlier (13). The plasmid used to express Gal4 fusion proteins in yeast is derived from pGBT9, a 2µ plasmid that expresses Gal41–147 from the ADH1 promoter. First, the TRP1 marker in pGBT9 was replaced by HIS3. Then, recombination cloning was used to remove the coding region of GAL4 encoding amino acids 95–147 and simultaneously insert a sequence encoding a single Myc epitope tag. The resulting plasmid is pPAD8. A plasmid containing the chicken CTCF cDNA was kindly provided by Rainer Renkawitz. Different segments of CTCF were amplified by PCR using this plasmid as a template, and inserted by conventional cloning techniques into pPAD8. All of the resulting constructs were verified by automated sequencing.
Yeast insulation assay
The assay was carried out as previously described (12,13), with the following minor variations. Yeast cells transformed with the plasmid to be tested were grown overnight at 30°C in 250 µl of selective medium in a 96-well culture plate. Ten microliters of the undiluted culture and of serial 10-fold dilutions (ranging from 10–1 to 10–5) were then spotted on the adequate selection plates. Each plasmid construct was tested in at least three independent experiments, with four independent cultures each time.
Protein extraction and western blotting
We verified the expression of the various chimeric proteins by western blotting: yeast cells harboring the different expression plasmids were grown overnight in 2 ml of selective medium. Proteins were then extracted following the protocol of Horvath and Riezman (14). Western blotting was carried out according to standard procedures using an antibody directed against the Myc tag.
RESULTS
A genetic insulation assay
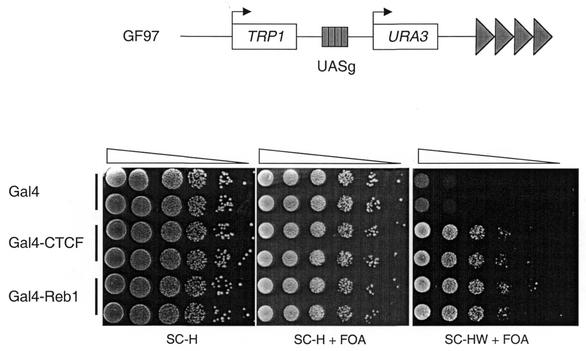
To monitor insulation we used a dual-reporter assay that we have previously described in detail (13). The relevant strain, GF97, contains the TRP1 and URA3 reporter genes inserted in proximity to telomere VIIL (Fig. 1, top). Four Gal4-binding sites (UASg) placed between the reporter genes are used to recruit chimeric proteins containing the Gal4 DNA-binding domain (Gal4DB). The chimeric proteins are expressed from a HIS3-marked plasmid.
Figure 1.
CTCF functions as an insulator in S.cerevisiae. (Top) The yeast strain used in the insulation assay, GF97. It has two reporter genes, TRP1 and URA3, in proximity to a telomere (dark triangles) which represses their expression. Protein fragments fused to the Gal4DB are recruited to the UASg sites located between the reporter genes. (Bottom) A representative insulation experiment. GF97 was transformed with the indicated plasmids and two clones of each construct were tested. Serial dilutions of the cells were spotted on the indicated test media. SC-H is the control medium that allows determination of the total cell count. Growth on SC-H+FOA reflects repression of URA3. Growth on SC-HW+FOA indicates repression of URA3 and simultaneous expression of TRP1, or in other words insulation of TRP1. Gal4–CTCF has no effect on URA3 expression but greatly increases the proportion of cells where TRP1 is insulated. The magnitude of this effect is comparable to that of Reb1, a natural yeast insulator.
In this assay, three parameters are measured. First, the cells are plated on medium that lacks only histidine (SC-H) to determine the total cell count. Secondly, the cells are plated on medium lacking histidine and containing 5-FOA (SC-H+ FOA). The number of cells growing on this medium is divided by the total number of cells to yield the fraction of cells that are FOA-resistant, referred to as the FOAr fraction. 5-FOA is a drug that kills only the cells expressing URA3, therefore, the FOAr fraction is the proportion of cells in which URA3 is repressed. Thirdly, the cells are plated on medium lacking histidine and tryptophan, and containing 5-FOA (SC-HW+ FOA). Again, the number of viable cells on these plates is divided by the total cell number to yield the fraction of cells that are simultaneously Trp+ and FOA-resistant. This ratio is the Trp+FOAr fraction. It is equal to the proportion of cells in the population that express TRP1 and simultaneously repress URA3.
In the absence of exogenous proteins, TRP1 and URA3 are subject to telomeric silencing and their expression is jointly repressed in a majority of cells. Therefore, the FOAr fraction is close to 1, and the Trp+FOAr fraction is close to 0. Recruitment of a transcriptional activation domain to the UASg increases the fraction of cells that express URA3. This is visualized as a decrease in the FOAr fraction. In contrast, recruitment of an insulation domain to the UASg protects TRP1 from telomeric silencing while leaving URA3 unaffected. This translates as an increase in the Trp+FOAr fraction, with no change in the FOAr fraction.
CTCF generates transcriptional barriers in S.cerevisiae
Strain GF97 was transformed with a vector expressing a fusion of Gal4DB to the full-length chicken CTCF protein, or the Gal4DB alone to serve as a control. The cells were first plated on medium lacking histidine (SC-H). Cells expressing Gal4DB and Gal4–CTCF grew equally well (Fig. 1). This means that expression of the Gal4–CTCF chimera is not detrimental to cell growth. The cells were then assayed on SC-H+FOA to monitor URA3 expression. Again cells transformed with either plasmid were indistinguishable. This means that Gal4–CTCF does not activate transcription of the URA3 reporter gene. Finally, the cells were spotted on SC-HW+FOA to detect insulation of TRP1. No cells expressing Gal4DB grew on this medium. In other words no cells express TRP1 while simultaneously repressing URA3 in these conditions. In sharp contrast, over 10% of the cells expressing Gal4–CTCF grew on SC-HW+FOA. Therefore, we conclude that Gal4– CTCF can shelter TRP1 from telomeric repression while leaving URA3 unaffected and behaves as an insulator.
We then sought to compare the strength of insulation by CTCF to that of an endogenous yeast insulator. Reb1 is a yeast transcription factor known to be involved in insulation (12,13). The insulation domain of Reb1 is located between its amino acids 1 and 405, and is the most potent that we have identified to date. We fused this domain to Gal4DB in the chimera Gal4–Reb1. We compared the activity of Gal4–CTCF to that of Gal4–Reb1. As can be seen in Figure 1, Gal4–CTCF is only slightly less active than Gal4–Reb1 in our assay.
From this set of experiments, we conclude that CTCF functions as a strong insulator in yeast. We then set out to use our assay to locate the insulation domain(s) of CTCF by deletion analysis.
Two domains of CTCF are necessary for insulation
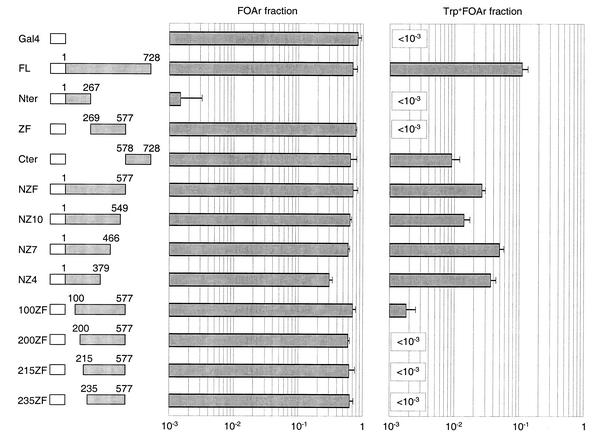
We first subdivided CTCF into three domains: the N-terminus (Nter, amino acids 1–267), the central region containing the zinc fingers (ZF, amino acids 269–577), and the C-terminus (Cter, amino acids 578–728).
Each of these regions was fused to Gal4DB and tested in strain GF97 as above. Again, insulation activity in the assay is visualized as an increase in the Trp+FOAr fraction.
As depicted in Figure 2, the Nter and ZF regions had no insulation activity. The Cter had some activity, but it was only ∼10% as high as that of the full-length protein. Expression of the Gal4–Nter chimera greatly decreased the fraction of cells that were FOA-resistant, meaning that Nter contains a transcriptional activation domain. In contrast, neither ZF nor Cter noticeably activated transcription. Expression of these chimeras and the ones mentioned later was verified by western blotting (data not shown). All were expressed to comparable levels.
Figure 2.
Two domains of CTCF are required for insulation. The indicated residues of CTCF were fused to the Gal4DB (white box) and tested in strain GF97. (Left) The FOAr fraction is the proportion of cells that grow in the presence of 5-FOA, or cells where URA3 is repressed. (Right) The Trp+FOAr fraction is the proportion of cells that express TRP1 and repress URA3, or cells where TRP1 is insulated. Two regions are necessary for insulation by CTCF: the first 100 amino acids and the C-terminus.
There are two possible explanations to the finding that only Cter has insulation potential, but that it is 10-fold less active than the full-length CTCF. One possibility is that CTCF contains only one insulation domain that is located in or around the Cter, and that it is truncated or improperly folded in our chimeras, resulting in poor activity. The other possibility is that Cter normally cooperates with another insulation domain in CTCF that is not detectable in this first set of constructs. If the former were true, then deletion of Cter should fully inactivate CTCF for insulation. If the latter were true, then deletion of Cter might result in only partial loss of insulation potential. Therefore, we proceeded to delete Cter from CTCF. The resulting construct is designated NZF. Gal4–NZF had clear insulating potential (Fig. 2). From this we conclude that two domains of CTCF are required for insulation: one in Cter, and another one within NZF. We tried to locate the second insulation domain with more precision and generated a series of deletions of NZF.
The DNA-binding domain of CTCF contains 11 zinc-finger motifs (15). We first engineered C-terminal deletions of NZF that removed one, four or seven of the zinc fingers. This yielded constructs NZ10, NZ7 and NZ4, respectively. All three were as potent for insulation as NZF (Fig. 2). Thus, we conclude that the insulation domain is contained within the first 379 amino acids of the protein. We then tested a series of NZF derivatives with N-terminal deletions that removed the first 100, 200, 215 or 235 amino acids. These constructs are named 100ZF, 200ZF, 215ZF and 235ZF, respectively. As can be seen in Figure 2, ablation of the first 100 amino acids of NZF resulted in almost complete loss of insulation activity. Larger deletions also had the same effect. Therefore, region 1–100 is necessary for insulation by NZF.
These experiments establish that two domains are necessary for insulation by CTCF in our assay: the first 100 amino acids of the protein, as well as the Cter.
Fine-mapping of the insulation domains
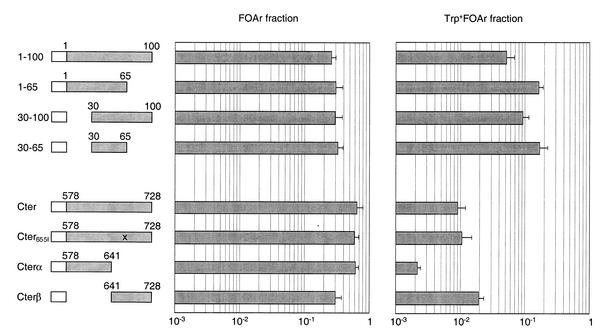
The first 100 amino acids of CTCF being necessary for insulation, we tested whether they might also be sufficient for this function. This domain was fused to Gal4DB and assayed in our strain, and indeed the resulting chimera was highly active, in fact almost as potent as full-length CTCF (Fig. 3, top). The fact that the region containing amino acids 1–100 of CTCF behaves as an insulator, while the larger region Nter does not, is due to the presence in Nter of a transactivation domain (see details in Discussion). We then proceeded to try and narrow down the insulation domain in region 1–100. Deletion of the first 30, or of the last 35 amino acids of this fragment had very little effect on insulation activity. Therefore, only the region comprising amino acids 30–65 was necessary for function. To test if it was also sufficient, we fused the 35 amino acids in question to Gal4DB. This chimera was in fact slightly more potent than amino acids 1–100, and about equally as active as full-length CTCF. The primary sequence of the domain is shown in Figure 4.
Figure 3.
Delineation of the insulation domains. The indicated residues of CTCF were fused to the Gal4DB and tested in strain GF97 as in Figures 1 and 2. (Top) Residues 30–65 of CTCF form a strong insulation domain. (Bottom) The AT-hook within Cter is dispensable for insulation. Amino acids 641–728 recapitulate the activity of Cter.
Figure 4.
The N-terminal insulation domain of CTCF. The 35 amino acid insulation domain found in chicken CTCF was aligned with the corresponding regions in the human, mouse, rat and Xenopus proteins. h, conserved hydrophobic amino acids, c, charged amino acids.
We also investigated in more detail the C-terminal insulation domain. The Cter peptide contains a sequence motif, KRRGRPPG, characteristic of an AT-hook. This is a DNA-binding motif that permits interaction with AT-rich stretches of DNA (16). To determine if this motif was involved in insulation, we inactivated it by a point mutation. The core sequence GRP was changed to GIP, which should result in loss of function of the AT-hook (16). The mutant chimera Cter655I had an insulation activity similar to that of Cter, and therefore, we conclude that the AT-hook is not involved in insulation by Cter. We then subdivided Cter into two smaller domains, Cterα (amino acids 578–641) and Cterβ (amino acids 641–728), and assayed the activity of both subregions. Cterα only has marginal insulation potential, whereas Cterβ seems to harbor all the activity of Cter.
These data show that CTCF harbors two autonomous insulation domains. The first one is small and very active and corresponds to amino acids 30–65. The second one is 5–10-fold less active and is located at the C-terminus of the protein, within residues 641–728.
DISCUSSION
Characteristics of the insulation domains
We have identified two insulation domains within CTCF. We were able to narrow down the C-terminal insulation domain to a region of approximately 85 residues. This region contains an AT-hook, but this motif is not necessary for insulation. No other sequence motifs are salient in this domain. The N-terminal insulation domain is very active and can be narrowed down to only 35 amino acids (Fig. 4). Its sequence does not include any recognizable protein motif, and is not predicted to have an organized tridimensional structure. The sequence of CTCF has been strikingly well conserved during vertebrate evolution, and correspondingly the N-terminal insulation domain is almost identical in the human, mouse, rat and chicken proteins (Fig. 4). Comparison with the recently described Xenopus CTCF (17) is more informative as this homolog is more evolutionarily distant. Twenty-one out of 35 residues are identical in the chicken and Xenopus proteins. This similarity, although high, is weaker that that observed over the total length of the protein (84% amino acid identity). Provided that the insulation function is conserved in Xenopus, this would mean that the primary sequence requirements for the insulation domain are relatively loose. This situation is generally reminiscent of that of transcriptional activation domains. Even domains that target the same protein in the transcriptional machinery only show very faint sequence resemblance (18). Also, some of the strongest known natural activators are short peptides that are unstructured in solution, such as the activation domain of the viral protein VP16 (19).
Insulation and activation domains within CTCF
It may seem paradoxical that the region containing amino acids 1–100 of CTCF has strong insulation activity, whereas the larger Nter region (amino acids 1–267) has none. The explanation for this situation is that Nter contains a strong transcriptional activation domain. This domain increases the transcription of URA3 to such an extent that only ∼1 in 1000 cells is resistant to FOA (see line Nter in Fig. 2). This of course precludes the detection of Trp+FOAr cells. Given that region 1–100 does not activate transcription (see Fig. 3), the activation domain must reside between residues 100 and 267 of CTCF. This correlates closely with results recently obtained in a mammalian system. Vostrov et al. (20) have found that CTCF transactivates the amyloid precursor protein (APP) promoter. A CTCF truncation variant lacking the first 249 amino acids still binds the promoter, but does no longer transactivate (20). This strongly argues for the presence of a transactivating domain within the first 249 residues. Of note, this transactivation domain has not been detected by other investigators, and its activity may be promoter- or cell type-specific (21).
A related intriguing point is that the Nter region strongly activates transcription, while the larger NZF domain and its derivatives do not. The most likely explanation is that the activation domain is masked by the zinc-finger domain through intramolecular folding. It is possible that, upon binding its recognition sites, CTCF unfolds and reveals its transactivation domain. Such a mechanism has been evidenced for several mammalian transcription factors (22,23).
Relevance to vertebrate cells
Here we describe two domains of CTCF that can block the spreading of repressive telomeric heterochromatin in S.cerevisiae. How does this relate to the function of CTCF in vertebrate cells?
First and foremost, our experience with a number of yeast and vertebrate proteins shows that the vast majority of peptides do not insulate in our assay (12,13,24, this work). It seems unlikely, then, that the presence of very potent insulation domains in CTCF is spurious. We do not know for certain that our yeast assay is functionally equivalent to the barrier assays used in vertebrates but it could be expected to identify domains that have barrier activity. It may seem paradoxical to find such domains in CTCF, which is thought to be an enhancer-blocker (9). However, two simple explanations for this situation can be put forward. First, it is possible that CTCF opposes telomeric heterochromatin by the same mechanisms that it blocks enhancers. How CTCF functions as an insulator is unknown, but some of the existing hypotheses are compatible with this idea (1). The second possibility is that CTCF does have barrier activity, but that this potential is not employed at the insulators examined so far. CTCF is a very versatile factor that activates transcription at certain promoters and represses it at others (15), and it could have an equally complex role in insulation. Experiments testing the activity in vertebrate cells of the two domains presented here will be necessary to directly address this question.
How do the insulation domains function? Future directions
Recent reports in yeast have shown that many transcriptional activation domains can also be insulation domains (7,13,24). In the case of CTCF, the two insulation domains we have identified do not activate transcription, and we hypothesize that they act by recruiting other proteins. One possibility we actively consider is that they could contact a structural component of the nucleus to raise a physical barrier against silencing (4,6).
Our yeast assay has allowed us to rapidly identify candidate domains for the insulation activity of CTCF. An added advantage of the system is that it opens up opportunities to undertake a genetic analysis of insulation. Future experiments will aim to identify mutant yeast strains in which CTCF is less, or more, active than in the wild-type strain. This approach should shed light on the mechanisms of insulation by CTCF in vertebrate cells.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Rainer Renkawitz for the gift of the chicken CTCF cDNA. We thank members of the Gilson laboratory for reagents and entertaining discussions, special thanks go to Geneviève Fourel for the gift of useful reagents and advice. P.-A.D. thanks Allison Bardin for support. This work was supported by the Ligue Nationale contre le Cancer.
REFERENCES
- 1.West A.G., Gaszner,M. and Felsenfeld,G. (2002) Insulators: many functions, many mechanisms. Genes Dev., 16, 271–288. [DOI] [PubMed] [Google Scholar]
- 2.Bi X. and Broach,J.R. (2001) Chromosomal boundaries in S. cerevisiae. Curr. Opin. Genet. Dev., 11, 199–204. [DOI] [PubMed] [Google Scholar]
- 3.Dhillon N. and Kamakaka,R.T. (2002) Breaking through to the other side: silencers and barriers. Curr. Opin. Genet. Dev., 12, 188–192. [DOI] [PubMed] [Google Scholar]
- 4.Donze D. and Kamakaka,R.T. (2002) Braking the silence: how heterochromatic gene repression is stopped in its tracks. Bioessays, 24, 344–349. [DOI] [PubMed] [Google Scholar]
- 5.Chung J.H., Whiteley,M. and Felsenfeld,G. (1993) A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 74, 505–514. [DOI] [PubMed] [Google Scholar]
- 6.Ishii K., Arib,G., Lin,C., Van Houwe,G. and Laemmli,U.K. (2002) Chromatin boundaries in budding yeast: the nuclear pore connection. Cell, 109, 551–562. [DOI] [PubMed] [Google Scholar]
- 7.Donze D. and Kamakaka,R.T. (2001) RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J., 20, 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell A.C., West,A.G. and Felsenfeld,G. (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell, 98, 387–396. [DOI] [PubMed] [Google Scholar]
- 9.Recillas-Targa F., Pikaart,M.J., Burgess-Beusse,B., Bell,A.C., Litt,M.D., West,A.G., Gaszner,M. and Felsenfeld,G. (2002) Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl Acad. Sci. USA, 99, 6883–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Struhl K. (1999) Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell, 98, 1–4. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy B.K. (2002) Mammalian transcription factors in yeast: strangers in a familiar land. Nature Rev. Mol. Cell Biol., 3, 41–49. [DOI] [PubMed] [Google Scholar]
- 12.Fourel G., Revardel,E., Koering,C.E. and Gilson,E. (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J., 18, 2522–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fourel G., Boscheron,C., Revardel,E., Lebrun,E., Hu,Y.F., Simmen,K.C., Muller,K., Li,R., Mermod,N. and Gilson,E. (2001) An activation-independent role of transcription factors in insulator function. EMBO Rep., 2, 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvath A. and Riezman,H. (1994) Rapid protein extraction from Saccharomyces cerevisiae. Yeast, 10, 1305–1310. [DOI] [PubMed] [Google Scholar]
- 15.Ohlsson R., Renkawitz,R. and Lobanenkov,V. (2001) CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet., 17, 520–527. [DOI] [PubMed] [Google Scholar]
- 16.Aravind L. and Landsman,D. (1998) AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res., 26, 4413–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burke L.J., Hollemann,T., Pieler,T. and Renkawitz,R. (2002) Molecular cloning and expression of the chromatin insulator protein CTCF in Xenopus laevis. Mech. Dev., 113, 95–98. [DOI] [PubMed] [Google Scholar]
- 18.Choi Y., Asada,S. and Uesugi,M. (2000) Divergent hTAFII31-binding motifs hidden in activation domains. J. Biol. Chem., 275, 15912–15916. [DOI] [PubMed] [Google Scholar]
- 19.Uesugi M., Nyanguile,O., Lu,H., Levine,A.J. and Verdine,G.L. (1997) Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science, 277, 1310–1313. [DOI] [PubMed] [Google Scholar]
- 20.Vostrov A.A., Taheny,M.J. and Quitschke,W.W. (2002) A region to the N-terminal side of the CTCF zinc finger domain is essential for activating transcription from the amyloid precursor protein promoter. J. Biol. Chem., 277, 1619–1627. [DOI] [PubMed] [Google Scholar]
- 21.Lutz M., Burke,L.J., Barreto,G., Goeman,F., Greb,H., Arnold,R., Schultheiss,H., Brehm,A., Kouzarides,T., Lobanenkov,V. et al. (2000) Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res., 28, 1707–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laget M.P., Defossez,P.A., Albagli,O., Baert,J.L., Dewitte,F., Stehelin,D. and de Launoit,Y. (1996) Two functionally distinct domains responsible for transactivation by the Ets family member ERM. Oncogene, 12, 1325–1336. [PubMed] [Google Scholar]
- 23.Li X.Y. and Green,M.R. (1996) Intramolecular inhibition of activating transcription factor-2 function by its DNA-binding domain. Genes Dev., 10, 517–527. [DOI] [PubMed] [Google Scholar]
- 24.Fourel G., Miyake,T., Defossez,P.A., Li,R. and Gilson,E. (2002) General regulatory factors as genome partitioners. J. Biol. Chem., 277, 41736–41743. [DOI] [PubMed] [Google Scholar]