Abstract
We characterised the human hSuv3p protein belonging to the family of NTPases/helicases. In yeast mitochondria the hSUV3 orthologue is a component of the degradosome complex and participates in mtRNA turnover and processing, while in Caenorhabditis elegans the hSUV3 orthologue is necessary for viability of early embryos. Using immunofluorescence analysis, an in vitro mitochondrial uptake assay and sub-fractionation of human mitochondria we show hSuv3p to be a soluble protein localised in the mitochondrial matrix. We expressed and purified recombinant hSuv3p protein from a bacterial expression system. The purified enzyme was capable of hydrolysing ATP with a Km of 41.9 µM and the activity was only modestly stimulated by polynucleotides. hSuv3p unwound partly hybridised dsRNA and dsDNA structures with a very strong preference for the latter. The presented analysis of the hSuv3p NTPase/helicase suggests that new functions of the protein have been acquired in the course of evolution.
INTRODUCTION
Our research is focused on the structure and function of human hSuv3p protein belonging to the class of putative ATP-dependent RNA helicases from the DExH-box family. Sequence database analyses imply that Suv3p is encoded by a very ancient gene and highly conserved during evolution. Orthologues have been found in, for example, Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens. In Saccharomyces cerevisiae the hSUV3 orthologue encodes a subunit of the yeast mitochondrial degradosome (also known as mtEXO), which exhibits 3′→5′ exoribonuclease activity in vitro (1). The degradosome complex consists of two proteins encoded by nuclear genes and is imported into mitochondria from the cytosol: the DSS1 gene codes for a protein with domains characteristic for RNase II activity (2) while the SUV3 gene encodes a protein with a putative RNA helicase activity (3). The SUV3 null mutant lacks mtEXO RNase activity, which leads to abnormal phenotypes associated with RNA metabolism, including accumulation of excised group I introns, instability of intron-containing and intronless mitochondrial transcripts and aberrant processing of the precursors of the mitochondrial transcripts. Therefore, SUV3 knock-out strains are respiratory incompetent, show no mitochondrial translation and their mitochondrial genomes are quickly lost (2–6).
Our recent data indicate that the yeast mitochondrial degradosome is associated with mitochondrial ribosomes and that RNA helicase activity of the complex precedes the 3′→5′ exoribonucleolytic activity. The physiological function of the degradosome is not intron-related, the complex ensures proper turnover of mitochondrial RNA (mtRNA) and is involved in mtRNA surveillance (7).
In A.thaliana the AtSuv3p protein has a predicted molecular weight of 63.6 kDa. Subcellular fractionation experiments with transgenic plants allowed localisation of AtSuv3p to mitochondria. The N-terminal domain of AtSuv3p contains the motifs characteristic of RNA helicases and exhibits a low endogenous ATPase activity in vitro which can be stimulated by the presence of mtRNA (8). In C.elegans the activity of the SUV3 orthologue was found to be necessary for survival of early embryos (9).
Previously we cloned and initially characterised the cDNA of the human orthologue of SUV3. The gene was named SUPV3L1 and herein is referred to as hSUV3. We assigned hSUV3 to human chromosome band 10q22.1 by in situ hybridisation (10). Sequence analysis showed that the hSuv3p protein possesses a putative mitochondrial targeting sequence. The human gene is expressed in all tissues analysed. The highest levels of expression were found in liver and the lowest in lung (11).
In this paper we present more detailed studies on human hSuv3p. The protein was found to differ significantly from its yeast orthologue Suv3p. Using immunofluorescence, an in vitro mitochondrial uptake assay and sub-fractionation of human mitochondria we show that hSuv3p is localised in the mitochondrial matrix and is not membrane bound, as in yeast. Unexpectedly, enzymatic studies revealed that hSuv3p, in addition to its previously predicted RNA helicase activity, has much stronger DNA unwinding activity.
MATERIALS AND METHODS
Materials
DNA oligonucleotides were synthesised by Dr M. Schreiber (Bernhard-Nocht Institute) or purchased from Sigma. RNA oligonucleotides were purchased from HHMI Biopolymer/Keck Foundation Biotechnology (Resource Laboratory, Yale University School of Medicine). [γ-32P]ATP (220 Tbq/mmol) and [γ-33P]ATP (110 Tbq/mmol) were from Hartmann Analytic. 125I-labelled protein A (1.23 GBq/mg) was from Amersham Pharmacia Biotech. All other chemicals were obtained from Sigma.
Construction of plasmids
The pchSUV3myc and pcΔmthSUV3myc plasmids used for expression of wild-type hSuv3p and its mutated form without the predicted mitochondrial targeting sequence in COS-1 and HeLa cells were constructed as follows. Fragments of the hSUV3 cDNA encoding the full-length hSuv3p protein (2358 bp, 786 amino acids) or the form lacking the predicted mitochondrial targeting sequence (2304 bp, 768 amino acids), respectively, were PCR amplified using the forward primer 5′-GCA TCT AGA CAC GAT GGC CTT CTC CCG TGC CCT ATT GTG G-3′ (for pchSUV3myc) or 5′-CTA GTC TAG AAT GGG CCA CCG GGC AGC CAT CTG C-3′ (for pcΔmthSUV3myc), both introducing a XbaI site (underlined). In both cases the reverse primer was 5′-CGT GAA TTC ACA GGT CCT CCT CGG AGA TCA GCT TCT GCT CGT CCG AAT CAG GTT CCT TCT TC-3′, introducing a c-myc (9E10) epitope coding sequence (italic) and EcoRI site (underlined). The pKK plasmid (11) containing a full-length cDNA clone of the hSUV3 gene served as a template. The resulting fragment was cloned into the XbaI and EcoRI sites of the pcDNA3.1(–) vector (Stratagene).
For the biochemical characterisation of the hSUV3 gene product and raising specific antibodies against hSuv3p, a fragment of the protein devoid of 22 amino acids from the N-terminus was expressed in Escherichia coli. The expression plasmid pQEΔmthSUV3his was constructed as follows. A 2292 bp fragment of the hSUV3 cDNA was amplified by PCR using the primers 5′-TAC CCA TGG GCA TCT GCT CTG CCC TTC G-3′ (forward), introducing a NcoI site (underlined), and 5′-CTG GGA TCC GTC CGA ATC AGG TTC CTT C-3′ (reverse), introducing a BamHI site (underlined), using the pKK plasmid as a template. The resulting fragment was cloned into the NcoI and BamHI sites of the pQE60 expression vector (Qiagen).
pQEΔmthSUV3G207Vhis encoding mutated protein hSuv3p(G207V) in which Gly207 was replaced by valine was constructed employing a QuickChange Site-Directed Mutagenesis Kit (Invitrogen) according to the manufacturer’s instructions and pQEΔmthSUV3his plasmid as a template. The following primers were used in site-directed mutagenesis: forward, 5′-TAA TAT TTC ATT CAG TCC CCA CAA ACA GTG-3′; reverse, 5′-CAC TGT TTG TGG GGA CTG AAT GAA ATA TTA-3′ (substituted nucleotide underlined).
The sequences of all resulting constructs were verified by sequencing of both strands using an ABI Prism 377DNA Sequencer.
Immunofluorescence experiments
Samples of 5 × 104 HeLa or COS-1 cells were plated per well in 6-well plates (Costar) with a coverslip placed at the bottom of the well and grown overnight in DMEM (Sigma) supplemented with 10% FCS and 4 mM glutamine. Immunofluorescence microscopy was performed as follows. Twenty-four hours after transfection Mitotracker CMX-Ros (300 nM) was added to the medium and incubated at 37°C for 30 min. Thereafter, the cells were washed three times with phosphate-buffered saline (PBS), fixed with 4% formaldehyde in PBS for 15 min at 20°C and then permeabilised with 1% Triton X-100 in PBS for 5 min. The cells were then incubated with anti-hSuv3p serum (diluted 1:200 in PBS with 10% FCS) (see below) for 1 h, washed in PBS and the bound antibody was detected with fluorescein 5-isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (1:200) (Molecular Probes). Specimens were examined using a MRC Bio-Rad confocal microscope.
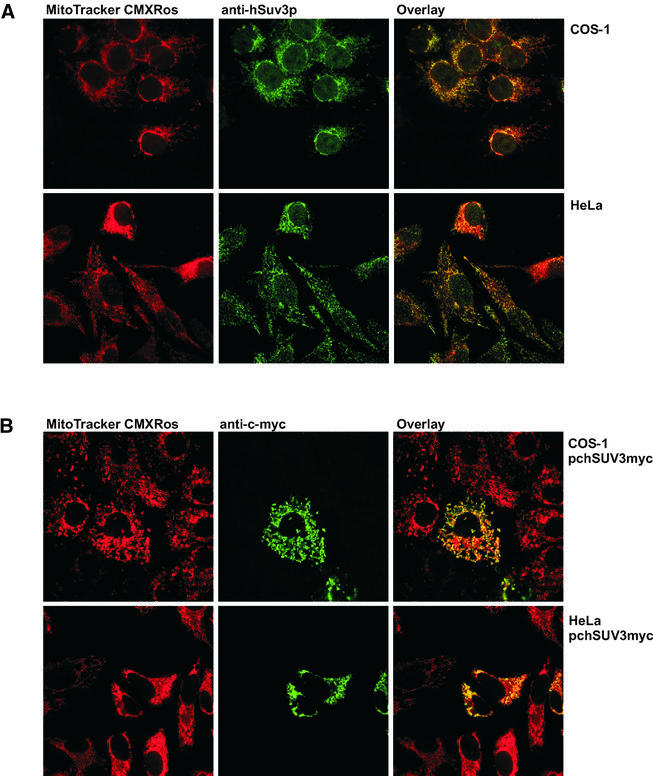
In order to determine the subcellular localisation of the overexpressed hSuv3p protein HeLa or COS-1 cells were plated as described above and transiently transfected with pchSUV3myc or pcΔmthSUV3myc using FuGene6 reagent (Roche). Detection of the protein was performed as described above using anti-c-myc 9E10 monoclonal antibody (1:200) (Santa Cruz Biotechnology), followed by incubation with FITC-conjugated goat anti-mouse IgG (1:200) (Molecular Probes).
Import of hSuv3p into isolated mitochondria
The in vitro import assay was performed essentially as described by Tokatlidis (12). Briefly, yeast mitochondria (100 µg protein) were incubated with 35S-labelled proteins (synthesised in vitro using the rabbit reticulocyte lysate system; Promega) for 15 min at 30°C in 200 µl of import buffer (IB) containing 100 mM HEPES, pH 7.1, 1.2 M sorbitol, 4 mM KH2PO4, 100 mM KCl, 20 mM MgCl2, 10 mM l-methionine and 2 mg/ml fatty acid-free bovine serum albumin supplemented with 5 mM NADH. Mitochondria were harvested by centrifugation and resuspended in 100 µl of IB supplemented with 0.1 mg/ml trypsin and incubated on ice. After 30 min trypsin was inhibited by addition of soy bean trypsin inhibitor to a concentration of 0.2 mg/ml and incubation for 15 min on ice. In order to generate mitoplasts by hypotonic swelling, mitochondrial pellets were resuspended in 30 µl of IB, diluted with 210 µl of 20 mM HEPES–KOH, pH 7.4, and incubated for 30 min on ice. After centrifugation mitoplasts were precipitated from the supernatant with TCA and resuspended in 100 µl of IB containing 30 µg/ml proteinase K. After 15 min incubation on ice proteinase K was inhibited by incubating with 2 mM phenylmethylsulphonyl flouride (PMSF) for 10 min on ice. For alkali extraction, mitoplasts were sedimented by centrifugation, resuspended in 100 µl of 100 mM Na2CO3, 2 mM PMSF (ice-cold) and incubated for 30 min on ice. The supernatant (matrix) and the pellet (membranes) were separated by centrifugation at 70 000 r.p.m. (TL100) for 30 min. The matrix proteins were TCA precipitated and all the fractions were resolved by SDS–PAGE, transferred to nitrocellulose membranes and visualised by autoradiography (Fuji Bas Station v.1.3). Subsequently the blots were analysed using antibodies specific to endogenous yeast mitochondrial proteins: anti-porin for the outer membrane, anti-mtHsp70p for the matrix and anti-Cytb2 for the inter membrane space. These antibodies were prepared against purified yeast mitochondrial proteins and originated from the laboratory collection of Dr G. Schatz (also available in the laboratory of K. Tokatlidis) (13).
Isolation and fractionation of mitochondria from HeLa cells
Mitochondria were isolated as described by Gaines (14) with the following modifications. Briefly, exponentially growing HeLa cells (conditions as described above) were trypsinised, washed twice in PBS and then twice with the washing buffer (1 mM Tris–HCl, pH 7.0, 0.13 M NaCl, 5 mM KCl and 7.5 mM MgCl2). The cell pellet (∼1.5 ml) was resuspended in 0.75 ml of 0.1× IB (4 mM Tris, pH 7.4, 2.5 mM NaCl and 0.5 mM MgCl2) and homogenised using a Potter homogeniser. The homogenate was immediately mixed with 0.17 ml of 10× IB. The unbroken cells and nuclei were removed by three consecutive low speed centrifugations (700 g for 3 min). The remaining supernatant was centrifuged at 18 000 g for 2 min. The pelleted proteins served as a crude mitochondrial fraction and the supernatant as a cytosolic fraction. The identity of the cytosolic and mitochondrial fractions was verified by the presence of specific marker enzymes. When the activity of lactate dehydrogenase in the starting cell homogenate was taken as 100%, 84–90% of the activity was recovered in the cytosolic fraction as measured according to the method described in Bergmeyer and Bernt (15). The recovery of the matrix enzyme citrate synthase in the mitochondrial fraction, measured as described in Graham and Rickwood (16), was in the range 75–82%.
For sub-fractionation experiments the crude mitochondrial fraction was further purified. The crude mitochondria were resuspended in TES buffer [10 mM Tris–HCl, pH 7.4, 1 mM EDTA, 0.25 M sucrose supplemented with protease inhibitor cocktail (Roche) and 2 mM PMSF], layered on a discontinuous sucrose gradient made by successive layering of 1.5 M and 1.0 M sucrose and centrifuged at 60 000 g for 1 h. The phase between the 1.5 and 1.0 M sucrose layers (mitochondrial fraction) was collected and washed twice with TES buffer.
The sub-fractionation of mitochondria and preparation of mitoplasts was described previously (17). Briefly, 3 mg of freshly prepared mitochondria in 1 ml of TES were sonicated and then centrifuged at 320 000 g for 1 h at 4°C. The supernatant was TCA precipitated and used as the mitochondrial soluble fraction; the pellet served as the mitochondrial membrane fraction. To obtain mitoplasts, mitochondria (3 mg) in 0.3 ml of TES were diluted 10-fold with buffer containing 5 mM Tris–HCl, pH 7.4, and 1 mM EDTA and centrifuged at 14 000 g for 20 min at 4°C. The pellet was resuspended in 3 ml of the same hypotonic buffer and left to stand on ice for 20 min. The suspension containing mitoplasts was centrifuged at 14 000 g for 20 min at 4°C and washed three times with 3 ml of the hypotonic buffer. All supernatants were pooled and proteins were TCA precipitated. Sub-fractions were additionally verified by immunoblotting with antibodies specific to human mitochondrial proteins: anti-Hsp60p (monoclonal antibody purchased from Sigma) and anti-cytochrome c (rabbit polyclonal antibody obtained from Santa Cruz Biotechnology).
Purification of hSuv3p
Escherichia coli strain DH5α transformed with pQEΔmthSUV3his or pQEΔmthSUV3G207Vhis expression plasmid (see Construction of plasmids) was cultured and induced with isopropyl-β-d-thiogalactoside according to the protocol for the QIAexpress Expression System (Qiagen).
In order to obtain the antigen for immunisation of rabbits the hSuv3p protein was purified under denaturing conditions as follows. Bacteria were lysed at 100°C using a buffer containing 6 M guanidine hydrochloride, 2% Triton-X, pH 8.0. After binding to Ni–NTA resin the protein was eluted at pH 4.2. The hSuv3p specific band was verified by immunoblotting with anti-hexahistidine (his) tag monoclonal antibody (not shown) and electroeluted from a polyacrylamide gel.
To study the biochemical properties of hSuv3p, the wild-type or mutated form of the protein was purified under non-denaturing conditions as follows. After induction the bacteria were collected by centrifugation and disrupted by sonication in lysis buffer (100 mM Tris–HCl, pH 7.5, 20% glycerol, 0.1% Triton X-100, 200 mM NaCl, 1 mM β-mercaptoethanol, 2 mM PMSF and 10 mM imidazole). The insoluble material was pelleted at 26 000 g and the supernatant was mixed with 3 ml of nickel-charged resin (Qiagen) equilibrated with buffer containing 20 mM Tris–HCl, pH 7.5, 10% glycerol, 0.05% Triton X-100 and 1 mM β-mercaptoethanol for 12 h at 4°C. The matrix was transferred onto a column and washed with the same buffer supplemented with 200 mM NaCl and 20 mM imidazole. The bound protein was eluted with the above buffer containing 0.5 M imidazole. The purity of the obtained protein was ∼70% as judged by SDS–PAGE. The eluted fraction was concentrated by ultrafiltration on a 30 kDa membrane (Millipore) and fractionated on a Superdex-200 column (Hi-Load; Amersham Pharmacia Biotech) equilibrated with TGT buffer (20 mM Tris–HCl, pH 7.5, 10% glycerol, 0.05% Triton X-100, 1 mM EDTA and 1 mM β-mercaptoethanol). Fractions containing the highest ATPase and helicase activities were pooled and analysed for the enzymatic properties of the NTPase/helicase (storage in 30% glycerol at –70°C). The hSuv3p was >95% pure as judged by densitometric analysis of Coomassie Blue stained polyacrylamide gels.
Raising antibodies
The purified hSuv3p protein (200 µg) was mixed with complete Freund’s adjuvant and used for the immunisation of rabbits. Booster injections were performed using incomplete Freund’s adjuvant, each of them about 1 month apart. After the fourth injection ∼50 ml of serum was taken, salted out twice using 50% ammonium sulphate and the resulting pellet resuspended and dialysed against PBS, pH 7.4. The immunoreactivty of the serum prepared against hSuv3p was verified by immunoblotting. The total homogenate of HeLa cells was immunoblotted and probed with the anti-hSuv3p serum. The antibodies reacted with an 87 kDa protein (Fig. 4A, lane 1). This corresponded to the molecular mass deduced from the hSUV3 cDNA sequence. No signals were visible when preimmune IgG was used (not shown).
Figure 4.
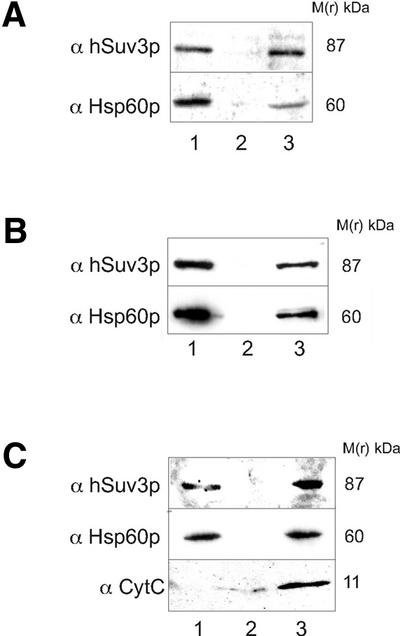
Localisation of the hSuv3p protein in the mitochondrial matrix of human mitochondria. (A) Crude mitochondrial fraction (lane 1), cytosolic fraction (lane 2) and total cell extract (lane 3) of HeLa cells were separated by SDS–PAGE, immunoblotted and probed with the anti-hSuv3p serum (α hSuv3p). (B) Soluble submitochondrial fraction (lane 1), membrane submitochondrial fraction (lane 2) and total extract of purified mitochondria of HeLa cells (lane 3) were immunoprobed with the anti-hSuv3p serum (α hSuv3p) as above. (C) Mitoplasts (lane 1), post-mitoplast supernatant (lane 2) and total extract of purified mitochondria of HeLa cells (lane 3) were separated by SDS–PAGE, immunoblotted and probed with the anti-hSuv3p serum (α hSuv3p). Subcellular and submitochondrial fractions were verified by immunoblotting with antibodies specific to the endogenous proteins localised in different mitochondrial subcompartments: anti-Hsp60p for the matrix (α Hsp60p) and in (C) anti-CytC for the intermembrane space (α CytC). Molecular masses are indicated on the left-hand side of the blot images.
ATPase assay
The ATPase activity was determined under conditions described previously (18,19). Briefly, the enzyme (2 pmol) was incubated in a reaction mixture (final volume 25 µl) containing 20 mM Tris–HCl, pH 7.5, 2 mM MgCl2, 1 mM β-mercaptoethanol, 10% glycerol, 0.01% Triton X-100, 0.1 mg/ml BSA, 25 nCi [γ-33P]ATP and ATP adjusted to the concentrations indicated in the figure legends (Fig. 8). The reaction was carried out for 30 min at 30°C and terminated by the addition of 0.75 ml of activated charcoal (2 mg/ml). After incubation for 30 min at 30°C followed by centrifugation at 10 000 g for 10 min, 200 µl aliquots of the supernatant were removed and subjected to scintillation counting.
Figure 8.
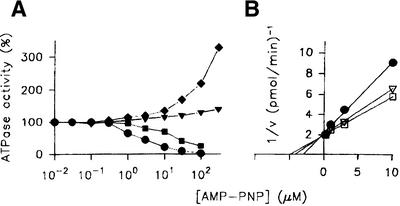
Modulation of the ATPase activity of hSuv3p by AMP-PNP with variations in ATP concentration. (A) Investigation of the ATPase activity as a function of increasing concentrations of AMP-PNP in the presence of different ATP concentrations. The demonstrated assays were performed at ATP concentrations of hSuv3p ATPase (41.9 µM) equal to (diamond) 100 × Km; (triangle) 1 × Km; (square) 0.001 × Km; and (circle) 0.00001 × Km. The activities of the enzyme measured for each ATP concentration in the absence of AMP-PNP was referred to as 100%. (B) Kinetic analysis of the inhibition of the hSuv3p ATPase at lower ATP concentrations. Data obtained at ATP concentrations equal to (circle) 0.00001 × Km, (open triangle) 0.00003 × Km and (open square) 0.0001 × Km were replotted according to the method of Dixon (37).
Preparation of substrates for the helicase reaction
The substrates for the helicase reaction were obtained by annealing two partially complementary DNA or RNA oligonucleotides with the sequences reported by Gallinari et al. (20). The release strand (26mer) was 5′-end-labelled with [γ-32P]ATP by using T4 polynucleotide kinase (MBI Fermentas) as recommended by the manufacturer. For the annealing reaction the labelled oligonucleotide was combined at a molar ratio of 1:10 with the template strand (40mer), denatured for 5 min at 96°C and slowly renatured as described previously (20). The resulting duplex was electrophoresed in a 15% native TBE polyacrylamide gel, visualised by autoradiography and extracted as described previously (21). The amount of the DNA or RNA duplex used as substrate for the NTPase/helicase was determined by an ethidium bromide fluorescent quantitation method described previously (22).
Helicase assay
The helicase activity was tested using 2 pmol (if not indicated otherwise) hSuv3p protein. Unwinding of the partially hybridised DNA or RNA substrate (4.7 pM) was allowed to occur in a reaction mixture (final volume 25 µl) containing 20 mM Tris–HCl, pH 7.5, 2 mM MgCl2, 1 mM β-mercaptoethanol, 10% glycerol, 0.01% Triton X-100, 0.1 mg/ml BSA and ATP adjusted to a concentration corresponding to the Km value determined for the ATPase reaction. The reaction was carried out for 30 min at 30°C and stopped by addition of 8 µl of termination buffer (100 mM Tris–HCl, pH 7.5, 20 mM EDTA, 0.5% SDS, 0.1% Triton X-100, 25% glycerol and 0.1% bromophenol blue). The samples were separated in a 15% Tris–borate–EDTA (TBE) polyacrylamide gel containing 0.1% SDS (23). The gels were dried and exposed to Kodak X-ray film at –70°C. Subsequently, the parts of the gels corresponding to the released strand and to not unwound substrate were cut out and 32P radioactivity was measured. Alternatively, the films were scanned and the radioactivity was quantified using GelImage software (Amersham Pharmacia Biotech).
Immunoblotting
After SDS–PAGE the proteins were transferred to nitrocellulose filters (BA 85, 0.45 µm; Schleicher & Schüll). The filters were incubated for 1 h with 1 mg/ml BSA in 25 mM Tris–HCl, pH 7.5, 150 mM NaCl and 0.05% Tween-80 (TTBS buffer) and for 2 h with antisera (1:500 in TTBS containing 10% glycerol and 3 mg/ml BSA). The filters were washed again with 1 mg/ml BSA in TTBS and the bound antibodies were detected by binding of [125I]protein A (0.05 µCi/ml) or with rabbit anti-mouse antibodies followed by [125I]protein A. The nitrocellulose filters were dried and subjected to autoradiography.
Other assays
Protein concentration was determined by SDS–PAGE as described previously (24) or using a Bio-Rad DC protein assay kit with bovine serum albumin as a standard, according to the manufacturer’s instructions. Kinetic parameters were determined by a non-linear regression analysis using ENZFITTER (BioSoft) and SIGMA PLOT (Jandel Corp.). Protein sequences of hSUV3 orthologues were aligned with Gapped BLAST (v.2.0.10) with SEG and COIL filtering (25).
RESULTS
Subcellular distribution of hSuv3p
In order to determine the subcellular distribution of the endogenous hSuv3p within mammalian cells we used a polyvalent anti-hSuv3p rabbit serum prepared as described in Materials and Methods. COS-1 and HeLa cells were stained with the primary anti-hSuv3p serum and visualised with FITC-conjugated secondary antibodies. Immunofluorescence microscopy showed that the signal originating from hSuv3p co-localised with the mitochondrial marker MitoTracker CMXRos in both COS-1 and in HeLa cells (Fig. 1A). This analysis also revealed faint nuclear staining patterns for both of the examined cell types. However, when a control staining with preimmune serum was carried out we could observe a weak signal with a cytoplasmatic and nuclear pattern (not shown). Therefore, we concluded that the majority of the hSuv3p protein localises in mitochondria. In order to exclude a possible double, nucleo-mitochondrial localisation, the hSuv3p protein was C-terminally tagged with c-myc epitope 9E10 and its subcellular localisation was studied in transiently transfected COS-1 and HeLa cells (Fig. 1B). The mitochondrial localisation was confirmed by using the mitochondria-specific dye MitoTracker CMXRos.
Figure 1.
Mitochondrial localisation of hSuv3p in mammalian cells. (A) COS-1 and HeLa cells were grown on coverslips, incubated with the mitochondria-specific dye MitoTracker CMXRos (300 nM), fixed with 4% formaldehyde and permeabilised with 1% Triton X-100. The cells were immunostained with anti-hSuv3p serum and visualised with FITC-conjugated secondary antibody. Fluorescent images of MitoTracker (red) and hSuv3p specific signal (green) were taken by a confocal microscope. Co-localisation of hSuv3p and mitochondria appear yellow in digitally overlaid images. (B) COS-1 and HeLa cells were grown on coverslips and transiently transfected with cDNA encoding c-myc-tagged hSuv3p (pchSUV3myc). After incubation with MitoTracker, fixation and permeabilisation as described in (A), the cells were immunostained with anti-c-myc monoclonal antibody, which was then visualised with FITC-conjugated antibody. Fluorescent images of MitoTracker (red) and c-myc-tagged hSuv3p (green) were taken by a confocal microscope. Co-localisation of hSuv3p and mitochondria appear yellow in digitally overlaid images.
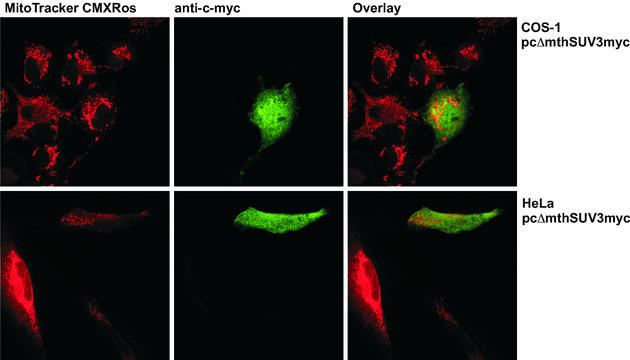
We used SignalP v.2.0 software (26) in order to predict the presence and localisation of signal peptide cleavage sites in the amino acid sequence of hSuv3p. The most probable cleavage site of the mitochondrial leader in hSuv3p was found between the residues at positions 18 and 19 (in the hSuv3p sequence …RQA↓GH…; cleavage site marked by arrow). We constructed hSuv3p truncated N-terminally of 18 amino acids and incorporated a c-myc tag at the C-terminus. When this tagged protein was transiently expressed in COS-1 and HeLa cells, co-localisation with the mitochondrial marker MitoTracker was significantly reduced (Fig. 2). It therefore appears that the N-terminal part of hSuv3p is responsible for targeting of the protein to human mitochondria whilst the protein lacking the mitochondrial targeting sequence is retained in the cytoplasm.
Figure 2.
N-terminal sequence is necessary for the mitochondrial import of the hSuv3p protein. COS-1 and HeLa cells were grown on coverslips and transiently transfected with cDNA encoding N-terminally truncated (the first 18 amino acids) hSuv3p with a c-myc tag at the C-terminus (pcΔmthSUV3myc). After incubation with the mitochondria-specific label MitoTracker CMXRos (300 nM), fixation with 4% formaldehyde and permeabilisation with 1% Triton X-100 the cells were immunostained with anti-c-myc monoclonal antibody. The c-myc-tagged N-terminally truncated hSuv3p was visualised with FITC-conjugated antibody (green); the mitochondria stained with MitoTracker are red. The hSuv3p protein devoid of the 18 N-terminal amino acids did not co-localise with the mitochondria in either COS-1 or HeLa cells.
Localisation of hSuv3p in the mitochondrial matrix
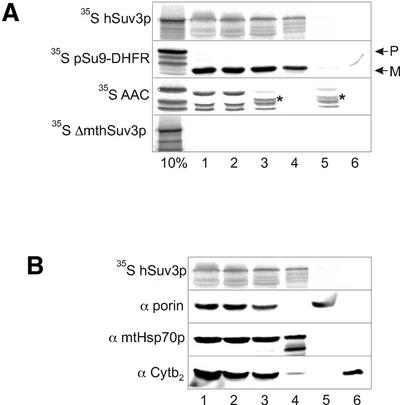
In order to determine the localisation of hSuv3p within mitochondria yeast mitochondria were used. Published data indicate that the transport machinery in yeast and human mitochondria do not differ significantly (27). Radiolabelled hSuv3p synthesised in vitro using the rabbit reticulocyte transcription and translation system was subjected to in vitro mitochondrial import followed by fractionation of the mitochondria into separate compartments. In parallel, mitochondrial uptake and fractionation of two well characterised mitochondrial proteins, pSu9–DHFR and AAC, was performed. pSu9–DHFR is a chimeric protein that contains the first 69 amino acids of Neurospora crassa Fo-ATP synthase subunit 9 fused to murine dihydrofolate reductase (DHFR). After the import of pSu9–DHFR the pre-protein is cleaved, liberating the mature form which localises in the mitochondrial matrix (28). AAC is an ADP/ATP carrier protein localised in the mitochondrial inner membrane. In contrast to pSu9–DHFR, AAC does not have a cleavable pre-sequence and its targeting information is contained within its internal sequence (29). When [35S]methionine-labelled hSuv3p was incubated with yeast mitochondria the protein was transported across the mitochondrial outer membrane and became inaccessible to trypsin. AAC and mature pSu9–DHFR were also imported and resistant to proteolysis. In contrast to this, hSuv3p without the mitochondrial targeting sequence (see above) was not imported into the mitochondria and hence was susceptible to the added protease (Fig. 3A, lane 1). To sub-fractionate mitochondria the outer membrane was disrupted by osmotic shock, which released components of the inter-membrane space. The resulting mitoplasts were additionally treated with proteinase K. Under these conditions full-length hSuv3p along with mature pSu9–DHFR and AAC remained associated with the mitoplast pellet and were protected from proteinase K enzymatic digestion (Fig. 3A, lanes 2 and 3). No hSuv3p was detected in the intermembrane space fraction (Fig. 3A, lane 6). Alkali extraction of the mitoplasts with Na2CO3 showed that hSUV3p was imported mainly into the mitochondrial matrix and is not embedded in the membrane, just like mature pSu9–DHFR (Fig. 3A, lane 4), whereas AAC was inserted into the inner membrane (Fig. 3A, lane 5). The submitochondrial fractionation after hSuv3p import was confirmed by immunoblotting using antibodies against endogenous marker mitochondrial proteins localised in different mitochondrial subcompartments: anti-porin for the outer membrane, anti-mtHsp70p for the matrix and anti-Cytb2 for the inter-membrane space (Fig. 3B) (13).
Figure 3.
The hSuv3p protein imported into isolated yeast mitochondria is localised in the mitochondrial matrix. (A) In vitro synthesised radiolabelled proteins, hSuv3p (wild-type), pSu9–DHFR (localised in the mitochondrial matrix), AAC (localised in the mitochondrial inner membrane) and ΔmthSuv3p (hSuv3p protein devoid of the 18 N-terminal amino acids), were incubated with isolated yeast mitochondria. After import, mitochondria were treated with protease (trypsin) to remove non-imported precursor (lane 1). In the next step the mitoplasts were prepared by osmotic shock (lane 2) and treated with proteinase K (lane 3). After alkali extraction of the mitoplasts with Na2CO3 the soluble fraction (lane 4) and the membrane pellets (lane 5) were analysed. The inter-membrane space fraction released after osmotic swelling was also examined (lane 6). All the protein samples were separated by SDS–PAGE, transferred onto nitrocellulose membranes and visualised by autoradiography. For pSu9–DHFR arrows indicate the precursor (P) and mature peptide cleaved by mitochondrial MPP (M). Asterisks show the protease-resistant fragment of the protein AAC inserted into the mitochondrial inner membrane. The 10% of the input is also shown. (B) The mitochondrial fractions transferred onto nitrocellulose membranes after the import of hSuv3p from (A) were verified by immunoblotting with antibodies specific to the endogenous proteins localised in different mitochondrial subcompartments: anti-porin for the outer membrane (α porin), anti-mtHsp70p for the matrix (α mtHsp70p) and anti-Cytb2 for the inter-membrane space (α Cytb2).
In order to confirm the localisation of hSuv3p in the mitochondrial matrix in human cells the sub-fractions of mitochondria from HeLa cells were examined for the presence of hSuv3p by immunoblotting using the anti-hSuv3p serum. The hSuv3p-specific band was detected predominantly in the crude mitochondrial fraction (Fig. 4A, lane 1). Subsequently purified human mitochondria were sub-fractionated into membrane and soluble fractions. When the submitochondrial fractions were examined by immunoblotting, the hSuv3p-specific band was detected almost exclusively in the soluble fraction, along with the mitochondrial matrix protein Hsp60p, which was used as a marker (30) (Fig. 4B, lane 3). Soluble mitochondrial fraction contained both matrix and inter-membrane space components. Therefore, to investigate whether mitochondrial hSuv3p is localised in the inter-membrane space or in the matrix, mitoplasts and the post-mitoplast supernatant were analysed by immunoblotting. hSuv3p was retained exclusively in the mitoplasts, similarly to Hsp60p matrix marker protein (Fig. 4C, lane 3). Cytochrome c, an inter-membrane space marker protein (31), was recovered in the post-mitoplast supernatant (Fig. 4C, lane 2). These results indicate that hSuv3p is localised in the matrix of human mitochondria.
Heterologous production and enzymatic properties of hSuv3p
In order to study the biochemical properties of hSuv3p protein, the cDNA encoding the polypeptide consisting of 764 amino acid residues (hSuv3p without the first 22 N-terminal amino acids) fused to a his tag at its C-terminus was expressed in E.coli. After disruption of the bacteria, the pool of enzyme present in the Triton X-100 solubilised fraction was purified by Ni2+ affinity chromatography under non-denaturing conditions. In addition to the expected 87 kDa protein, the imidazole-eluted fraction contained an induction-specific protein that migrated at ∼55 kDa and some background of bacterial proteins (Fig. 5A, lane 2). The 87 and 55 kDa polypeptides were not present in a similar fraction prepared from non-induced bacteria or from induced bacteria harbouring empty pQE60 plasmid (not shown).
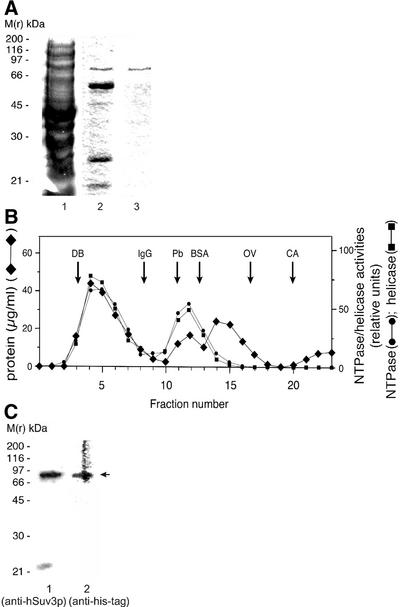
Figure 5.
Purification of hSuv3p from E.coli. (A) SDS–PAGE analysis of purification steps. Triton X-100 soluble fraction (lane 1); Ni2+ affinity chromatography (lane 2); Superdex-200 column (lane 3). The samples were separated by SDS–PAGE and the proteins stained with Coomassie Blue. M(r) kDa, molecular mass markers in kilodaltons. (B) Gel filtration purification of hSuv3p. The distribution of protein and of NTPase and helicase activities of the hSuv3p preparation obtained by Ni2+ affinity chromatography on an analytical Superdex-200 column is shown. The NTPase and the helicase assays were performed as described in Materials and Methods. In the case of the helicase assay a dsDNA substrate was used. The column was calibrated with dextran blue (DB) (2000 kDa) and with the following marker proteins: IgG (160 kDa); PB, phosphorylase b (97 kDa); BSA (66 kDa); OV, ovalbumin (45 kDa); CA, carbonic anhydrase (30 kDa). (C) Aliquots of pooled fractions from the Superdex-200 column displaying ATPase and helicase activity were removed and precipitated with TCA. Samples were separated by SDS–PAGE followed by immunoblotting with anti-hSuv3p antiserum (lane 1) and with anti-his tag antibody (lane 2). The nitrocellulose filters were autoradiographed for 24 h. M(r) kDa, molecular mass markers in kilodaltons. The arrow indicates the position of hSuv3p.
In order to separate the 87 and 55 kDa polypeptides and to remove the contaminating proteins gel filtration chromatography on Superdex-200 was employed. The elution profile was monitored by ATPase and helicase assays (see below). Both enzymatic activities migrated in two peaks: the first one at the void volume of the column and the second one corresponding to proteins migrating at 80–90 kDa (Fig. 5B). The void volume fraction analysed by SDS–PAGE contained a mixture of both 55 and 87 kDa proteins (not shown). The second peak of the ATPase and helicase activities consisted of a homogeneous preparation of a protein with an apparent molecular mass of 87 kDa, which is in agreement with the predicted molecular mass of monomeric form hSuv3p-his protein (86.4 kDa) (Fig. 5A, lane 3).
The identity of the purified protein was verified by immunoblotting with anti-hSuv3p antiserum and with anti-his tag antibody (Fig. 5C, lanes 1 and 2, respectively). The identity of the 55 kDa protein could also be determined. The protein was reactive with both antibodies, i.e. with anti-hSuv3p serum and with anti-his tag antibody, in immunoblots (data not shown). Since the his tag was attached to the C-terminus of hSuv3p protein we concluded that the 55 kDa protein represented the N-terminally degraded fragment of the intact 87 kDa polypeptide.
The above purification procedure was applied to the E.coli strain expressing the hSuv3p mutant with the replacement of Gly207 by valine [named hSuv3p(G207V)] within the N-terminus of the Walker A motif (32). An analogous replacement performed on previously described NTPase/helicases abolished their ATPase activity (33,34). As expected, for the hSuv3p(G207V) mutant protein neither ATPase nor helicase activity for either double-stranded (ds)DNA or dsRNA could be detected after gel filtration chromatography. Figure 6 presents data obtained with the dsDNA substrate.
Figure 6.
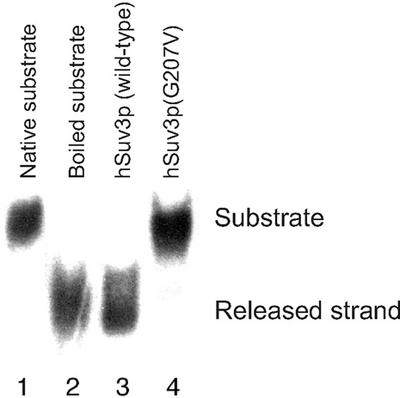
Lack of the NTPase and helicase activity of hSuv3p mutated in the Walker A motif. An aliquot of 2 pmol of homogeneously purified hSuv3p wild-type enzyme (lane 3) and its mutated form hSuv3p(G207V) (lane 4) was allowed to react with a dsDNA substrate (4.7 pM) as described in Materials and Methods. The substrate and released strand were separated in a TBE polyacrylamide gel and visualised by exposure of the dried gel to X-ray film for 24 h.
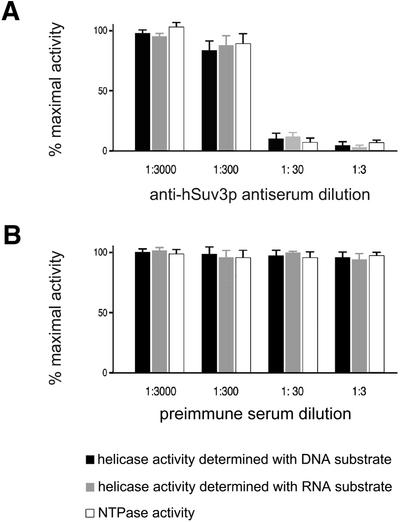
The anti-hSuv3p antibody reacting with the 87 kDa protein in the immunoblot was capable of inhibiting the enzymatic activities associated with the enzyme when diluted 1:30. The helicase activity was determined with both dsDNA and dsRNA substrate (Fig. 7A). The negative sera failed to affect these enzymatic activities (Fig. 7B).
Figure 7.
Depletion of enzymatic activities of hSuv3p NTPase/helicase by anti-hSuv3p antibodies. Aliquots of the purified hSuv3p NTPase/helicase (2 pmol) were incubated with anti-hSuv3p serum (A) or with preimmune serum (B) at 30°C for 30 min, then adjusted to the concentrations indicated in the figure. Thereafter the unwinding or ATPase reactions were started and the samples were processed as described in Materials and Methods. Values for the enzymatic activities are shown as means ± SEMs for representative experiments performed in triplicate.
The above results confirm that the purified recombinant 87 kDa wild-type hSuv3p protein showing anti-hSuv3p and anti-his tag immunoreactivity represented the only NTPase/helicase present in the enzyme preparation.
Characterisation of the biochemical properties of the hSuv3p NTPase/helicase
Analysis of the amino acid sequence of hSuv3p protein revealed motifs that are typical for helicases and suggested that the protein might be an RNA helicase (35,36). Thus, further experiments were performed to characterise the NTPase activity and unwinding activities of the protein. These investigations were performed with the 87 kDa polypeptide present in the fractions obtained from the Superdex-200 column corresponding to the monomeric form of the protein.
The ATPase activity of the enzyme was monitored in a standard assay by determination of the 33Pi released from [γ-33P]ATP due to enzyme-mediated hydrolysis as described in Materials and Methods. Using this method we estimated the optimum reaction conditions for the ATPase activity of hSuv3p NTPase/helicase. The optimal ATP hydrolysis was at 30°C. The monovalent cations Na+ and K+ did not exert any effect on the ATPase activity of the enzyme up to a concentration of 150 mM. Higher salt concentrations inhibited ATP hydrolysis with IC50 values of 280 and 200 mM for Na+ and K+, respectively. The ATPase activity of the hSuv3p NTPase/helicase was tested as a function of increasing concentrations of the divalent ions Mg2+ and Mn2+. The two metal cations revealed optimal activity of the protein at 1–5 mM for Mg2+ and 0.2–1 mM for Mn2+. Under the optimised reaction conditions the Lineweaver–Burk plot was linear up to ATP concentrations of 100 µM and yielded a Km value of 41.9 µM. At saturating concentrations of ATP 1 mol of the enzyme hydrolysed 0.32 mol ATP per second.
An interesting phenomenon was observed when the ATPase activity of the enzyme was measured in the presence of 1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide-5′-triphosphate (ribavirin-TP). Ribavirin-TP was previously reported to inhibit hepatitis C virus (HCV) NTPase/helicase (19). At lower ATP concentrations (<<Km) ribavirin-TP exerted a strong inhibitory effect on the ATPase activity of the hSuv3p NTPase/helicase. Reduction of the ATP concentration increased the inhibitory effect of ribavirin-TP. Thus, at an ATP concentration of 1 × 10–5 × Km (42 nM) an IC50 of 3–5 µM was measured. On the other hand, increasing the ATP concentration in the reaction mixture reduces the inhibitory effect of ribavirin-TP. At ATP concentrations corresponding to the Km value ribavirin-TP does not inhibit and even activates ATP hydrolysis. Increasing the ATP concentration could further enhance this activating effect. For example, at an ATP concentration equal to 100 × Km (4.2 mM) and 300 µM ribavirin-TP an activation of 250% was measured.
Analogous results were obtained with a range of slowly hydrolysed or non-hydrolysable nucleoside 5′-triphosphate analogues, like adenosine 5′-γ-thiotriphosphate (ATP-γ-S), 5′-adenylimidodiphosphate (AMP-PNP) and β,γ-methyleneadenosine 5′-triphosphate (AMP-PCP). All the compounds tested were potent inhibitors of the enzyme at lower ATP concentrations, while having an activating effect at higher ATP concentrations with the following order of efficiency: ATP-γ-S < AMP-PCP < AMP-PNP. Figure 8A presents the modulation of the ATPase activity by AMP-PNP measured at different ATP concentrations. Analysis of the AMP-PNP-mediated inhibition, measured at strongly reduced ATP concentrations, by mean of a graphic method (37) revealed pure competitive inhibition with regard to ATP (Fig. 8B).
A common property of numerous NTPase/helicases is their activation and even dependency of the ATPase activity on RNA, DNA or homopolymeric polynucleotides (36 and references therein). Under optimal reaction conditions we analysed the effect of various ribo- and deoxyribonucleotides on the ATPase activity of the hSuv3p NTPase/helicase. However, in contrast to the enzymes described previously the ATPase activity of hSuv3p protein is only moderately affected by the polynucleotides tested; the highest activation was observed with poly(U) (140% of control) and DNA (120% of control) with optimum concentrations of 330 and 120 µM of nucleotide base, respectively.
The unwinding activity of hSuv3p protein was monitored and characterised with RNA and DNA substrates. Both substrates consisted of two partially hybridised oligonucleotides; a 26mer, the release strand, and a 40mer, the template strand. The same substrates were previously used to characterise the helicase activity of West Nile virus (WNV) and HCV NTPase/helicases (20,23,38). The hSuv3p enzyme was found to be capable of unwinding DNA/DNA and RNA/RNA homoduplexes (Fig. 9, lanes 3 and 10). To compare the catalytic efficiency of the unwinding activity of hSuv3p NTPase/helicase towards RNA and DNA substrates we performed a strand displacement experiment in which decreasing concentrations of enzyme were added at the constant non-saturating concentration of 4.7 pM of RNA or DNA substrate. Although the hSuv3p NTPase/helicase separated the dsDNA with an ∼10 000 higher activity compared to dsRNA, it unwound 95% of the DNA substrate but 100% of RNA substrate (Fig. 9).
Figure 9.
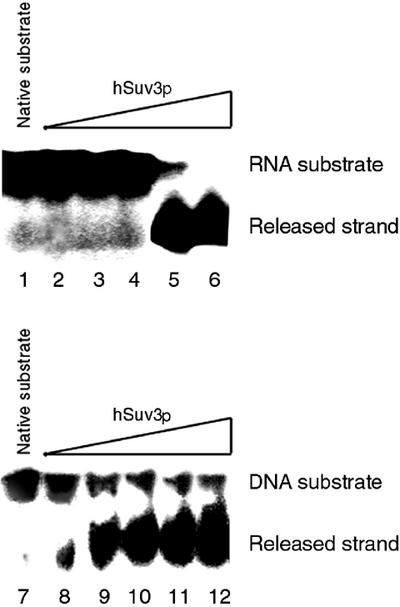
Comparison of efficacy of RNA and DNA unwinding reaction mediated by hSuv3p as a function of decreasing enzyme concentration. The reaction mixture was composed as described in Materials and Methods with various hSuv3p enzyme concentrations used: 0.66 fM (lanes 2 and 8); 66 fM (lanes 3 and 9); 6.6 pM (lanes 4 and 10); 0.66 nM (lanes 5 and 11); 66 nM (lanes 6 and 12). The substrate and released strand were separated in a TBE polyacrylamide gel and visualised by exposure of the dried gel to X-ray film for 24 h. The results shown are representative of three independent experiments.
DISCUSSION
Proteins involved in mitochondrial mRNA processing and degradation in humans have yet to be characterised. In S.cerevisiae RNA turnover is mediated by a 3′→5′ exoribonuclease complex known as the mitochondrial degradosome, mtEXO (39). Previously we cloned the cDNA coding for the human orthologue of the putative yeast RNA NTPase/helicase Suv3p, a component of the yeast mitochondrial degradosome (11). Both yeast and human proteins display high homology (35% identity, 53% similarity over 567 residues), but sequence similarity does not allow prediction of the precise function of the human orthologue. The properties of the human protein may have been modified during the course of evolution and in humans hSuv3p might be now associated with different molecular mechanisms.
There are indications that the metazoan SUV3 gene is indispensable for proper cellular function. The data obtained in a large-scale analysis of SUV3 function in C.elegans (47% identity and 65% similarity over 669 residues on the protein level between C.elegans and H.sapiens) by high throughput RNAi showed that inactivation of the gene caused embryo development to arrest 24 h post-inactivation (9). In addition, no rearrangements have been found in the band 10q22.1 of human chromosome 10, to which the hSUV3 gene was assigned (40). This could be an indication that hSuv3p in its intact functional form is also essential for human cells.
In order to investigate the possible function of hSuv3p we decided first to analyse the precise cellular localisation of hSuv3p in mammalian cells and to study the enzymatic activity of the protein in vitro.
Most mitochondrial proteins are encoded by the nuclear genome and thus have to be synthesised in the cytosol and then imported into mitochondria. Protein translocation across and into the mitochondrial membranes is a multistep process facilitated by the coordinated action of specialised translocation systems in the outer and inner membranes of mitochondria (41). Previous analysis of hSuv3p for the presence of a mitochondrial targeting sequence indicated that hSuv3p might be imported into mitochondria (11). However, the localisation of the protein in vivo in human cells remained to be shown. Immunofluorescence microscopy clearly demonstrates that hSuv3p is localised in mitochondria of human and simian cells. The N-terminal part of hSuv3p was also shown to be necessary for mitochondrial import, since a mutant form of hSuv3p lacking the 18 residues at the N-terminus was not targeted to the mitochondria.
The classical mitochondrial targeting N-terminal pre-sequences consist of positively charged, hydrophobic and hydroxylated amino acids that can form an amphiphilic α-helix and function as a matrix-targeting signal. However, pre-sequence-like signals are also used by several pre-proteins that are destined for the three other mitochondrial subcompartments: the outer membrane, the inter-membrane space and the inner membrane. In these cases the pre-sequences are complemented by other sorting signals that lead to a specific arrest of the pre-proteins in the respective compartment (stop-transfer signals) (13). The presence of a functional mitochondrial targeting presequence on the N-terminus and the lack of other predicted sorting signals (e.g. a hydrophobic membrane anchor) suggested that hSuv3p is imported to the mitochondrial matrix space. Two sets of experiments support this point: (i) an import assay into isolated yeast mitochondria followed by separation of mitochondrial subcompartments; (ii) immunoblotting of human mitochondrial subfractions with hSuv3p specific antiserum. Using yeast mitochondria to study the import of human proteins has been reported previously (42,43).
A number of yeast mitochondrial proteins have their homologues in human mitochondria. Thus, S.cerevisiae is considered as a particularly suitable simple organism widely used for studying the function of human proteins (44). However, because of differences in mitochondrial DNA organisation and expression between human and yeast mitochondria, it cannot be assumed a priori that given orthologues play identical biological functions. For example, in mitochondrial DNA replication orthologic DNA polymerases (45) but unrelated DNA helicases are involved in yeast and humans. Human mitochondria contain a T7-related DNA helicase (46) that has no orthologue in yeast.
The results presented suggest important differences between the human and yeast SUV3 gene products. hSuv3p was shown to be a soluble protein localised in the human mitochondrial matrix space, whereas our recent experiments have revealed that in yeast mitochondria the complex consisting of Suv3p helicase and Dss1p RNase is associated with mitochondrial ribosomes (37), which are thought to be associated with the inner membrane (47). Additionally, no orthologue of Dss1p has been found in the human genome and we were not able to complement the yeast SUV3 knock-out by its human orthologue (not shown), although hSuv3p is properly imported into yeast mitochondria in vitro. These results suggest that in humans, in spite of high conservation through evolution, hSuv3p might be involved in different mitochondrial processes than its yeast counterpart.
For most proteins defined as NTPase/helicases on the basis of their amino acid sequences, no biochemical studies have been performed. Up to now there were also no data about the enzymatic activity of hSuv3p. Analysis of the amino acid sequence of the protein revealed a range of conserved motifs, which are associated with NTP and/or polynucleotide binding characteristic of NTPase/helicases (35,48,49). The best conserved are motifs I and II, the so-called ‘Walker motifs A and B’, that are present in all NTPase/helicases as well as in a wide variety of other NTP-utilising proteins. The functions of these motifs, which compose the NTP-binding pocket of the proteins, are association of the terminal phosphate groups of NTP (Walker motif A) and chelatation of the Mg2+ of the Mg-NTP complex (Walker motif B) (32). There are a number of reasons to classify hSuv3p in superfamily 1 (SFI) of NTPase/helicases: (i) the amino acid sequence of the hSuv3p Walker A motif (G-P-T-N-S-G-K-T) is characteristic of DNA SFI NTPase/helicases (35,36,48); (ii) the presence of a hydrophobic isoleucine residue within the Walker B motif (35); (iii) lack of a typical SAT motif, unique to SF2 NTPase/helicases. Membership of a particular superfamily has, however, no clear predictive value regarding the key properties of the enzymes (35,36).
The hSuv3p enzyme utilises both RNA and DNA substrates with a strong preference for the latter. To our knowledge there are only a few NTPase/helicases that show an exclusive RNA or DNA specificity. The explanation for this lack of specificity might be the fact that the interaction between the protein molecule and the DNA or RNA substrate is mainly mediated by phosphate groups and not by the nucleotide base or sugar moieties (50–52). Although hSuv3p protein was previously referred to as a putative RNA NTPase/helicase (11), the sequence characteristics mentioned above characterise the protein rather as a DNA enzyme. Our enzymatic data show that hSuv3p unwinds dsDNA 104 times more efficiently than dsRNA. This seems to be in contrast to yeast Suv3p protein, which was shown to function as an RNA helicase participating in RNA degradation and 3′ and 5′ processing of mtRNA. The observed reaction rates for the enzyme/substrate ratios used in our experiments are similar to published results obtained for in vitro helicase assays (53). It is obvious, however, that characteristics of a recombinant protein in an in vitro assay may differ from those in vivo. We do not know yet if hSuv3p protein forms a complex with another protein(s) in mitochondria and what its physiological function is. The observed strong preference for dsDNA substrates might indicate the participation of hSuv3p in DNA replication, but more research is needed to prove this hypothesis.
The NTPase/helicase nature of hSuv3p was verified by site-directed mutagenesis experiments in which Gly207 was replaced by valine. The Gly207 residue represents the N-terminus of the Walker A motif (see above). As mentioned above, the glycine residue in the first position of the Walker A motif is highly conserved between DNA NTPase/helicases of SF1 and numerous ATPases and GTPases, whereas RNA NTPase/helicases have in most cases an alanine in this position (36,54,55). Substitution of alanine by glycine in the translation factor Tif1 from S.cerevisiae was shown to have no effect on the growth rate (33). An analogous exchange performed on the eukaryotic translation initiation factor 4A (eIF-4A) RNA NTPase/helicase even increased its ATP binding capacity (34). Such an effect could be expected since both amino acids have uncharged and unbranched side chains. However, the introduction of the isopropyl moiety of valine in this position causes dramatic inhibition of the functions of the RNA helicase activity in the translation factor Tif1 from S.cerevisiae and is lethal for the cells (33). The respective substitution dramatically reduces ATP binding by the eIF-4A RNA NTPase/helicase (34). Also, for hSuv3p replacement of the glycine of the Walker A motif with valine abolished both the ATPase and helicase activities of the enzyme.
In the helicase assays the hSuv3p enzyme was added in a vast excess over the substrate, therefore one could speculate that the unwinding reaction mediated by hSuv3p would be non-enzymatic. This possibility is, however, excluded by the experiments with a mutated protein, hSuv3p(G207V), which could not utilise ATP. The mutated protein was unable to separate the dsDNA or dsRNA substrates. Additionally, corroborating this are our observations with a second mutant enzyme in which the Walker motif B (32), DEIQ, was replaced by DAIQ [hSuv3p(E292A) mutant]. The protein was not able to hydrolyse ATP and to separate dsDNA or dsRNA used as substrates (manuscript in preparation).
The majority of the NTPase/helicases characterised to date exhibit polynucleotide-dependent and polynucleotide-independent NTPase activities (56). The enzymes differ dramatically in the extent of nucleic acid-induced stimulation of their NTPase activity. For example, poly(rU) enhanced the kcat value of HCV ATPase up to 30-fold (56). In contrast, the ATPase activities of bacterial helicases such as Rep and helicase II are increased over 1000-fold by single-stranded DNA (57). In contrast, the hSuv3p ATPase activity displays rather modest stimulation by polynucleotides (1.4-fold). The weak response of hSuv3p to nucleic acids is, however, not unique. There are several known viral NTPase/helicases whose ATPase activity is stimulated only weakly by polynucleotides (23,35). This apparent difference might result from different reaction conditions used; differences in purity of the enzyme preparation or source of the enzyme (19,37,56,58–60). On the other hand it is possible that in vivo a specific RNA or DNA cofactor(s) exists, as for the ATPase activity of protein Slt22p (61), that in the case of hSuv3p remains to be discovered.
Non-hydrolysable ATP analogues are effective inhibitors of the NTPase activity of the hSuv3p enzyme, although the inhibition occurred only at low ATP concentrations. Higher concentrations of ATP abolished the inhibition and even stimulated the hydrolytic activity of the enzyme. A possible explanation of this phenomenon is that hSuv3p protein has two binding sites for nucleosides and nucleotides, as previously postulated for the HCV enzyme (62). The first one is equipped with Walker motifs (where the ATP normally binds) and is, therefore, more specific for nucleoside 5′-triphosphate substrates. The second could be an allosteric binding site less specific for nucleosides or nucleotides. According to this hypothesis, at low ATP concentrations (less than the Km value of the specific NTPase reaction) ribavirin-TP, as well as other nucleoside 5′-triphosphate analogues, competes against ATP for the first site containing Walker motifs and thus causes competitive inhibition of the NTPase. At high concentrations of ATP along with ribavirin-TP or nucleoside 5′ triphosphate analogues the second site becomes more and more occupied and there is a consequent conformational change occurring in the NTPase, which ultimately leads to its activation. A similar pattern of interaction of ATP, ribavirin-TP and NTPase/helicase was previously described by us for some members of SFII family of enzymes, like HCV and WNV NTPase/helicase (19,23).
In conclusion, we biochemically characterised the nuclear encoded RNA and DNA helicase present in the mitochondrial matrix of human mitochondria. Our results indicate that the human enzyme significantly differs from its yeast orthologue. While from numerous studies in different laboratories the involvement of yeast Suv3p in RNA turnover seems to be very well proven, the human protein shows a strong preference for dsDNA and is present exclusively in the mitochondrial matrix. Further research is in progress to answer the question whether hSuv3p protein is part of the mitochondrial RNA degrading machinery or has acquired new features during evolution.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge Prof. Sir Aaron Klug for critical reading of the manuscript. We also wish to thank Prof. Ewa Bocian and Zofia Helias-Rodziewiecz for their help in immunoflourescence microscopy analysis and Katarzyna Stanco and Piotr Szczeglow for help in in vitro mutagenesis experiments and raising antibodies. We are particularly grateful to Jonathan Papworth for suggestions and critical review of the manuscript and Dr Stephanie Agius for valuable discussions. This work was supported by Polish Committee for Scientific Research (KBN) grants KBN 4PO4A 01818 and KBN 4PO4A 078 22 and University of Warsaw grant BW 2000. M.M. and J.P. were supported by EMBO short-term fellowships (ASTF9882 and ASTF 5-02, respectively). Additionally, J.P. was supported by a Marie Curie Training grant (MC QLK3-GH-99-50415-02). Financial support of the Medical Research Council, UK, and the Centre of Excellence in Molecular Biotechnology, Poland, is also acknowledged.
REFERENCES
- 1.Margossian S.P., Li,H., Zassenhaus,H.P. and Butow,R.A. (1996) The DexH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell, 84, 199–209. [DOI] [PubMed] [Google Scholar]
- 2.Dmochowska A., Golik,P. and Stepien,P.P. (1995) The novel nuclear gene DSS-1 of Saccharomyces cerevisiae is necessary for mitochondrial biogenesis. Curr. Genet., 28, 108–112. [DOI] [PubMed] [Google Scholar]
- 3.Stepien P.P., Margossian,S.P., Landsman,D. and Butow,R.A. (1992) The yeast nuclear gene suv3 affecting mitochondrial post-transcriptional processes encodes a putative ATP-dependent RNA helicase. Proc. Natl Acad. Sci. USA, 89, 6813–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conrad-Webb H., Perlman,P.S., Zhu,H. and Butow,R.A. (1990) The nuclear SUV3-1 mutation affects a variety of post-transcriptional processes in yeast mitochondria. Nucleic Acids Res., 18, 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stepien P.P., Kokot,L., Leski,T. and Bartnik,E. (1995) The suv3 nuclear gene product is required for the in vivo processing of the yeast mitochondrial 21s rRNA transcripts containing the r1 intron. Curr. Genet., 27, 234–238. [DOI] [PubMed] [Google Scholar]
- 6.Dziembowski A., Malewicz,M., Minczuk,M., Golik,P., Dmochowska,A. and Stepien,P.P. (1998) The yeast nuclear gene DSS1, which codes for a putative Rnase II, is necessary for the function of the mitochondrial degradosome in processing and turnover of RNA. Mol. Gen. Genet., 260, 108–114. [DOI] [PubMed] [Google Scholar]
- 7.Dziembowski A., Piwowarski,J., Hoser,R., Minczuk,M., Dmochowska,A., Siep,M., Van der Speck,H., Grivell,L. and Stepien,P.P. (2002) The yeast mitochondrial degradosome: its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 8.Gagliardi D., Kuhn,J., Spadinger,U., Brennicke,A., Leaver,C.J. and Binder,S. (1999) An RNA helicase (AtSUV3) is present in Arabidopsis thaliana mitochondria. FEBS Lett., 458, 337–342. [DOI] [PubMed] [Google Scholar]
- 9.Maeda I., Kohara,Y., Yamamoto,M. and Sugimoto,A. (2001) Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol., 11, 171–176. [DOI] [PubMed] [Google Scholar]
- 10.Dmochowska A., Stankiewicz,P., Golik,P., Stepien,P.P., Bocian,E., Hansmann,I. and Bartnik,E. (1998) Assignment of SUPV3L1 to human chromosome band 10q22.1 by in situ hybridization. Cytogenet. Cell Genet., 83, 84–85. [DOI] [PubMed] [Google Scholar]
- 11.Dmochowska A., Kalita,K., Krawczyk,M., Golik,P., Mroczek,K., Lazowska,J., Stepien,P.P. and Bartnik,E. (1999) A human putative Suv3-like RNA helicase is conserved between Rhodobacter and all eukaryotes. Acta Biochim. Pol., 46, 155–162. [PubMed] [Google Scholar]
- 12.Tokatlidis K. (2000) Directing proteins to mitochondria by fusion to mitochondrial targeting signals. Methods Enzymol., 327, 305–317. [DOI] [PubMed] [Google Scholar]
- 13.Glick B.S., Brandt,A., Cunningham,K., Muller,S., Hallberg,R.L. and Schatz,G. (1992) Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell, 69, 809–822. [DOI] [PubMed] [Google Scholar]
- 14.Gaines G.L. III (1996) In organello RNA synthesis system from HeLa cells. Methods Enzymol., 264, 43–49. [DOI] [PubMed] [Google Scholar]
- 15.Bergmeyer H.U. and Bernt,E. (1974) In Bergmeyer,H.U. (ed.), Methods of Enzymatic Analysis. Academic Press, New York, NY.
- 16.Graham J.M. and Rickwood,D. (1997) Subcellular Fractionation— A Practical Approach. Oxford University Press, Oxford, UK.
- 17.Ostrowski J., Wyrwicz,L., Rychlewski,L. and Bomsztyk,K. (2002) Heterogeneous nuclear ribonucleoprotein K protein associates with multiple mitochondrial transcripts within the organelle. J. Biol. Chem., 277, 6303–6310. [DOI] [PubMed] [Google Scholar]
- 18.Borowski P., Kuehl,R., Mueller,O., Hwang,L.-H., Schulze zur Wiesch,J. and Schmitz,H. (1999) Biochemical properties of a minimal functional domain with ATP-binding activity of the NTPase/helicase of hepatitis C virus. Eur. J. Biochem., 266, 715–723. [DOI] [PubMed] [Google Scholar]
- 19.Borowski P., Mueller,O., Niebuhr,A., Kalitzky,M., Hwang,L.-H., Schmitz,H., Siwecka,M.A. and Kulikowski,T. (2000) ATP-binding domain of NTPase/helicase as target for hepatitis C antiviral therapy. Acta Biochim. Pol., 47, 173–180. [PubMed] [Google Scholar]
- 20.Gallinari P., Brennan,D., Nardi,C., Brunetti,M., Tomei,L., Steinkühler,C. and De Francesco,R. (1998) Multiple enzymatic activities associated with recombinant NS3 of hepatitis C virus. J. Virol., 72, 6758–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai C.-L., Chi,W.-K., Chen,D.-S. and Hwang,L.-H. (1996) The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J. Virol., 70, 8477–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J., Fritsch,F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Borowski P., Niebuhr,A., Mueller,O., Bretner,M., Felczak,K., Kulikowski,T. and Schmitz,H. (2001) Purification and characterization of West Nile virus NTPase/helicase. Evidence for dissociation of the NTPase and helicase activities of the enzyme. J. Virol., 75, 3220–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hames B.D. and Rickwood,D. (1990) Gel Electrophoresis of Proteins: A Practical Approach, 2nd Edn. Oxford University Press, Oxford, UK.
- 25.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 27.Bauer M.F. and Neupert,W. (2001) Import of proteins into mitochondria: a novel pathomechanism for progressive neurodegeneration. J. Inherit. Metab. Dis., 24, 166–180. [DOI] [PubMed] [Google Scholar]
- 28.Rojo E.E., Stuart,R.A. and Neupert,W. (1995) Conservative sorting of F0-ATPase subunit 9: export from matrix requires delta pH across inner membrane and matrix ATP. EMBO J., 14, 3445–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfanner N. and Neupert,W. (1987) Distinct steps in the import of ADP/ATP carrier into mitochondria. J. Biol. Chem., 262, 7528–7536. [PubMed] [Google Scholar]
- 30.Martin J. (1997) Molecular chaperones and mitochondrial protein folding. J. Bioenerg. Biomembr., 29, 35–43. [DOI] [PubMed] [Google Scholar]
- 31.Gonzales D.H. and Neupert,W. (1990) Biogenesis of mitochondrial c-type cytochromes. J. Bioenerg. Biomembr., 22, 753–768. [DOI] [PubMed] [Google Scholar]
- 32.Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmid S.R. and Linder,P. (1991) Translation initiation factor 4A from Saccharomyces cerevisiae. Analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol. Cell. Biol., 11, 3463–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pause A. and Sonenberg,N. (1992) Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J., 11, 2643–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadare G. and Haenni,A. (1997) Virus-encoded RNA helicases. J. Virol., 71, 2583–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lüking A., Stahl,U. and Schmidt,U. (1998) The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol., 33, 259–296. [DOI] [PubMed] [Google Scholar]
- 37.Dixon M. (1952) The determination of enzyme inhibitor constants. Biochem. J., 55, 170–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borowski P., Lang,M., Niebuhr,A., Haag,A., Schmitz,H., Schulze zur Wiesch,J., Choe,J., Siwecka,M.A. and Kulikowski,T. (2001) Inhibition of the helicase activity of HCV NTPase/helicase by 1-β-D-ribofuranosyl-1,2,4-triazole-3 carboxamide-5′ triphosphate (ribavirin-TP). Acta Biochim. Pol., 48, 739–744. [PubMed] [Google Scholar]
- 39.Dziembowski A. and Stepien,P.P. (2001) Genetic and biochemical approaches for analysis of mitochondrial degradosome from Saccharomyces cerevisiae. Methods Enzymol., 342, 367–378. [DOI] [PubMed] [Google Scholar]
- 40.Deloukas P., French,L., Meitinger,T. and Moschonas,N.K. (2000) Report of the third international workshop on human chromosome 10 mapping and sequencing 1999. Cytogenet. Cell Genet., 90, 1–12. [DOI] [PubMed] [Google Scholar]
- 41.Pfanner N. and Geissler,A. (2001) Versatility of the mitochondrial protein import machinery. Nature Rev. Mol. Cell Biol., 2, 339–349. [DOI] [PubMed] [Google Scholar]
- 42.Renold A., Koehler,C.M. and Murphy,M.P. (2000) Mitochondrial import of the long and short isoforms of human uncoupling protein 3. FEBS Lett., 465, 135–140. [DOI] [PubMed] [Google Scholar]
- 43.Seki N., Moczko,M., Nagase,T., Zufall,N., Ehmann,B., Dietmeier,K., Schafer,E., Nomura,N. and Pfanner,N. (1995) A human homolog of the mitochondrial protein import receptor Mom19 can assemble with the yeast mitochondrial receptor complex. FEBS Lett., 375, 307–310. [DOI] [PubMed] [Google Scholar]
- 44.Foury F. and Kucej,M. (2002) Yeast mitochondrial biogenesis: a model system for humans? Curr. Opin. Chem. Biol., 6, 106–111. [DOI] [PubMed] [Google Scholar]
- 45.Lecrenier N. and Foury,F. (2000) New features of mitochondrial DNA replication system in yeast and man. Gene, 246, 37–48. [DOI] [PubMed] [Google Scholar]
- 46.Spelbrink J.N., Li,F.Y., Tiranti,V., Nikali,K., Yuan,Q.P., Tariq,M., Wanrooij,S., Garrido,N., Comi,G., Morandi,L. et al. (2001) Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nature Genet., 28, 223–231. [DOI] [PubMed] [Google Scholar]
- 47.Fox T.D. (1996) Translational control of endogenous and recoded nuclear genes in yeast mitochondria: regulation and membrane targeting. Experientia, 52, 1130–1135. [DOI] [PubMed] [Google Scholar]
- 48.Gorbalenya A.E., Koonin,E.V., Donchenko,A.P. and Blinov,V.M. (1989) Two related superfamilies of putative helicases involved in replication, recombination, repair, and expression of DNA and RNA genomes. Nucleic Acids Res., 17, 4713–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorbalenya A.E. and Koonin,E.V. (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol., 3, 419–429. [Google Scholar]
- 50.Yao N., Hesson,T., Cable,M., Hong,Z., Kwong,A., Le,H. and Weber,P. (1997) Structure of the hepatitis C virus RNA helicase domain. Nature Struct. Biol., 4, 463–467. [DOI] [PubMed] [Google Scholar]
- 51.Kim J.L., Morgenstern,K., Griffith,J., Dwyer,M., Thomson,J., Murcko,M., Lin,C. and Caron,P. (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure, 6, 89–100. [DOI] [PubMed] [Google Scholar]
- 52.Theis K., Chen,P.J., Skorvaga,M., Van Houten,B. and Kisker,C. (1999) Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J., 18, 6899–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y. and Liu,Z.-R. (2002) The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J. Biol. Chem., 277, 12810–12815. [DOI] [PubMed] [Google Scholar]
- 54.Gorbalenya A.E., Koonin,E.V. Donchenko,A.P. and Blinov,V.M. (1988) A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett., 235, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorbalenya A.E. and Koonin,E.V. (1989) Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res., 17, 8413–8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Porter D. (1998) A kinetic analysis of the oligonucleotide-modulated ATPase activity of the helicase domain of the NS3 protein from hepatitis C virus. J. Biol. Chem., 273, 14247–14253. [DOI] [PubMed] [Google Scholar]
- 57.Wong I., Moore,K., Bjornson,K.P., Hsieh,J. and Lohman,T.M. (1996) ATPase activity of Escherichia coli Rep helicase is dramatically dependent on DNA ligation and protein oligomeric states. Biochemistry, 35, 5726–5734. [DOI] [PubMed] [Google Scholar]
- 58.Morgenstern K.A., Landro,J.A., Hsiao,K., Lin,C., Gu,Y., Su,M.S. and Thomson,J.A. (1997) Polynucleotide modulation of the protease, nucleoside triphosphatase, and helicase activities of a hepatitis C virus NS3-NS4A complex isolated from transfected COS cells. J. Virol., 71, 3767–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin L. and Peterson,D.L. (1995) Expression, isolation, and characterization of the hepatitis C virus ATPase/RNA helicase. Arch. Biochem. Biophys, 323, 47–53. [DOI] [PubMed] [Google Scholar]
- 60.Suzich J., Tamura,J., Palmer-Hill,F., Warrener,P., Grakoui,A., Rice,C., Feinstone,S. and Collett,M. (1993) Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related Pestivirus and Flavivirus enzymes. J. Virol., 67, 6152–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu D., Nouraini,S., Field,D., Tang,S.J. and Friesen,J.D. (1996) An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature, 381, 709–713. [DOI] [PubMed] [Google Scholar]
- 62.Porter D.J.T. (1998) Inhibition of the hepatitis C virus helicase-associated ATPase activity by the combination of ADP, NaF, MgCl2, and poly(rU). J. Biol. Chem., 273, 7390–7396. [DOI] [PubMed] [Google Scholar]