Abstract
The promoter of the rat aldolase B (AldB) gene functions in vivo as an origin of DNA replication in the cells in which transcription of the gene is repressed. Previously, we identified two closely related DNA-binding proteins, AlF-C1 and AlF-C2, which repressed the AldB gene promoter. We also reported that the binding site of these proteins, site C, is one of the required DNA elements of the AldB gene origin/promoter for autonomously replicating activity in transfected cells. In the present study, we show that AlF-C1 and AlF-C2 bind directly to Orc1, a subunit of the origin recognition complex (ORC). Deletion analyses revealed a functional domain in AlF-C2 for binding to Orc1, which is located separately from the DNA-binding domain. In addition, we found a novel protein-interacting domain in Orc1 required for the binding of AlF-C2, which was conserved in human, mouse and Chinese hamster, but not in Drosophila, frog and yeast. Thus, it is assumed that in mammalian cells, sequence- specific DNA-binding proteins are involved in recruiting ORC to regulate replication initiation and/or transcription repression.
INTRODUCTION
It has been generally accepted that origins of DNA replication in higher eukaryotes do not share common specific sequences. Nevertheless, the positions of origins are not distributed randomly on the chromosomal DNA (1). This origin spacing might involve recognition of specific sites by the origin recognition complex (ORC), a six-subunit protein, the role of which is to mark and activate origins (2). ORC subunits are highly conserved from yeast to plant to mammalian cells, indicating the conserved functions throughout the species (3). In budding yeast, ORC directly binds to a specific sequence called autonomously replicating sequence (ARS) consensus sequence (ACS) (4). However, in higher eukaryotes there is no direct evidence, to our knowledge, that ORC binds directly to specific DNA sequences while it is necessarily required for replication initiation. Therefore, what directs ORC to specific sites spaced at appropriate intervals is currently important to understand mechanisms operating in initiation of DNA replication. The aim of the present study is to investigate this point.
The promoter of the rat aldolase B (AldB) gene acts bifunctionally in vivo, but these functions are mutually exclusive; it functions as an origin of DNA replication in the rat hepatoma cells in which the gene is repressed, while in differentiated hepatic cells it functions as a cell type-specific transcription promoter (5,6). Our previous works identified two closely related proteins, AlF-C1 and AlF-C2, which bind to site C in the origin/promoter and are involved in the repression of the AldB gene (7). In addition, we showed that the origin/promoter fragment directs autonomous replication when transfected in Cos1 cells in a plasmid form. Site C is one of the necessarily required DNA elements for such an autonomously replicating activity (8). Therefore, we are interested in the association of AlF-C1 and AlF-C2 with ORC that might lead to the initiation of replication. In this report, we show that AlF-C1 and AlF-C2 bind directly to Orc1, a subunit of ORC, and discuss that in mammalian cells, sequence-specific DNA-binding proteins might be involved in recruiting ORC to regulate replication initiation and/or transcription repression.
MATERIALS AND METHODS
Plasmids
The plasmids carrying human ORC subunits were constructed as follows. The BamHI–NotI fragment of pGEX-ORC1 (9) was subcloned into the BamHI–NotI site of pcDNA3.1/HisC (Invitrogen) (referred to as pcDNA3.1/Orc1), the SmaI–NotI fragment of pGEX-ORC2 (9) into the EcoRV–NotI site of pcDNA3.1/HisA (Invitrogen) (pcDNA3.1/Orc2), the BamHI– NotI fragment of pGEX-ORC4 (9) into the BamHI–NotI site of pcDNA3.1/HisC (pcDNA3.1/Orc4) and the EcoRI-NotI fragment of pGEX-ORC5 (9) into the EcoRI–NotI site of pcDNA3.1/HisC (pcDNA3.1/Orc5). Orc1 deletion mutant, Orc(1-208), was prepared by ligating NdeI–MscI fragment of pcDNA3.1/Orc1 to NdeI- and EcoRV-digested pcDNA3.1/HisC. Orc(1-278) was prepared by ligating NdeI–BglII fragment of pcDNA3.1c/Orc1 to NdeI- and BamHI-digested pcDNA3.1/HisC. Orc(210-862) was prepared by ligating MscI–XbaI fragment of pcDNA3.1/Orc1 to EcoRI (blunt-ended with Klenow fragment)- and XbaI-digested pcDNA3.1/HisC. Orc(210-511) was prepared by ligating EcoRI insert of Orc(210-862) to EcoRI-digested pcDNA3.1/HisC. Orc(512-862) was prepared by self-ligating EcoRI-digested Orc(210-862). Orc(1-239) was prepared by PCR using BamOrc(1-19) and Orc(717-693) as primers on the template pcDNA3.1/Orc1 under the following conditions: preheating at 95°C for 3 min, 15 cycles of 96°C for 1 s, 55°C for 30 s and 72°C for 30 s with pfu polymerase (Promega). PCR products were digested with BamHI and inserted to the BamHI- and EcoRV-digested pcDNA3.1/HisC. The EcoRI fragment from AlF-C1 and AlF-C2 expression plasmids (7) were ligated to pGEX-5X-3 (Amersham) (named pGEX/AlF-C1 and pGEX/AlF-C2, respectively). AlF-C2(1-206) was prepared by ligating EcoRI–DraI fragment of pGEX/AlF-C2 to EcoRI–SmaI-digested pGEX-6P-3 (Amersham). AlF-C2(1-179) was prepared by ligating EcoRI–MscI fragment of pGEX/AlF-C2 to EcoRI- and SmaI-digested pGEX-6P-3. The EcoRI fragment of pGEX/AlF-C2 was ligated to EcoRI site of pcDNA3.1/HisB (pcDNA3.1b/AlF-C2). AlF-C2(207-284) was prepared by ligating DraI–NotI fragment of pcDNA3.1b/AlF-C2 to SmaI–NotI-digested pGEX-6P-2 (Amersham). The AlF-C2 cDNA fragments, P240-284, P249-284, P264-284, P207-272, P207-263 and P207-238 were amplified by PCR using AlF-C2(207-284) as a template with primer sets pGEX3′/BC718-734, BC745-764/EC855-840, BC790-812/EC855-840, pGEX5′/EC816-790, pGEX5′/EC789-766 and pGEX5′/EC716-695, respectively (listed in Table 1). P240-284 was digested with BamHI and NotI, and inserted into the BamHI–NotI site of pGEX-6P-1 (Amersham) to generate AlF-C2(240-284). P249-284 was digested with EcoRI, and inserted into the BamHI (blunt-ended with Klenow fragment)–EcoRI site of pGEX-6P-1 [AlF-C2(249-284)]. P264-284 was digested with BamHI and EcoRI, and inserted into the BamHI–EcoRI site of pGEX-6P-1 [AlF-C2(264-284)]. The PCR products P207-272, P207-263 and P207-238 were digested with BamHI and inserted in to the BamHI–SmaI site of pGEX-6P-1 to generate the plasmids AlF-C2(207-272), AlF-C2(207-263) and AlF-C2(207-238), respectively. The following AlF-C2 cDNA fragments were amplified by PCR using pGEX-AlF-C2 as a template; P1-159 with primer sets pGEX5′/C477-451, P1-146 with pGEX5′/C438-418, P76-159 with C226-243/C477-451 and P67-159 with C198-216/C477-451. The PCR products P1-159 and P1-146 were digested with EcoRI and inserted in to the EcoRI–SmaI site of pGEX-6P-3 to generate AlF-C2(1-159) and AlF-C2(1-146), respectively. P76-159 and P67-159 were digested with EcoRI and inserted into the EcoRI–SmaI site of pGEX-6P-1 to generate AlF-C2(76-159) and AlF-C2(67-159), respectively. AlF-C2(59-159) was prepared by ligating EheI–XhoI fragment of AlF-C2(1-159) into SmaI- and XhoI-digested pGEX-6P-2. The oligonucleotide primers used for PCR amplification were listed with their Tm in Table 1.
Table 1. Oligonucleotide primers used for PCR.
| Name | Sequence | Tm |
|---|---|---|
| BamOrc(1-19) | 5′-CGGGATCCATGGCACACTACCCCACAA-3′ | 66.6 |
| Orc(717-693) | 5′-TTAAAGCTCCAGCCTCTTTCTGGCTCTT-3′ | 62.0 |
| pGEX5′ | 5′-GGGCTGGCAAGCCACGTTTGGTG-3′ | 65.0 |
| pGEX3′ | 5′-CCGGGAGCTGCATGTGTCAGAGG-3′ | 65.0 |
| BC718-734 | 5′-CGGGATCCCAGCAACAGCAGTATGG-3′ | 65.0 |
| BC745-764 | 5′-CGGGATCCGAGGAAATCGCAATCGAG-3′ | 65.2 |
| BC790-812 | 5′-CGGATCCGGTAGTACAAATTACGGGAAGAG-3′ | 64.6 |
| EC855-840 | 5′-GGAATTCAGTATGGCTTGTAG-3′ | 54.6 |
| EC816-790 | 5′-GGAATTCTGGCTCTTCCCGTAATTTGTACTACC-3′ | 64.3 |
| EC789-766 | 5′-GGAATTCTGACCTCCACCACTGCCTCGGTT-3′ | 67.4 |
| EC716-695 | 5′-GGAATTCTTACACCTCTTTGGGCTGGGCAAC-3′ | 67.0 |
| C477-451 | 5′-TTTCTTCACAGGGTCCTTCTTCATAGC-3′ | 60.5 |
| C438-418 | 5′-GAGGGTCAATGACACGACCATC-3′ | 58.5 |
| C226-243 | 5′-GGAATTCATGTTCGTTGGTGGTCTG-3′ | 60.5 |
| C198-216 | 5′-GGAATTCAGCAAGAACGAGGAGGAC-3′ | 62.1 |
Expression and purification of glutathione S-transferase (GST) fusion proteins
Recombinant AlF-C1, AlF-C2 and various AlF-C2 deletion mutants from pGEX vector were each expressed as a fusion protein with GST by culturing Escherichia coli BL21(DE3, pLysS) cells carrying pGEX/AlF-C1, pGEX/AlF-C2 and pGEX/AlF-C2 deletion mutants with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 0.5–3 h at 20–37°C. The cells were collected and suspended in phosphate-buffered saline (PBS) containing 0.5% NP-40 and frozen at –80°C. The cell suspension was thawed on ice, treated with 12 U/ml DNase I (Takara) for 10 min. The cell lysate was cleared by centrifugation at 14 000 g for 30 min and mixed with glutathione–Sepharose 4B (Amersham) for 1 h on a rotary shaker at 4°C. The beads were washed four times with PBS containing 0.5% NP-40 before elution with 50 mM Tris–HCl (pH 9.6) containing 20 mM reduced glutathione. The eluted fraction was dialyzed against 20 mM Tris–HCl (pH 7.5) containing 20% glycerol, 20 mM NaCl, 0.2 mM EDTA, 1 mM dithiothreitol and 0.1 mM phenylmethylsulfonyl fluoride. Protein concentrations were determined by comparison with bovine serum albumin as a standard on Coomassie Blue stained protein gels.
Protein–protein interaction assay
Aliquots (10 µg) of the purified GST-fusion proteins or GST were first applied to 25 µl of glutathione–Sepharose 4B (Amersham) in a binding buffer containing 20 mM Tris–HCl (pH 7.5), 80 mM KCl, 2 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail and 0.1% NP-40 for 1 h at 4°C. The protease inhibitor cocktail was prepared as a 1000× concentrated stock that contained 10 mg/ml each of leupeptin, chymostatin, pepstatin A and antipain in dimetyl sulfoxide. The beads were washed three times with 500 µl of the binding buffer. Recombinant Orc1, Orc2, Orc4, Orc5 and various Orc1 deletion mutants from pcDNA3.1 vector (Invitrogen) were used as a template for in vitro transcription and translation with TNT T7-coupled reticulocyte lysate system (Promega). Aliquots (10 µl) of the lysate were diluted with 90 µl of the binding buffer containing 80–250 mM KCl, and mixed with the beads that were bound one GST-fused AlF-C1 or AlF-C2 proteins on a rotary shaker for 16 h at 4°C. The beads were washed four times with 1 ml of the same buffer as above. The proteins bound to the beads were eluted by boiling in SDS sample buffer (10), separated on a 10% SDS–polyacrylamide gel, blotted with anti-T7 tag antibody (Novagen), and detected by horseradish peroxidase-conjugated anti-mouse Ig in an ECL system (Amersham).
Electrophoretic mobility shift assay (EMSA)
The GST-fused proteins (200 ng) and 3 ng of 32P-labeled site C probe (labeled with [α-32P]dCTP by Klenow fragment) were incubated in a final volume of 15 µl containing 10 mM Tris–HCl (pH 7.9), 100 mM KCl, 0.5 mM EDTA, 2 mM dithiothreitol, 0.5 mg/ml RNase A, 10% glycerol and 33 µg/ml poly(dI–dC) for 40 min on ice. The DNA–protein complexes were resolved on 5% polyacrylamide gels in TBE buffer. The probe and competitors used are: site A, 5′-CAAT CAGATTATTGAATAAACACCTTC-3′; site C, 5′-GTG AGCCTGATTACAAAGATTGGCTGTTCAC-3′.
RESULTS
AlF-C1 and AlF-C2 proteins interact with Orc1
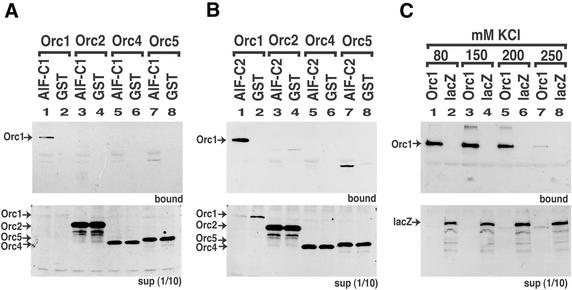
To examine whether AlF-C proteins interact with ORC, we carried out a GST pull-down assay. AlF-C1 and AlF-C2 proteins were synthesized in E.coli as fusion proteins of GST and affinity-purified by glutathione–Sepharose. We employed these fusion proteins as a bait for probing T7-tagged human ORC subunits translated in vitro. Binding assays were carried out in 80 mM KCl, and the proteins bound to glutathione–Sepharose via GST–AlF-C1 or GST–AlF-C2 were eluted and detected by western blotting using an anti-T7 tag antibody (Fig. 1A and B). Under this condition, non-specific binding of T7-tagged lacZ to GST–AlF-C1 was not observed (Fig. 1C, lane 2). Of the four ORC subunits tested, Orc1 specifically bound to both AlF-C1 and AlF-C2 proteins but not to GST alone (Fig. 1A and B, lanes 1 and 2). Orc2 and Orc4 bound to neither AlF-C1 nor AlF-C2 (Fig. 1A and B, lanes 3 and 5). A weak interaction of Orc5 with AlF-C proteins was observed (Fig. 1A and B, lane 7). However, the amount of Orc5 in the supernatant was much greater than that of Orc1. Significantly greater quantities of Orc1 in the bound fraction (Fig. 1A and 1B, lanes 1 and 7) suggested that the affinity of Orc1 to AlF-C is much higher than that of Orc5. We then examined the interaction of GST–AlF-C1 and T7-Orc1 under increasing concentrations of KCl (Fig. 1C) to examine the stability of interaction. At 80–200 mM KCl, stable binding of Orc1 to AlF-C1 was detected (lanes 1, 3 and 5). However, the only detectable amount of binding was observed at 250 mM KCl (lane 7). Under these conditions, non-specific binding of T7-tagged lacZ to GST–AlF-C1 was not observed (Fig. 1C, lanes 2, 4, 6 and 8). These results clearly indicated that Orc1 directly bound to AlF-C1 and AlF-C2 proteins with a relatively strong binding affinity.
Figure 1.
Orc1 interacted with AlF-C1 and AlF-C2. AlF-C1 (A) and AlF-C2 (B) were expressed as GST-fusion proteins in E.coli and purified. The GST-fusion proteins (lanes 1, 3, 5 and 7), or GST alone (lanes 2, 4, 6 and 8), were bound to the glutathione beads. The fusion-protein-bound glutathione beads were incubated with in vitro translated T7-tagged Orc1 (lanes 1 and 2), Orc2 (lanes 3 and 4), Orc4 (lanes 5 and 6) and Orc5 (lanes 7 and 8). After extensive washing, the protein bound to the beads were separated on an SDS–PAGE and blotted with an anti-T7 antibody (upper panel). The 1/10 volume of unbound fractions (supernatant) was also analyzed (lower panel). Arrows indicate the positions of Orc1, Orc2, Orc4 and Orc5. (C) The GST-fused AlF-C1 protein and T7-tagged Orc1 (lanes 1, 3, 5 and 7), or lacZ (lanes 2, 4, 6 and 8) were incubated with 80 mM (lanes 1 and 2), 150 mM (lanes 3 and 4), 200 mM (lanes 5 and 6) and 250 mM (lanes 7 and 8) KCl, and their binding activities were tested as in (A). Arrows indicate the positions of Orc1 and lacZ.
Orc1 domain responsible to AlF-C2 binding
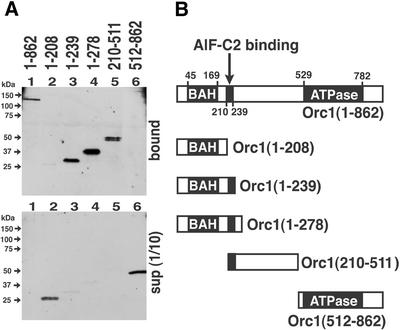
To define the domain of Orc1 required for the interaction with AlF-C2, a series of T7-tagged Orc1 deletion mutants were translated in vitro, and the products were applied to the GST–AlF-C2-coupled glutathione beads. The Orc1 mutant proteins bound to the beads via GST–AlF-C2 were eluted and detected as described in Figure 1 (Fig. 2). As shown in Figure 2A (lane 1), full length Orc1 protein [Orc1(1-862)] bound to AlF-C2. Orc1(1-208) containing BAH (bromo-adjacent homology) domain (11), did not bind to AlF-C2 (Fig. 2A, lane 2), whereas Orc1(1-239), Orc1(1-278) and Orc1(210-511) bound (Fig. 2A, lanes 3–5). Orc1(512-862), which contains Cdc6 homology region and NTP-binding domain (Fig. 2B), did not bind to AlF-C2 (Fig. 2A, lane 6). These results indicated that amino acids 210–239 in Orc1 were required for the interaction with AlF-C2 (Fig. 2B). Comparison of amino acid sequences of Orc1 from different species revealed that the amino acids 210–239 were conserved among human, mouse and Chinese hamster, but not in yeast, Xenopus laevis and Drosophila (Fig. 3).
Figure 2.
Determination of the Orc1 domain responsible for binding AlF-C2. (A) The wild-type (lane 1) and various mutants of Orc1 (lanes 2–6), represented in (B), were translated in vitro as T7-tagged proteins and incubated with glutathione beads coupled with GST–AlF-C2. Proteins bound to the beads were separated by SDS–PAGE and blotted with anti-T7 antibody (upper panel). The 1/10 volume of unbound fractions was also analyzed (lower panel). (B) Schematic representation of Orc1 and its truncation mutants. The BAH domain, ATPase (AAA+ family of ATPase) homology domain and AlF-C2 binding domain are shown with amino acid residue number.
Figure 3.
Amino acid sequence alignment of AlF-C2-binding domain in Orc1 from human, mouse and Chinese hamster. Shaded boxes display the identical amino acid sequences. Numbers denotes amino acid sequence number obtained from GenBank database, human Orc1 (accession no. Q13415), mouse Orc1 (accession no.Q9Z1N2) and Chinese hamster Orc1 (accession no. Q9JI69).
AlF-C2 domain responsible to Orc1 binding
AlF-C1 and AlF-C2 share an identical amino acid sequence except for a 47-residue-insertion in AlF-C1 (7). As described above, both proteins bound to Orc1 with strong affinity. In order to clarify AlF-C1 and AlF-C2 domains required for binding to Orc1, the AlF-C2 deletion mutants were similarly examined by in vitro pull-down assays (Fig. 4).
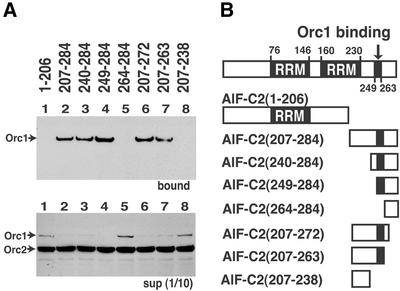
Figure 4.
Determination of the region of AlF-C2 that binds to Orc1. (A) Various deletion mutants of AlF-C2, schematically presented in (B), were prepared as GST-tagged proteins and bound to glutathione beads. T7-tagged Orc1 and Orc2 were incubated with glutathione beads that were coupled with GST–AlF-C2 deletion mutants. Proteins bound to the beads were separated by SDS–PAGE and blotted with anti-T7 antibody (upper panel). The 1/10 volume of unbound fractions was also analyzed (lower panel). (B) Schematic representation of AlF-C2 and its truncation mutants. The RRM and Orc1-binding domain are shown with amino acid residue number.
A series of deletion mutants of AlF-C2 were prepared as GST-tagged proteins (Fig. 4B), and each mutant protein was individually bound to glutathione beads. To these beads, T7-tagged Orc1 (full length) was added and the amounts of Orc1 bound to the mutant AlF-C2 proteins were determined by western blotting using anti-T7 tag antibody. In these experiments, in vitro translated T7-tagged Orc2 was included along with Orc1 protein in the binding reactions, as Orc2 could not bind to AlF-C1 and AlF-C2. Interaction of Orc2 with any of the AlF-C2 deletion mutants was not observed (Fig. 4A, upper panel), indicating that non-specific binding of in vitro translated Orc2 protein to AlF-C2 did not occur under our experimental condition. The results further confirm specific interaction between Orc1 and AlF-C proteins.
Deletion of C-terminal amino acids 207–284 [AlF- C2(1-206)] abolished the Orc1 binding (Fig. 4A, lane 1). In contrast, AlF-C2(207-284) containing C-terminal amino acids 207–284 (Fig. 4B) showed binding to Orc1 (Fig. 4A, lane 2). Then, a series of binding assays for N-terminal deletion mutants of AlF-C2(207-284) were performed (Fig. 4B). AlF-C2(240-284) and AlF-C2(249-284) bound to Orc1 (Fig. 4A, lanes 3 and 4), but AlFC2(264-284) did not (Fig. 4A, lane 5). The results indicated that amino acid sequence 249–263 in AlF-C2 was a prerequisite for the interaction with Orc1 (Fig. 4B). We also constructed a series of C-terminal deletion mutants from AlF-C2(207-284) (Fig. 4B). AlF-C2(207-272) and AlF-C2(207-263) bound to Orc1 (Fig. 4A, lanes 6 and 7). However, AlFC2(207-238) abolished the Orc1-binding activity (Fig. 4A, lane 8). Taking these results together, amino acids 249–263 in AlF-C2 were responsible for the interaction with Orc1.
AlF-C2 domain responsible for sequence-specific DNA binding
AlF-C1 and AlF-C2 have been identified as sequence-specific DNA-binding proteins (7). To define the region of AlF-C proteins responsible for bind site C DNA, a series of deletion mutants of AlF-C proteins were prepared as GST-fusion proteins (Fig. 5A) and these fusion proteins were subjected to EMSA with site C DNA as a probe (Fig. 5B).
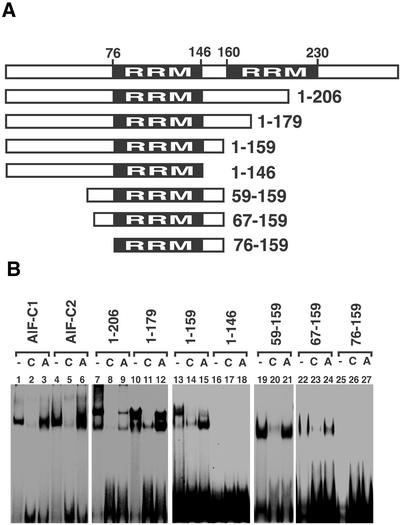
Figure 5.
Determination of the sequence-specific DNA-binding domain of AlF-C2. (A) Schematic representation of AlF-C2 and its truncation mutants. The RRM are shown with amino acid residue number. (B) EMSA using site C DNA as a probe. 32P-Labeled site C DNA was mixed with 200 ng of various mutants of GST–AlF-C2-fusion protein in the absense (lanes 1, 4, 7, 10, 13, 16, 19, 22 and 25) or presence of 100-fold molar excess amounts of non-labeled site C DNA (lanes 2, 5, 8, 11, 14, 17, 20, 23 and 26) or unrelated DNA[site A DNA from the AldB gene promoter (8); lanes 3, 6, 9, 12, 15, 18, 21, 24 and 27).
As shown in Figure 5B, both AlF-C1 and AlF-C2 proteins bind to site C DNA (lanes 1 and 4). The GST tag alone yielded no shifted band (data not shown). In competitive binding assays, the addition of 100-fold molar excess amount of non-labeled site C DNA abolished the shifted band (Fig. 5B, lanes 2 and 5), while addition of the same amount of site A DNA of the AldB promoter did not (Fig. 5B, lanes 3 and 6). Therefore, both AlF-C1 and AlF-C2 bound sequence-specifically to site C DNA. It should be noted here that, in our previous paper, we described that the AlF-C2 protein expressed in E.coli without tag bound to site C only weakly (7). In this work, we expressed GST-tagged AlF-C1 and AlF-C2 as soluble proteins in E.coli. In the former study, AlF-C proteins without tags were expressed in the inclusion body of E.coli, solubilized with guanidine–HCl and renatured by dialysis. Thus, it is possible that denaturation and renaturation caused lowered binding activity of AlF-C2 to site C.
A series of deletion mutants of AlF-C2 were prepared as GST-fusion proteins and subjected to EMSA. AlF-C2(1-206), AlF-C2(1-179) and AlF-C2(1-159) bound to site C DNA (lanes 7, 10 and 13). The addition of non-labeled site C DNA efficiently diminished the shifted bands (lanes 8, 11 and 14), while site A DNA did so only partially (lanes 9, 12 and 15). In contrast, AlF-C2(1-146) did not bind to site C (lanes 16). Thus, the amino acids 147–159 were indispensable for sequence-specific binding to site C. Protein motif search with Blocks (12) identified HMG14/HMG17 non-histone chromatin protein family motif (Block name: IPB000079) in 129–159 and histone H5 signature (Block name: PR00624E) in 141–155. Amino acids 129–159 of AlF-C2 were rich in basic residues (32%). Moreover, 46% (6/13) were basic in 147–159. Thus, basic amino acids in 147–159 might be important for binding to DNA.
To clarify the role of the N-terminal region, we constructed N-terminal truncation mutants from AlF-C2(1-159) (Fig. 5A). AlF-C2(59-159) and AlF-C2(67-159) bound to site C (lanes 19 and 22) and their sequence-specific binding was confirmed by competitive assays (lanes 20–21 and 23–24). In contrast, AlF-C2(76-159) did not bind to site C (lane 25). Therefore, the amino acids 67–75 in AlF-C2 were also important for binding to site C. Protein motif search with Blocks identified a 7 kDa DNA-binding protein motif (Block name: IPB003212B) in amino acids 71–96. The 7 kDa DNA-binding proteins were small abundant DNA-binding proteins in thermoacidophilic archaeon (13).
Taking these results together, a stretch of amino acids 67–159 in AlF-C proteins is responsible for the interaction with site C DNA, and amino acids 67–75 and 147–159 are indispensable. In the region from position 76 to 146, the Pfam protein families’ database search (14) identified the RNA recognition motifs (RRM). AlF-C proteins contain two RRM motifs at 76–146 and 160–230 (Fig. 5A). However, deletion of the latter RRM did not affect the DNA-binding activity.
DISCUSSION
In this work, we found that Orc1 directly bound to novel growth-regulated DNA-binding proteins, termed AlF-C1 and AlF-C2, which represses the AldB gene. AlF-C1 and AlF-C2 are members of a hnRNP family that have two RNA-binding domains (RRM and also RBD) (7). Although hnRNPs were first described as a major group of chromatin-associated RNA-binding proteins (15), recent studies revealed that they are involved in various biological functions, including repression of viral DNA replication (16), repression of estrogen- (17) and vitamin D-induced transcription (18) and transcriptional regulation of various genes (19–24). In addition, hnRNP, which binds telomeric DNA and is involved in mammalian telomere biogenesis, has been reported (25). The scaffold attachment factor A (SAF-A) and P130, components of the nuclear scaffold, are also members of hnRNP (26,27). Another hnRNP, MSSP, which binds to an origin of replication in the c-myc gene, interacts with DNA polymerase α and stimulates its activity (28). Usually, hnRNP proteins contain two to three RRM, and amino acids outside of RRM are variable (29,30). The variable region seems to be important to confer different functions of hnRNP proteins.
AlF-C1 and AlF-C2 are novel members of hnRNP family, in that they have separate domains to bind DNA sequence-specifically, to bind Orc1, and to repress transcription. These features of hnRNP have not been reported to date. We demonstrated that a stretch of amino acids 67–159 in AlF-C2 (and AlF-C1) was required for the interaction with site C DNA. Although this region contains RRM (amino acids 76–146), our study clearly indicated that RRM alone was not sufficient for DNA binding, but required flanked variable regions. However, both of the two RRMs in AlF-C2 are not required. This is contrasting to the MSSP protein, which requires two RRMs for all of its functions (31).
To the binding of Orc1, the amino acid sequence 249–263 of AlF-C proteins was shown to be responsible, which was not found in other hnRNPs except for closely related AlF-C proteins found in rat, mouse and human. This sequence is rich in Gly and Arg. Interestingly, a similar sequence was also found in the Epstein–Barr (EB) virus EBNA1 protein that binds to the replication origin of the viral DNA and activates DNA replication. Deletion of Gly- and Arg-rich sequence in EBNA1 protein resulted in decreased replication activity (32). In addition, it has recently been shown by immunoprecipitation experiments, that EBNA1 and Orc1 are co-precipitated as a complex (33). Although direct Orc1-EBNA1 interaction was not strictly proven, it might be possible that Gly- and Arg-rich sequences in EBNA1 and AlF-C proteins are important for interaction with Orc1.
Selection of replication origins in Saccharomyces cervisiae is mediated by the direct binding of ORC (34). In contrast, studies of metazoan ORCs have been less clear as to specific ORC-binding sequences (2). Recently, chromatin immunoprecipitation assays with antibodies against human Orc1 and Orc2 demonstrated that an Orc1- and Orc2-binding region on the chromatin coincided with an origin of DNA replication (35). However, direct binding of any metazoan ORC subunits to origin DNA has yet to be reported.
The way in which ORC interacts with chromatin differs between yeast and metazoan cells. Yeast ORC is stably associated with the chromatin throughout the cell cycle (36,37), whereas the interaction of metazoan ORC with chromatin changes during the cell cycle. Metazoan Orc1 is selectively released from chromatin during M phase (38) and S phase (39). Ubiquitination of the released mammalian Orc1 is sequestered from re-association (40,41). ORC activity is restored during M to G1 phase, which is concomitant with the reappearance of Orc1 tightly bound to chromatin, and with formation of pre-RCs at specific genomic sites (38,42,43). Therefore, mammalian Orc1 is thought to play a key role in such a cell cycle-regulated ORC activity for replication initiation.
Comparison of Orc1 among 11 different species, including single cell to Metazoa, revealed 52% similarity in the C-terminal regions that contain homologies with Cdc6 protein and the AAA+ family of ATPases, while exhibited 35% overall similarity (44). Presumably, the highly conserved C-terminal region of Orc1 is that involved in its functions conserved in all those species. The BAH domain which is identified in eukaryotic DNA methyltransferases and several proteins involved in transcription regulation, locates in the N-terminal region of Orc1 (11). In contrast, the middle part of the Orc1 proteins is highly variable among species, and thus it is conceivable that the middle part functions in species-specific mode of origin recognition. Accordingly, we found an Orc1 domain responsible for the interaction with AlF-C2 in the highly variable region. The amino acid sequence of this domain is conserved among human, mouse and Chinese hamster, but not in Drosophila, X.laevis and yeast (Fig. 3). This sequence is also conserved in rat (data not shown). Likewise, the amino acid sequence required for binding to Orc1 was perfectly conserved among rat, human and mouse AlF-C homologs (7). Therefore, it is considered that the interaction of Orc1 and AlF-C or AlF-C homologs occurs only in mammals.
It has been reported that human Orc2, Orc3, Orc4 and Orc5 form a core complex and that the interaction of Orc1 with the core complex is labile or cell-cycle dependent (45,46). Although we showed the direct interaction of Orc1 with AlF-C proteins (Fig. 1), we do not know at present whether AlF-C proteins can bind to Orc1 and subsequently associate with Orc2–5 complex, as such described above. Further experiments in vitro and in vivo are needed to answer the question to this point.
Concerning the origin selection by ORC, the ARS consensus sequence plays a large part in S.cerevisiae (34). In Schizosaccharomyces pombe, Orc4 is solely responsible for selection of specific ORC-binding sites; it binds to AT-rich sequences in the origin through its nine AT-hook motifs (47,48). In this view, the presence of an AT-rich sequence juxtaposed to the AlF-C-binding site at the rat AldB gene origin is of great interest; the AT-rich sequence has been shown to be required for ARS activity of the origin sequence (8). It is also interesting to consider that an unknown AT-hook protein might also be involved in recruiting ORC to the origin region of the rat AldB gene.
Unlike yeast S.cerevisiae, origins of DNA replication do not share common specific sequences in mammalian cells. If mammalian ORC does not have the ability to select origins based solely on its own affinity for specific consensus DNA sequence, how does it become localized to origins of replication? As mentioned above, a role for sequence-specific DNA-binding factors in the recruitment of ORC to specific genomic loci is currently suggested. It has been reported that the viral transcription factor EBNA1 binds to the EB virus origin of replication and recruits ORC in human cells (33,49,50). If such a mechanism does operate in mammalian genomic DNA, it is likely that a number of different DNA-binding factors would be involved in recruiting ORC to their selected origins. It is interesting to note here that the AlF-C1 and AlF-C2 proteins bound to site C in the AldB origin/promoter (7), which was one necessarily required element for autonomous replication in transfected cells (8). The association of AlF-C proteins with Orc1 implied that AlF-C proteins play a role in the recruitment of ORC to the replication origins in mammalian cells, similar to EBNA1 protein.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported in part by a Grant-in-aid for scientific research from The Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Gilbert D.M. (2001) Making sense of eukaryotic DNA replication origins. Science, 294, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell S.P. (2002) The origin recognition complex: from simple origins to complex functions. Genes Dev., 16, 659–672. [DOI] [PubMed] [Google Scholar]
- 3.Gavin K.A., Hidaka,M. and Stillman,B. (1995) Conserved initiator proteins in eukaryotes. Science, 270, 1667–1671. [DOI] [PubMed] [Google Scholar]
- 4.Diffley J.F. (2001) DNA replication: building the perfect switch. Curr. Biol., 11, R367–R370. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y., Tsutsumi,R., Yamaki,M., Nagatsuka,Y., Ejiri,S. and Tsutsumi,K. (1994) Initiation zone of DNA replication at the aldolase B locus encompasses transcription promoter region. Nucleic Acids Res., 22, 5385–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyagi S., Zhao,Y.P., Saitoh,Y., Tamai,K. and Tsutsumi,K. (2001) Replication of the rat aldolase B locus differs between aldolase B-expressing and non-expressing cells. FEBS Lett., 505, 332–336. [DOI] [PubMed] [Google Scholar]
- 7.Yabuki T., Miyagi,S., Ueda,H., Saitoh,Y. and Tsutsumi,K. (2001) A novel growth-related nuclear protein binds and inhibits rat aldolase B gene promoter. Gene, 264, 123–129. [DOI] [PubMed] [Google Scholar]
- 8.Miyagi S., Zhao,Y., Saitoh,Y. and Tsutsumi,K. (2000) An overlapping set of DNA elements in the rat aldolase B gene origin/promoter regulates transcription and autonomous replication. Biochem. Biophys. Res. Commun., 278, 760–765. [DOI] [PubMed] [Google Scholar]
- 9.Takayama M.A., Taira,T., Tamai,K., Iguchi-Ariga,S.M. and Ariga,H. (2000) ORC1 interacts with c-Myc to inhibit E-box-dependent transcription by abrogating c-Myc-SNF5/INI1 interaction. Genes Cells, 5, 481–490. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 11.Callebaut I., Courvalin,J.C. and Mornon,J.P. (1999) The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett., 446, 189–193. [DOI] [PubMed] [Google Scholar]
- 12.Henikoff J.G., Greene,E.A., Pietrokovski,S. and Henikoff,S. (2000) Increased coverage of protein families with the blocks database servers. Nucleic Acids Res., 28, 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mai V.Q., Chen,X., Hong,R. and Huang,L. (1998) Small abundant DNA binding proteins from the thermoacidophilic archaeon Sulfolobus shibatae constrain negative DNA supercoils. J. Bacteriol., 180, 2560–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- 16.Wang D. and Parrish,C.R. (1999) A heterogeneous nuclear ribonucleoprotein A/B-related protein binds to single-stranded DNA near the 5′ end or within the genome of feline parvovirus and can modify virus replication. J. Virol., 73, 7761–7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H., Hu,B., Gacad,M.A. and Adams,J.S. (1998) Cloning and expression of a novel dominant-negative-acting estrogen response element-binding protein in the heterogeneous nuclear ribonucleoprotein family. J. Biol. Chem., 273, 31352–31357. [DOI] [PubMed] [Google Scholar]
- 18.Chen H., Hu,B., Allegretto,E.A. and Adams,J.S. (2000) The vitamin D response element-binding protein. A novel dominant-negative regulator of vitamin D-directed transactivation. J. Biol. Chem., 275, 35557–35564. [DOI] [PubMed] [Google Scholar]
- 19.Leverrier S., Cinato,E., Paul,C., Derancourt,J., Bemark,M., Leanderson,T. and Legraverend,C. (2000) Purification and cloning of type A/B hnRNP proteins involved in transcriptional activation from the Rat spi 2 gene GAGA box. Biol. Chem., 381, 1031–1040. [DOI] [PubMed] [Google Scholar]
- 20.Michelotti E.F., Michelotti,G.A., Aronsohn,A.I. and Levens,D. (1996) Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol., 16, 2350–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miau L.H., Chang,C.J., Shen,B.J., Tsai,W.H. and Lee,S.C. (1998) Identification of heterogeneous nuclear ribonucleoprotein K (hnRNP K) as a repressor of C/EBPbeta-mediated gene activation. J. Biol. Chem., 273, 10784–10791. [DOI] [PubMed] [Google Scholar]
- 22.Tolnay M., Baranyi,L. and Tsokos,G.C. (2000) Heterogeneous nuclear ribonucleoprotein D0 contains transactivator and DNA-binding domains. Biochem. J., 348 (Pt 1), 151–158. [PMC free article] [PubMed] [Google Scholar]
- 23.Dempsey L.A., Hanakahi,L.A. and Maizels,N. (1998) A specific isoform of hnRNP D interacts with DNA in the LR1 heterodimer: canonical RNA binding motifs in a sequence-specific duplex DNA binding protein. J. Biol. Chem., 273, 29224–29229. [DOI] [PubMed] [Google Scholar]
- 24.Fuentes-Panana E.M., Peng,R., Brewer,G., Tan,J. and Ling,P.D. (2000) Regulation of the Epstein–Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J. Virol., 74, 8166–8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaBranche H., Dupuis,S., Ben-David,Y., Bani,M.R., Wellinger,R.J. and Chabot,B. (1998) Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nature Genet., 19, 199–202. [DOI] [PubMed] [Google Scholar]
- 26.Gohring F., Schwab,B.L., Nicotera,P., Leist,M. and Fackelmayer,F.O. (1997) The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target in apoptotic nuclear breakdown. EMBO J., 16, 7361–7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hibino Y., Ohzeki,H., Sugano,N. and Hiraga,K. (2000) Transcription modulation by a rat nuclear scaffold protein, P130 and a rat highly repetitive DNA component or various types of animal and plant matrix or scaffold attachment regions. Biochem. Biophys. Res. Commun., 279, 282–287. [DOI] [PubMed] [Google Scholar]
- 28.Niki T., Galli,I., Ariga,H. and Iguchi-Ariga,S.M. (2000) MSSP, a protein binding to an origin of replication in the c-myc gene, interacts with a catalytic subunit of DNA polymerase alpha and stimulates its polymerase activity. FEBS Lett., 475, 209–212. [DOI] [PubMed] [Google Scholar]
- 29.Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 30.Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- 31.Takai T., Nishita,Y., Iguchi-Ariga,S.M. and Ariga,H. (1994) Molecular cloning of MSSP-2, a c-myc gene single-strand binding protein: characterization of binding specificity and DNA replication activity. Nucleic Acids Res., 22, 5576–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H., Kapoor,P. and Frappier,L. (2002) Separation of the DNA replication, segregation and transcriptional activation functions of Epstein–Barr nuclear antigen 1. J. Virol., 76, 2480–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schepers A., Ritzi,M., Bousset,K., Kremmer,E., Yates,J.L., Harwood,J., Diffley,J.F. and Hammerschmidt,W. (2001) Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J., 20, 4588–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- 35.Ladenburger E.M., Keller,C. and Knippers,R. (2002) Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol., 22, 1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang C. and Stillman,B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev., 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- 38.Natale D.A., Li,C.J., Sun,W.H. and DePamphilis,M.L. (2000) Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G(1) transition in mammals. EMBO J., 19, 2728–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kreitz S., Ritzi,M., Baack,M. and Knippers,R. (2001) The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem., 276, 6337–6342. [DOI] [PubMed] [Google Scholar]
- 40.Li C.J. and DePamphilis,M.L. (2002) Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol., 22, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendez J., Zou-Yang,X.H., Kim,S.Y., Hidaka,M., Tansey,W.P. and Stillman,B. (2002) Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell, 9, 481–491. [DOI] [PubMed] [Google Scholar]
- 42.Sun W.H., Coleman,T.R. and DePamphilis,M.L. (2002) Cell cycle-dependent regulation of the association between origin recognition proteins and somatic cell chromatin. EMBO J., 21, 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li C.J., Bogan,J.A., Natale,D.A. and DePamphilis,M.L. (2000) Selective activation of pre-replication complexes in vitro at specific sites in mammalian nuclei. J. Cell Sci., 113 (Pt 5), 887–898. [DOI] [PubMed] [Google Scholar]
- 44.Bogan J.A., Natale,D.A. and Depamphilis,M.L. (2000) Initiation of eukaryotic DNA replication: conservative or liberal? J. Cell Physiol., 184, 139–150. [DOI] [PubMed] [Google Scholar]
- 45.Dhar S.K., Delmolino,L. and Dutta,A. (2001) Architecture of the human origin recognition complex. J. Biol. Chem., 276, 29067–29071. [DOI] [PubMed] [Google Scholar]
- 46.Vashee S., Simancek,P., Challberg,M.D. and Kelly,T.J. (2001) Assembly of the human origin recognition complex. J. Biol. Chem., 276, 26666–26673. [DOI] [PubMed] [Google Scholar]
- 47.Kong D. and DePamphilis,M.L. (2001) Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol. Cell. Biol., 21, 8095–8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J.K., Moon,K.Y., Jiang,Y. and Hurwitz,J. (2001) The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc. Natl Acad. Sci. USA, 98, 13589–13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dhar S.K., Yoshida,K., Machida,Y., Khaira,P., Chaudhuri,B., Wohlschlegel,J.A., Leffak,M., Yates,J. and Dutta,A. (2001) Replication from oriP of Epstein–Barr virus requires human ORC and is inhibited by geminin. Cell, 106, 287–296. [DOI] [PubMed] [Google Scholar]
- 50.Chaudhuri B., Xu,H., Todorov,I., Dutta,A. and Yates,J.L. (2001) Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein–Barr virus. Proc. Natl Acad. Sci. USA, 98, 10085–10089. [DOI] [PMC free article] [PubMed] [Google Scholar]