Abstract
Intron conservation, intron gain or loss and putative intron sliding events were determined for a set of three genes (SPO11, MRE11 and DMC1) involved in basic aspects of recombination in eukaryotes. These are ancient genes and present in nearly all of the major kingdoms. MRE11 is of bacterial origin and can be found in all kingdoms. DMC1 is a specialized homolog of the bacterial RecA protein, whereas the SPO11 gene is of archaebacterial origin. Only unique homologs of SPO11 are found in animals and fungi whereas three distantly related SPO11 copies are present in plant genomes. A comparison of the respective intron positions and phases of all genes was performed, demonstrating that a quarter of the intron positions were perfectly conserved over more than 1 000 000 000 years. Regarding the remaining three quarters of the introns we found insertions to be about three times more frequent than deletions. Aligning the introns of the three different SPO11 homologs of Arabidopsis thaliana we propose a conclusive model of their evolution. We postulate that at least one duplication event occurred shortly after the divergence of plants from animals and fungi and that a respective homolog has been retained in a protist group, the apicomplexa.
INTRODUCTION
To gain insight into the evolution of the recombination machinery of eukaryotes we performed an approach refered to as ‘comparison of intron positions across kingdoms’ (CIPAK). A similar approach of intron comparison across kingdom borders was recently used (1). The authors could thereby identify ancient introns in the large gene family of DEAD-box helicases and elucidate the evolution of the intron/exon structure of these genes in Arabidopsis, Caenorhabditis and Drosophila.
To perform a CIPAK analysis, genes should be used that fulfill the following prerequisites: (i) they should be of ancient origin and both cDNA and genomic sequences should be available from at least three kingdoms; (ii) the genes/proteins should not be too strictly conserved as selective pressure would hinder the investigation of intron evolution; (iii) the sequence conservation should be high enough to enable an unambiguous alignment of orthologs over a reasonable sequence length.
DNA recombination is one of the basic features common to all organisms. We chose three genes involved in this process that met the above criteria. They are ancient and widespread within the kingdoms and show a moderate sequence conservation (30–60% over the full protein length) but unambiguous alignments in most cases. The unambiguous alignment of intron positions is important because intron sliding events are rare and often misinterpreted due to ambiguous sequence alignments (2,3). All three genes harbor many more introns (14–22) than an average gene of Arabidopsis (approximately five introns) (4), a phenomenon observed in most recombination-related genes investigated so far (5–7). For all of these genes a strong functional con servation during evolution was demonstrated. The Spo11 protein introduces DNA double-strand breaks during meiosis in higher organisms (8). Spo11 has descended from archaebacteria, where it represents a subunit of an atypical type II topoisomerase (9). The Mre11 protein in higher organisms is part of a tripartite complex (Mre11, Rad50 and Xrs2/Nbs1) with an exo- or endonuclease function depending on the DNA substrate (10). Mre11 is most likely the homolog of the bacterial SbcD protein, which is also part of a complex, (SbcCD) and seems to fulfill the same nuclease function (11,12). Dmc1 is, besides Rad51, one of the two main eukaryotic homologs of RecA and forms complexes with DNA prior to meiotic chromosome synapsis (13,14).
MATERIALS AND METHODS
Presumptions and definitions
Regarding the actual hypothesis of eukaryotic evolution (15,16), we consider that animals and fungi diverged after the diversification of plants and animals. This means: (i) if only one group or organism of one kingdom possesses an intron we regard it as a gain event because one gain event is more likely than several independent intron loss events; (ii) if an intron is missing only in plants we consider it as an indecisive case because it is not possible to attribute it as intron loss in plants or as intron gain in animals/fungi after diversification from plants; (iii) all introns which are present in at least the plant and animal or plant and fungi kingdoms residing unambiguously at the same position were considered as ancient introns which must have been present in the last common ancestror of plants and animals (LCA-PA). As reference organisms for the respective kingdoms we used Arabidopsis thaliana (plants), Homo sapiens (animals) and Coprinus cinereus (fungi).
Finally, we compared all intron positions of A.thaliana with the putative exon/intron borders of Oryza sativa using the genomic sequence data of rice. This analysis demonstrated that A.thaliana and O.sativa, despite their diversification 150 000 000–200 000 000 years ago, possess an identical exon/intron structure of the respective genes. The only exception, an intron present in A.thaliana but not in O.sativa, is discussed below.
Protein alignments
All alignments were done with the DNASTAR package from Lasergene using the MEGALIGN module. The Lipman Pearson alignments method was performed with a ktuple of 1 or 2 and a gap penalty of 2 or 4. In all combinations of these parameters the alignment output was the same and mostly unambiguous. Multiple alignments for the Spo11 proteins were done with CLUSTALW (17).
Phylogenetic trees
Phylogenetic analyses were conducted with PAUP* 4.0b8a (18) to further test the intron-derived evolutionary hypotheses. In the cladistic analysis (Fitch parsimony, MP) TBR branch swapping, ACCTRAN character state opitmization and 250 random addition sequences were used. In the phenetic analysis mean character distances were calculated and the neighbor-joining cluster algorithm (NJ) was used. Statistical support for the branches was calculated with either 500 (parsimony) or 5000 (phenetic) bootstrap re-samples, with identical settings as before.
RESULTS
Analysis of the individual genes: SPO11
Comparing the intron positions of the three reference organisms (A.thaliana, H.sapiens and C.cinereus) we constructed a scaffold gene including all 27 different intron positions of the 41 introns detected in these organisms (Fig. 1, lower). We used only AtSPO11-1 in the scaffold gene because it is the true functional homolog of SPO11 from other organisms, whereas AtSPO11-2 and AtSPO11-3 are extra-functional genes which are not present in organisms other than plants.
Figure 1.
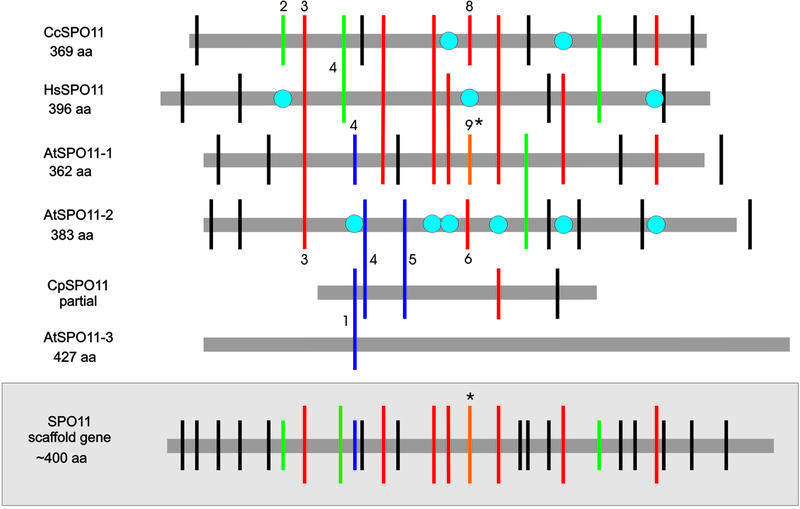
Schematic alignment of all introns of SPO11 from different organisms in relation to their protein sequences. The introns are differentiated by colored bars as follows: conserved introns are shown as red bars; gained introns as black bars; introns occurring only in animals and fungi as green bars; lost introns as blue circles; putative sliding events as orange bars marked with an asterisk; special cases as blue bars (see text for details). Numbers are only given for introns which are mentioned especially in the text. In the lower part (gray shaded) a scaffold gene only containing the different intron positions of AtSPO11-1, HsSPO11 and CcSPO11 is shown. For the sake of simplicity the intron positions of AtSPO11-2 and AtSPO11-3 were not included in the scaffold gene. Accession numbers: A.thaliana 1–3, AJ251989, AJ251990 and AJ297842; H.sapiens, AF149310; C.cinereus, AF214638; C.parvum, CVMUMN 5807, Contig 1824, available at: finished and unfinished genomes of eukaryotes from the NCBI (http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/genom_table_cgi?organism=euk).
The LCA-PA contained at least 7 of the 27 intron positions of the scaffold gene which are conserved between either plants and animals or plants and fungi (Fig. 1, upper, red bars). We could also consider intron 9 of AtSPO11-1 (corresponding to intron 8 of C.cinereus) as conserved but slid (probably by a sliding event of –1 nt and a 3 or 6 nt long insertion, respectively). This intron is located in C.cinereus 4 nt and in AtSPO11-2 7 nt upstream of its position in AtSPO11-1 (Fig. S1, Supplementary Material). If we regard only the seven clear-cut intron positions as conserved and thus being present in the LCA-PA, we can altogether classify 15 cases as intron gain events after diversification of plants and animals (of 41 introns) and five cases as intron loss (Table 1). Six additional cases are indecisive (Fig. 1, green bars) because they could be due to an intron gain event in animals/fungi after the plant–animal but before the animal–fungi diversification or due to a loss event in plants. Intron 2 of C.cinereus is shown as a green bar and not as a black bar (Fig. 1, upper) because it was found in the same position in Caenorhabditis elegans (Fig. S1). Finally, intron 4 of A.thaliana (Fig. 1, blue bar) is not included in the calculation and will be discussed later.
Table 1. Classification of the fate of introns within three genes involved in DNA recombination in eukaryotes.
| Gene | Intron no. | Intron gain | Intron loss | Putative intron sliding |
|---|---|---|---|---|
| AtSPO11-1 | 15 | 6 | 0 | 1 |
| HsSPO11 | 12 | 4 | 3 | |
| CcSPO11 | 14 | 5 | 2 | |
| Total | 41 | 15 | 5 | 1 |
| Conserved | 18 | |||
| Indecisive | 6 | |||
| AtDMC1 | 14 | 8 | 0 | |
| HsDMC1 | 12 | 7 | 3 | |
| CcDMC1 | 11 | 6 | 3 | |
| PoDMC1 | 18 | 10 | 0 | 1 |
| Total | 55 | 31 | 6 | 1 |
| Conserved | 18 | |||
| Indecisive | 5 | |||
| AtMRE11 | 22 | 12 | 0 | |
| HsMRE11 | 18 | 5 | 0 | – |
| CcMRE11 | 10 | 3 | 6 | – |
| Total | 50 | 20 | 6 | 0 |
| Conserved | 24 | |||
| Indecisive | 6 | |||
| Total | Gained | 60a of 146 | Lost 17 | Putative sliding events 2 |
| Conserved | 60 of 146 | |||
| Indecisive | 16 of 146 |
Indecisive, only conserved in animals and fungi. At, Arabidopsis thaliana; Hs, Homo sapiens; Cc, Coprinus cinereus; Po, Pleurotus ostreata.
aThe total number of gained introns is lower in comparison to all calculated single gain events (66 introns) because C.cinereus and P.ostreata share six introns which presumably result from only one gain event each.
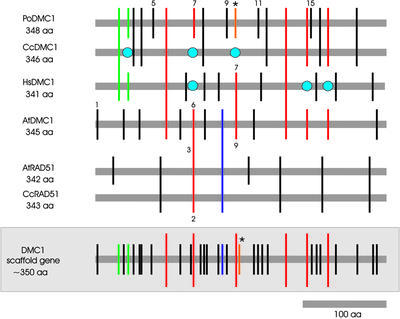
Analysis of the individual genes: DMC1
In the case of DMC1 we constructed a scaffold gene (Fig. 2, lower) with 34 different intron positions (of a total of 55 introns; see Table 1) from the four examined organisms (A.thaliana, H.sapiens, C.cinereus and Pleurotus ostreata). Six of the introns were in conserved positions (comprising 18 individual introns) and were present at least in plants and one more kingdom, indicating that they are descended most probably from the LCA-PA. In total we found 31 cases of intron gain and only six cases of intron loss (Table 1). One putative case of intron sliding, intron 10 of P.ostreata, which corresponds to intron 9 of A.thaliana and intron 7 of H.sapiens, respectively, could be detected (Fig. 2, upper, and Fig. S2). Homo sapiens and A.thaliana harbor this intron in phase 2 at the same position and P.ostreata 7 nt upstream, which indicates a sliding of –7 nt (probably by a sliding event of –1 nt and a 6 nt long insertion).
Figure 2.
Schematic alignment of all introns of DMC1 and RAD51 from different organisms in relation to their protein sequences. The introns are differentiated by colored bars as in Figure 1. The gray shaded scaffold gene contains only the different intron positions of AtDMC1, HsDMC1, CcDMC1 and PoDMC1. Accession numbers: A.thaliana DMC1, U76670; H.sapiens DMC1, D64108 + locus 4914531; C.cinereus DMC1, AB036801; P.ostreata DMC1, AJ311528; A.thaliana RAD51, AJ001100; C.cinereus RAD51, U21905.
Intron 1 of A.thaliana is an excellent example for a recent case of intron gain. This intron is unique to A.thaliana, neither occurring in another plant (O.sativa) nor in animals or fungi. This clearly demonstrates that introns were indeed invading genomic sequences in recent times. Introns 2, 3, 5, 6, 9 and 11 of C.cinereus, which are identical in position to introns 3, 4, 8, 12, 16 and 18 of P.ostreata, were gained most likely after diversification of animals and fungi because they appear only in fungi and not in animals or plants.
Comparison of DMC1 and RAD51
We compared the intron structure of two RAD51 genes (from A.thaliana and C.cinereus) to each other and to DMC1 homologs because it was postulated that both genes arose by a duplication event in the LCA-PA (19). This comparison could clearly sustain the hypothesis, which was originally based on protein homology, and the resulting phylogenetic trees. All five introns found in the C.cinereus RAD51 gene (Fig. 2) are identical in position and phase to the RAD51 gene of A.thaliana, clearly indicating that RAD51 was also duplicated before the divergence of plants and fungi. Furthermore, one of the A.thaliana and C.cinereus RAD51 introns (Fig. 2, red bar, introns 3 and 2, respectively) is identical to a conserved intron of the DMC1 gene (Fig. 2, intron 6 of A.thaliana and intron 7 of P.ostreata, respectively), demonstrating again the ancient relationship between DMC1 and RAD51.
Analysis of the individual genes: MRE11
Comparing homologs of the MRE11 gene of the three reference organisms we postulate a scaffold gene (Fig. 3, lower) with 33 different intron positions (resulting from 50 individual introns, see also Fig. S3). Ten introns are in conserved positions (all in all 24 individual introns) and thus were already present before plant and animal divergence (Fig. 3, upper). MRE11 seems to be the most conserved gene of the three investigated concerning the intron positions. Apparently 20 cases of intron gain occurred and only six of loss during evolution. Interestingly, all six cases of intron loss (Fig. 3, blue circles) are restricted to one organism, namely C.cinereus. Six cases are indecisive because these introns are located in human and fungi only (Fig. 3, green bars). No putative intron sliding event could be detected.
Figure 3.
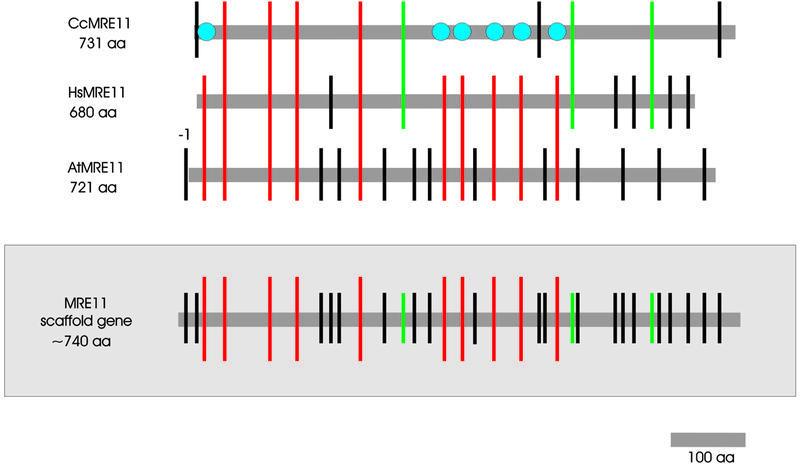
Schematic alignment of all introns of MRE11 from different organisms in relation to their protein sequences. The introns are differentiated by colored bars as in Figure 1. The gray shaded scaffold gene contains all different intron positions of AtMRE11, HsMRE11 and CcMRE11. Accession numbers: A.thaliana, AJ243822; H.sapiens, U37359; C.cinereus, AF178433.
Putative intron sliding
Regarding the introns of AtSPO11-1 and AtSPO11-2 we found that intron 9 (homologous to intron 6 of AtSPO11-2 and intron 8 of C.cinereus) is either a sliding event (+1 nt; see Fig. 1, asterisk) or the genetic position of this intron resembles a hotspot of intron gain. One explanation for this phenomenon is that the original intron position was the same as it is now in C.cinereus and AtSPO11-2 (intron phase = 0) and the sliding event occurred recently in AtSPO11-1. An alternative explanation would be that the nucleotide stretch where this intron is located resembles a hotspot of intron gain and loss. In this case the intron position would have been acquired nearby by three independent gain events in the respective organisms (H.sapiens, C.cinereus and A.thaliana).
We detected a second putative sliding event regarding intron 10 of the P.ostreata DMC1 gene. This intron is 7 nt downstream in comparison to introns 7 and 9 of H.sapiens and A.thaliana, respectively. In both A.thaliana and H.sapiens the intron is in phase 2, but in P.ostreata it is in phase 1 integrated. The closer relationship between fungi and animals argues for a real sliding event. An alternative explanation would be that plants, fungi and animals have acquired this intron three times independently or that fungi have lost it but gained a new one just 7 nt downstream.
Evaluation of the role of intron gain, loss and sliding
From 94 different intron positions present in the three scaffold genes at least 23 (24% of the intron positions, 60 of the individual introns) were identical or highly conserved (only 3 of the 23 conserved intron positions showed a slight in-frame variation of ±3 nt; see Figs S1–S3) in their position in plants and animals or plants and fungi and thus should be regarded as already present in the LCA-PA.
In total we found 60 cases of intron gain of 146 introns (41% of all; Table 1), counting all possible occurring introns in the investigated organisms. In contrast there were only 17 cases of intron loss and only two cases of putative intron sliding out of 99 possible positions (2%). For the identification of possible intron sliding events we took into account only the positions where an intron was present nearby in at least two organisms. Both putative intron sliding events resulted in a shift of more than –1 or +1 nt. Thus, we have to imply that in addition to the sliding event an insertion/deletion occurred in these two cases. Alternatively, a more complex scheme of two or three independent intron gain events can serve as an explanation for the occurrence of the putative sliding events.
The evolution of the SPO11 genes in eukaryotes
Plants are so far the only kingdom in which more than one SPO11 homolog could be found in the genome. These homologs of A.thaliana did not arise by recent duplications (20,21). Therefore, we were especially interested in the evolution of AtSPO11-1, AtSPO11-2 and AtSPO11-3 and addressed this question using a combination of the intron position data of the across kingdom alignment and a classical phylogenetic analysis.
AtSPO11-1 possesses 15 introns, AtSPO11-2 11 and AtSPO11-3 only one intron (see Fig. 1). AtSPO11-2 shares with AtSPO11-1 two of the nine conserved introns and one which is only present in AtSPO11-1 and AtSPO11-2 (Fig. 1, blue bar). Only one of the remaining eight introns of AtSPO11-2 can be aligned clearly to any other SPO11 gene, namely to intron 2 of the SPO11 gene of Cryptosporidium parvum (Fig. 1, blue bar). Cryptosporidium parvum is a human intestinal parasite belonging to a sister group of the dinoflagellates, the apicomplexa (22). The apicomplexa are a very diverse group of protozoan intracellular parasites which evolved very early after the plant–animal divergence (22,23). This points clearly to an early duplication of SPO11 shortly after the diversification of plants and animals. Subsequently, both genes must have had a different fate because the position and number of introns between AtSPO11-1 and AtSPO11-2 changed drastically. They share only three introns and AtSPO11-2 seems to have lost six (Fig. 1, blue circles) of the nine otherwise conserved introns but has gained seven introns at other positions (introns 1, 2, 5 and 8–11). At the same time AtSPO11-1 has lost only three introns [introns 4 and 5 of AtSPO11-2 of C.parvum (Fig. 1, blue bars) and intron 4 of H.sapiens] and gained five new ones (Fig. 1, small black bars).
In our opinion the occurrence of introns 4 and 5 of AtSPO11-2 only in plants and C.parvum but nowhere else is best explained by the postulated duplication of an ancestral gene of SPO11 shortly after the divergence of plants and animals but before the evolution of the apicomplexa. In the case of C.parvum we have no information on whether both genes are still present or if there is only the described gene left. Nevertheless, the known C.parvum gene seems to be a mixture of AtSPO11-1 and AtSPO11-2, each gene sharing two identical intron positions. A protein alignment of the partial Spo11 sequence of C.parvum with all other Spo11 proteins showed the highest identity with AtSpo11-2 (Fig. 5).
Figure 5.
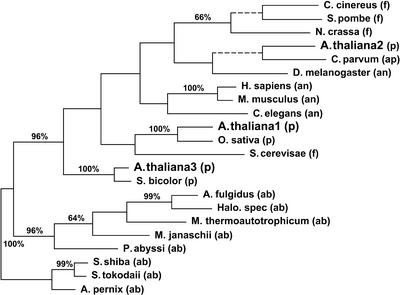
One of three most parsimonious trees (length 1671 steps, CI 0.6924, RI 0.5577) resulting from a Fitch parsimony analysis of 244 aligned amino acid positions of a conserved part of the SPO11 genes under study. Dashed branches will collapse in the strict consensus tree. Bootstrap values (>50%) are given along the branches. Ab, archaebacteria; an, animals; ap, apicomplexa; f, fungi; p, plants.
The third SPO11 gene
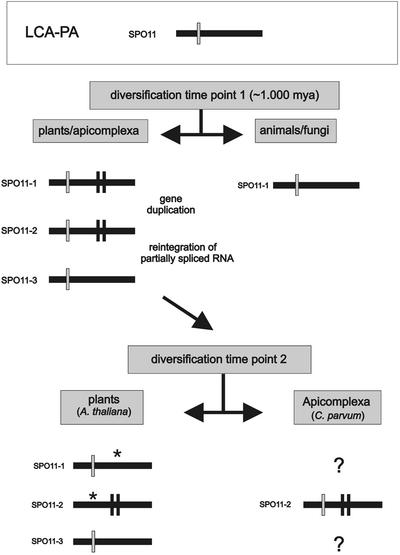
Also surprising is the presence of a third SPO11 homolog in plants. It is important to consider that plants also contain, besides the two extra SPO11 homologs, a homolog of subunit B of the archaebacterial topoisomerase. This homolog (AtTop6B) is able to interact with AtSpo11-2 and AtSpo11-3 (21). Thus, it seems that the existence of functional enzyme complexes prevents the loss of these genes in plants. Considering this, the loss of the B subunit in the animal and yeast lines might have made the extra Spo11 homologs functionally obsolete. Taking all data into account, we currently favor the following scenario (Fig. 4). As described, a duplication event of AtSPO11-1 occured shortly after plant and animal diversification. Subsequently, a partially spliced mRNA of AtSPO11-2, containing at least intron 4 of AtSPO11-1 (identical to C.parvum intron 1), which is not present in the recent AtSPO11-2, re-integrated by a reverse transcription mechanism into the genome resulting in AtSPO11-3 (Fig. 4, upper). The following facts support this hypothesis: (i) the single intron of AtSPO11-3 is in an identical position to intron 4 of AtSPO11-1 and intron 1 of C.parvum (Fig. 1, blue bar); (ii) both AtSPO11-2 and AtSPO11-3 possess in their catalytical domains a phenylalanine-tyrosine and not a tyrosine dyad, as is the case for all other SPO11 except that of yeast (21); (iii) both AtSPO11-2 and AtSPO11-3 are able to interact in vitro with subunit B (AtTOP6B) but AtSPO11-1 does not; (iv) phylogenetic analysis of the protein alignment (Fig. 5) places AtSpo11-2 and C.parvum Spo11 closer together (the clade also includes the D.melanogaster protein) than all other proteins (see below). The latter three points strongly suggest that it was SPO11-2 mRNA which re-integrated and not SPO11-1. We cannot rule out the possibility that the described duplication events of SPO11 genes occurred even before the divergence of plants and animals. However, this would imply that the ‘extra’ copies of SPO11 would have been lost in the animal/fungi lineage later on. We regard this scenario as less likely as two independent events of gene loss are required as an explanation.
Figure 4.
Postulated model of the evolution of SPO11 in the main eukaryotic kingdoms of life. Only three introns which are relevant for the model are shown. The introns lost during evolution are marked by asterisks. The numbering of the SPO11 genes from 1 to 3 is in accordance with their appearance in plants today. LCA-PA, last common ancestor of plants and animals.
Phylogenetic analysis of SPO11
Phylogenetic analysis of the amino acid alignment resulted in nearly identical tree topologies with phenetic (NJ) and cladistic (MP) algorithms (Fig. 5, only MP analysis shown). Arabidopsis thaliana Spo11 sequences uniformly occur in three different clades of the trees, the AtSpo11-1 protein together with O.sativa, AtSpo11-2 with C.parvum and D.melanogaster and AtSpo11-3 with the similar protein of Sorghum bicolor. The main difference between both analysis algorithms is caused by the position of the Saccharomyces cerevisae sequence. In the MP analysis S.cerevisae forms a weakly supported cluster with the Spo11-1 proteins of plants (Fig. 5), whereas in the NJ analysis S.cerevisae is a sister group of the Neurospora crassa clade, again with weak statistical support. The changed position of S.cerevisae in the NJ analysis results in a clade combining plant Spo11-1 proteins in a weakly supported cluster with the animal ones, except that of D.melanogaster.
DISCUSSION
Intron evolution
The analysis of our data supports the ‘weak intron sliding’ hypothesis (2) and argues against intron sliding as a mechanism responsible for a reasonable number of evolutionary differences in intron positions. Even in distantly related species we found only two putative cases of intron sliding out of 99 possible ones. Thus, intron sliding seems not to be a prominent pathway of intron evolution. Regarding the debate on whether introns are early or late (24–31) we found indications for both theories. It is very likely that a large part (41%) of the investigated introns were inserted late in evolution, but additionally a reasonable number of intron positions are quite conserved (23%) and must have been present at least in the LCA-PA. We are not sure whether these introns were already present in the last universal common ancestor (LUCA). At least, they are older than 1 000 000 000 years, the approximate time point of plant and animal divergence and the development of the apicomplexa (22,32,33). This finding contradicts the intron late theory, which claims that most introns evolved during the last billion years and almost no intron is of ancient origin (30). In the meantime this view has been challenged by the findings of several ancient introns in Euglena and microsporidia and also parts of the spliceosomal machinery in trichomonads and diplomonads (see 34 for a review). As shown in our study, a reasonable number of ancient introns exist in genes of the eukaryotic recombination machinery. Therefore, our results support the hypothesis of Phillipe and co-workers that the last common ancestror of extant eukaryotes possessed spliceosomal introns (34). We tend to a symbiotic view of the intron theories: some introns seem to be very old and others are quite recent (35). Intron evolution is most probably a mechanism in flux between gain and loss and not a static act of sequential intron gains or losses. Interestingly, for the three genes analyzed in this study, intron gain events seem to be nearly three times more frequent as intron loss events. However, one has to keep in mind that such comparisons strongly depend on the organisms investigated, e.g. the case of the two closely related Basidiomycetes. Pleurotus ostreata clearly favors intron gain, whereas C.cinereus does not and possesses the lowest number of introns in the DMC1 gene.
CIPAK as a tool to elucidate evolutionary events
Using CIPAK we were able to suggest that the duplication date of the three SPO11 homologs in plants occurred shortly after the divergence of the LCA into plants and animals. This was not possible unambiguously by comparison of the protein similarity. The analysis enabled us to suggest a conclusive model of the evolution of the three SPO11 genes present in plants today. Furthermore, we could confirm the earlier hypothesis of Yeager Stassen et al. (19) that DMC1 and RAD51 are two early (at least in the LCA-PA) duplicated homologs of RecA, at the level of the intron positions. This is to our knowledge the first report that comparison of intron positions can clearly support phylogenetic trees. In the case of SPO11 the phylogenetic trees alone were not able to resolve all evolutionary stages unambiguously. This is due to the fact that the amino acid sequences compared were rather diverse and a relatively low number of shared sequence positions was outnumbered by several autapomorphic characters in the single sequences. The resolution of this CIPAK seems to be more fine scaled over long time periods than the analysis of protein homology. For example, the phylogenetic tree of SPO11 proteins used alone would suggest that AtSPO11-2 and SPO11 of C.cinereus are more closely related than C.cinereus SPO11 and AtSPO11-1. This is definitely not the case as AtSPO11-1 and C.cinereus SPO11 both perform the same function (initiation of meiosis) (36,37). The CIPAK analysis showing five of the seven conserved introns shared between AtSPO11-1 and C.cinereus SPO11 and only two between AtSPO11-2 and C.cinereus SPO11 seems to resemble the evolutionary relationship of the genes more precisely. The more genomes are elucidated and the more EST data that appear, the more efficiently a refined technique like CIPAK can be performed. Especially in cases where the results of phylogenetic approaches are not sufficient to resolve the evolutionary distances (e.g. due to long branch artifacts or species with a high number of fast evolving characters), CIPAK could give additional information.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We want to thank Ingo Schubert and Anja Ipsen for carefully and critically reading the manuscript. This work was partly funded by the DFG (Pu137/6).
REFERENCES
- 1.Boudet N., Aubourg,S., Toffano-Nioche,C., Kreis,M. and Lecharny,A. (2001) Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis and Drosophila. Genome Res., 11, 2101–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoltzfus A., Logsdon,J.M.,Jr, Palmer,J.D. and Doolittle,W.F. (1997) Intron “sliding” and the diversity of intron positions. Proc. Natl Acad. Sci. USA, 94, 10739–10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogozin I.B., Lyons-Weiler,J. and Koonin,E.V. (2001) Intron sliding in conserved gene families. Trends Genet., 16, 430–432. [DOI] [PubMed] [Google Scholar]
- 4.Deutsch M. and Long,M. (1999) Intron–exon structures of eukaryotic model organisms. Nucleic Acids Res., 27, 3219–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartung F. and Puchta,H. (1999) Isolation of the complete cDNA of the Mre11 homologue of Arabidopsis (accession no. AJ243822) indicates conservation of DNA recombination mechanisms between plants and other eucaryotes (PGR 99-132). Plant Physiol., 121, 312. [Google Scholar]
- 6.Hartung F. and Puchta,H. (2000) Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res., 28, 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartung F., Plchova,H. and Puchta,H. (2000) Molecular characterisation of RecQ homologues in Arabidopsis thaliana. Nucleic Acids Res., 28, 4275–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keeney S., Giroux,C.N. and Kleckner,N. (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell, 88, 375–384. [DOI] [PubMed] [Google Scholar]
- 9.Bergerat A., de Massy,B., Gadelle,D., Varoutas,P.C., Nicolas,A. and Forterre,P. (1997) An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature, 386, 414–417. [DOI] [PubMed] [Google Scholar]
- 10.Paull T.T. and Gellert,M. (1998) The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell, 1, 969–979. [DOI] [PubMed] [Google Scholar]
- 11.Sharples G.J. and Leach,D.R.F. (1995) Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol., 17, 1215–1217. [DOI] [PubMed] [Google Scholar]
- 12.Connelly J., de Leau,E.S. and Leach,D.R.F. (1999) DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res., 27, 1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop D.K., Park,D., Xu,L. and Kleckner,N. (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- 14.Bishop D.K. (1994) RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell, 79, 1081–1092. [DOI] [PubMed] [Google Scholar]
- 15.Sogin M.L. (1991) Early evolution and the origin of eukaryotes. Curr. Opin. Genet. Dev., 1, 457–463. [DOI] [PubMed] [Google Scholar]
- 16.Dacks J.B. and Doolittle,W.F. (2001) Reconstructing/deconstructing the earliest eukaryotes: how comparative genomics can help. Cell, 107, 419–425. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swofford D.L. (2001) PAUP* Version 4.0b8a. Sinauer, Sunderland, MA.
- 19.Yeager Stassen N., Logsdon,J.M.,Jr, Vora,G.J., Offenberg,H.H., Palmer,J.D. and Zolan,M.E. (1997) Isolation and characterization of rad51 orthologs from Coprinus cinereus and Lycopersicon esculentum and phylogenetic analysis of eukaryotic recA homologs. Curr. Genet., 31, 144–157. [DOI] [PubMed] [Google Scholar]
- 20.Grant D., Cregan,P. and Shoemaker,R.C. (2000) Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl Acad. Sci. USA, 97, 4168–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartung F. and Puchta,H. (2001) Molecular characterization of homologues of both subunits A (SPO11) and B of the archaebacterial topoisomerase 6 in plants. Gene, 271, 81–86. [DOI] [PubMed] [Google Scholar]
- 22.Fast N.M., Kissinger,J.C., Roos,D.S. and Keeling,P.J. (2001) Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol., 18, 418–426. [DOI] [PubMed] [Google Scholar]
- 23.Striepen B., White,M.W., Li,C., Guerini,M.N., Malik,S.-B., Logsdon,J.M.,Jr, Liu,C. and Abrahamsen,M.S. (2002) Genetic complementation in apicomplexan parasites. Proc. Natl Acad. Sci. USA, 99, 6304–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doolittle W.F. (1978) Genes in pieces: where they ever together? Nature, 272, 581–582. [Google Scholar]
- 25.Gilbert W., Marchionni,M. and McKnight,G. (1986) On the antiquity of introns. Cell, 46, 151–153. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert W. (1987) The exon theory of genes. Cold Spring Harbor Symp. Quant. Biol., 52, 901–905. [DOI] [PubMed] [Google Scholar]
- 27.Cavalier-Smith T. (1985) Selfish DNA and the origin of introns. Nature, 315, 283–284. [DOI] [PubMed] [Google Scholar]
- 28.Palmer J.D. and Logsdon,J.M.,Jr (1991) The recent origins of introns. Curr. Opin. Genet. Dev., 1, 470–477. [DOI] [PubMed] [Google Scholar]
- 29.Logsdon J.M. Jr, and Palmer,J.D. (1994) Origin of introns–early or late? Nature, 369, 526–527. [DOI] [PubMed] [Google Scholar]
- 30.Logsdon J.M. Jr, (1998) The recent origins of spliceosomal introns revisited. Curr. Opin. Genet. Dev., 8, 637–648. [DOI] [PubMed] [Google Scholar]
- 31.Jean L., Long,M., Young,J., Pery,P. and Tomley,F. (2001) Aspartyl proteinase genes from apicomplexan parasites: evidence for evolution of the gene structure. Trends Parasitol., 17, 491–498. [DOI] [PubMed] [Google Scholar]
- 32.Gajadhar A.A., Marquardt,W.C., Hall,R., Gunderson,J., Ariztia-Carmona,E.V. and Sogin,M.L. (1991) Ribosomal RNA sequences of Sarcocystis muris, Theileria annulata and Crypthecodinium cohnii reveal evolutionary relationships among apicomplexans, dinoflagellates and ciliates. Mol. Biochem. Parasitol., 45, 147–154. [DOI] [PubMed] [Google Scholar]
- 33.Doolittle R.F., Feng,D.-F., Tsang,S., Cho,G. and Little,E. (1996) Determining divergence times of the major kingdoms of living organisms with a protein clock. Science, 271, 470–477. [DOI] [PubMed] [Google Scholar]
- 34.Philipe H., Germot,A. and Moreira,D. (2000) The new phylogeny of eukaryotes. Curr. Opin. Genet. Dev., 10, 596–601. [DOI] [PubMed] [Google Scholar]
- 35.Rzhetsky A. and Ayala,F.J. (1999) The enigma of intron origins. Cell. Mol. Life Sci., 55, 3–6. [Google Scholar]
- 36.Grelon M., Vezon,D., Gendrot,G. and Pelletier,G. (2001) AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J., 20, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merino S.T., Cummings,W.J., Acharya,S.N. and Zolan,M.E. (2000) Replication-dependent early meiotic requirement for Spo11 and Rad50. Proc. Natl Acad. Sci. USA, 97, 10477–10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.