Summary
Human processed pseudogenes are copies of cellular RNAs reverse transcribed and inserted into the nuclear genome by the enzymatic machinery of L1/LINE1 non-LTR retrotransposons. While it is generally accepted that germline expression is crucial for heritable retroposition of cellular mRNAs, almost nothing is known about the influences of RNA stability, mRNA quality control, and compartmentalization of translation on retroposition of processed pseudogenes. Here we found that frequently retroposed human mRNAs are derived from stable transcripts with translation-competent functional reading frames resistant to nonsense-mediated RNA decay. They are preferentially translated on free cytoplasmic ribosomes and encode soluble proteins. Our results indicate that interactions between mRNAs and L1 proteins seem to occur at free cytoplasmic ribosomes.
Keywords: mRNA surveillance, NMD, retropseudogene, retrotransposition, soluble proteins, membrane proteins
L1 elements were very active throughout mammalian evolution and 17% of the human genome is covered by recognizable copies of L1 [1,2]. Active copies still retropose in the human genome [3], cause insertional inactivation of human genes [2], and seem to stimulate recombinogenic genomic breaks by nicking of host DNA [4]. L1s also transpose other elements in trans, namely processed pseudogenes [5,6], Alu SINEs [7], and probably also SVA SINEs [8].
Processed pseudogenes (PPs), also known as retropseudogenes, are copies of cellular RNAs reverse transcribed and inserted into the genome [9]. Similarly to mRNAs, PPs typically contain polyA tails and lack promoters and introns. With rare exceptions [10], processed pseudogenes in the human genome seem to result from L1 retroposition activity. PPs are derived from normal protein-coding mRNAs, alternatively spliced mRNAs [11], antisense transcripts [12], non-protein-coding RNAs [13], incompletely spliced hnRNAs [14], or mRNAs derived from other repetitive elements [15]. Retroposed pseudogenes represent an important source of new genes called retrogenes [16–18] and retropositions of PPs into introns can also transfer new protein-coding domains into existing genes (exon shuffling) [19].
Depending on the search methodology and stringency, the estimated number of PPs in the human genome is from 8,000 up to 100,000, if truncated copies are also considered [20–24]. Yet, only about 10% of cellular genes have one or more retroposed copies in the genome [21,23]. While the effects of germline expression [23,25,26] and genomic stability [23,27,28] have been studied previously, little is known about the influence of RNA stability and translation on retroposition of PPs.
Effects of RNA stability and translational control
We compared several properties of human protein-coding mRNAs with the number of processed pseudogenes derived from them. We used the database of PPs developed by Zhang and coworkers [23]. After removing some redundancy (see Supplementary Methods), the set contains 7,464 pseudogenes derived from a subset of the 23,337 nonredundant protein-coding genes (0.32 pseudogenes/gene). Using strict parameters (>70% coverage of the protein sequence, >99% amino acid identity), we assigned mRNAs obtained from studies of mRNA stability and translation localization to the set of 23,337 genes and corresponding pseudogenes (Supplementary Table 1).
To study the effect of mRNA stability on retroposition of PPs, we combined the set of PPs with decay rates of human mRNAs determined by Yang and coworkers [29]. These authors inhibited transcription in hepatocellular carcinoma HepG2 cells line and Bud8 primary fibroblasts, and used microarrays to measure mRNA levels before, and after inhibition. Figure 1a shows the comparison of their derived mRNA decay rate to the number of PPs. Rapidly decaying RNA produced far less PPs than stable RNA or the genome average. Stable transcripts, on the other hand, have significantly more pseudogenes than the genome average.
Figure 1.
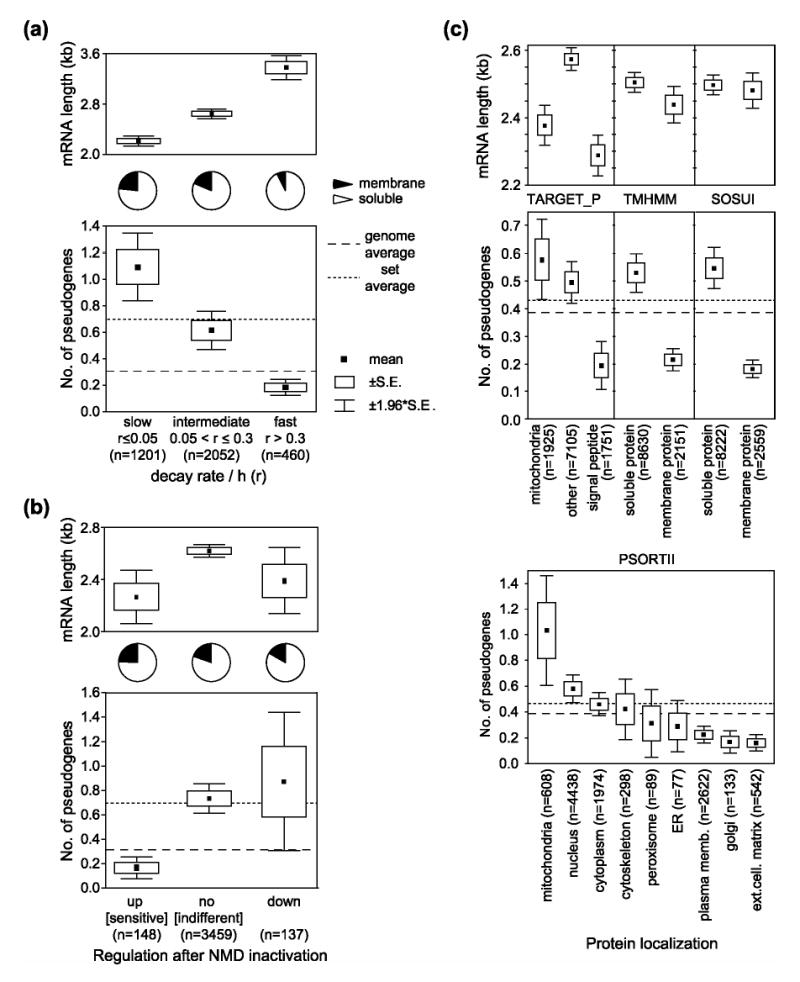
Effects of mRNA properties on retroposition of processed pseudogenes. The first plot (a) shows the number of processed pseudogenes per gene against its mRNA stability (r). The data on mRNA stability [29] were divided into several categories. For each group of genes, we calculated mean number of pseudogenes, standard error (S.E.), and the 1.96*S.E. interval, which for a normal distribution corresponds to 95% confidence interval. The numbers in parentheses reflect the sample size (number of genes). The dashed lines mark the average number of pseudogenes per gene, and per set analyzed. The upper part of the figure shows additional properties of the mRNA categories, namely average length and proportion of soluble to membrane proteins based on SOSUI predictions [42]. (b) The effect of NMD sensitivity [33] on retroposition of processed pseudogenes. The first category is up-regulated after NMD inactivation (NMD-sensitive). Some mRNAs seem to be down-regulated after NMD inactivation. The remaining mRNAs do not show any significant change after NMD inactivation (NMD-indifferent). (c) The effect of translation compartmentalization on retroposition of processed pseudogenes. Protein localizations predicted by four different methods were obtained from the H-Invitational database [36]. Results for each of these prediction methods are shown in separate boxes. The total number of compared nonredundant mRNAs is in each case 10781.
Stability of pol II transcripts depends on a variety of mechanisms including mRNA quality control, also known as mRNA surveillance [30]. Notably, nonsense mediated RNA decay (NMD) targets mRNAs with premature termination codons (PTC) and prevents synthesis of potentially negatively dominant truncated proteins [30,31]. PTC-containing mRNAs are degraded by several pathways [32].
We were interested in the possible influence of NMD on retroposition of PPs. Mendell et al. [33] used short interfering RNA (siRNA) to inhibit the Upf1/Rent1 factor essential for NMD. The authors compared RNA levels in HeLa cells before, and 72 hours after, the application of Upf1 siRNA. While the majority of mRNAs were relatively insensitive to NMD inhibition (NMD-indifferent), the authors found that 197 transcripts were consistently up-regulated and 176 down-regulated in duplicate experiments. As expected, many transcripts up-regulated after NMD inhibition are typical NMD-sensitive substrates such as mRNAs with upstream open reading frames or alternative splice variants with PTCs [33].
We compared the number of PPs between NMD-indifferent transcripts, and mRNAs up-regulated after NMD inhibition (Figure 1b). Potential NMD-sensitive mRNAs have significantly fewer PPs than the majority of NMD-indifferent RNAs (p=0.005, Mann-Whitney U Test). Transcripts down-regulated after NMD inactivation, have, on the other hand, slightly more PPs than NMD-indifferent (not significant). These results indicate that translation-competent mRNAs with intact ORFs are preferentially retroposed by L1 and that resistance to mRNA quality control during the pioneer round of translation is important for efficient retrotransposition.
Next, we focused on localization of translation. In general, mRNAs encoding soluble proteins are translated on free cytoplasmic ribosomes. On the other hand, translation of transcripts coding for transmembrane and secreted proteins is started in the cytoplasm and finished on the endoplasmic reticulum (ER). The transfer of the mRNA/ribosome complex to the ER depends on recognition of the signal peptide by the signal recognition particle [34,35]. We were interested in whether compartmentalization of translation has any effect on retroposition of PPs. The site of protein synthesis can be estimated by the presence of a signal peptide. Proteins containing this short motif are likely to be synthesized on ER-bound ribosomes, while secreted proteins without the signal peptide are translated on free cytoplasmic ribosomes.
Various strategies have been employed to detect the signal peptide and predict protein localization from amino acid sequences. We obtained four different predictions of protein localization from the H-Invitational database of human full-length mRNAs [36]. All the prediction methods consistently showed that soluble proteins have, on average, statistically significantly (not shown) more PPs than signal peptide-containing proteins (Figure 1c). Transcripts encoding mitochondrial proteins have the most retroposed copies in the genome, followed by mRNAs for nuclear and cytoplasmic proteins. The lowest numbers of PPs have transcripts encoding proteins excreted to the extracellular matrix, or found in the Golgi apparatus or plasma membrane. This strongly suggests that not only translation, but also localization of translation is important for efficient retroposition.
Our analysis revealed several factors influencing the retroposition of processed pseudogenes: (1) mRNA stability, (2) NMD sensitivity, and (3) localization of translation. We tested whether these effects represent a coincidental dependence between the sets used in different analyses. Figures 1a and 1b show proportions of soluble and membrane proteins for all categories of transcripts. Slowly decaying (stable) mRNAs encode proportionately fewer soluble proteins and more membrane proteins, than unstable transcripts (Figure 1a). Based on the proportion of soluble proteins we would therefore expect that stable mRNAs would have less, not more PPs than unstable transcripts. Similarly, the analysis of transcripts up-regulated after NMD inactivation (Figure 1b) shows only a small proportional decrease of soluble proteins in the set. Thus the decreased number of pseudogenes derived from up-regulated transcripts is not a consequence of a biased proportion of soluble proteins in the set.
We also analyzed the effect of mRNA length, since there is a bias for preferential detection of PPs derived from short mRNAs [37]. Figures 1b and 1c clearly show that the observed influences of NMD and translational localization on retroposition are not due to detection bias for pseudogenes derived from shorter mRNAs. However, in the case of mRNA stability, the mRNA length is negatively correlated with the number of pseudogenes (Figure 1a) and a partial correlation analysis confirmed that the mRNA length is a significant factor affecting the number of PPs. When we controlled for mRNA length, the partial correlation between pseudogene number and mRNA stability decreased, but still remained significant (not shown).
Finally, one should note that we did not compare actual rates of retroposition, but numbers of PP copies in the human genome. The number of genomic PPs is affected by at least two additional factors: germline expression and genomic stability. The genomic distribution of PPs is similar to L1 integration patterns [23,27,28] indicating relative stability of these elements in the genome. Only PPs retroposed in the germline can be transmitted to the next generation and permit fixation in the population. Therefore, strong germline expression seems to be necessary for efficient retroposition by L1, but it is rather difficult to measure. Contamination by somatic cells, or stress linked to separation from somatic cells, is likely to affect the obtained expression profiles. Changes associated with cellular transformation compromise reliability of expression profiles obtained from cancer cell lineages. In this context it is worth noting that, while the effects of genomic stability and germ line expression are difficult to verify, our results are directly testable using retrotransposition assays in cultured mammalian cells [38].
Implication for L1-mediated retrotransposition
How do our results fit the model of L1 retrotransposition? Analyses of human mutations caused by L1 retrotransposition [39] and in retrotransposition assays [6,38,40] indicated that L1 elements with intact functional open reading frames (ORFs) are preferentially transposed. L1 mRNAs with ORFs inactivated by mutations or other cellular mRNAs are retroposed much less efficiently. The apparent preference of L1 for intact ORFs can be explained by a cis model of retroposition [41]. Immediately after translation, the second protein (ORF2) interacts with the first available mRNA polyA tail, which is the polyA of the same mRNA that encoded ORF2. By this mechanism, mutated L1 mRNAs are eliminated, because they cannot encode functional proteins.
This model and the apparent lack of sequence specificity for L1 mRNA [41] imply an important conclusion: to ensure optimal interaction between the L1 mRNA and ORF2 protein, the interaction takes place immediately after protein synthesis, which occurs on the ribosome. This further implies that the chance of an mRNA being retroposed increases if it is: (1) stable in the cytoplasm, (2) resistant to NMD, since transcripts close to ribosomes may be scanned by mRNA quality control, and (3) translated on free ribosomes outside the ER, because none of the L1 proteins contain the signal peptide sequence. Our results strongly support this model of L1 retroposition. To be efficiently retroposed by L1, an mRNA must be in the right place (near ribosomes where L1 mRNAs are translated), at the right time (when L1 are expressed).
In conclusion, not only germ-line expression, but also RNA stability, translational competence and localization contributed strongly to retroposition of processed pseudogenes during human evolution.
Supplementary Material
Acknowledgments
We thank John Moran and members of his group for comments on the manuscript. This work was supported by National Institutes of Health grant 2 P41 LM006252-07A1 and by the Center for Applied Genomics (1M0520).
Footnotes
Teaser: The first study demonstrating that mRNA stability, quality control and compartmentalization of translation have a major impact on retroposition of processed pseudogenes.
References
- 1.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 2.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 3.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babcock M, et al. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome Res. 2003;13:2519–2532. doi: 10.1101/gr.1549503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci U S A. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esnault C, et al. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 7.Dewannieux M, et al. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 8.Ostertag EM, et al. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiner AM, et al. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 10.Jamain S, et al. Transduction of the human gene FAM8A1 by endogenous retrovirus during primate evolution. Genomics. 2001;78:38–45. doi: 10.1006/geno.2001.6642. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Gerstein M. Identification and characterization of over 100 mitochondrial ribosomal protein pseudogenes in the human genome. Genomics. 2003;81:468–480. doi: 10.1016/s0888-7543(03)00004-1. [DOI] [PubMed] [Google Scholar]
- 12.Ejima Y, Yang L. Trans mobilization of genomic DNA as a mechanism for retrotransposon-mediated exon shuffling. Hum Mol Genet. 2003;12:1321–1328. doi: 10.1093/hmg/ddg138. [DOI] [PubMed] [Google Scholar]
- 13.Jurka J, et al. Small cytoplasmic Ro RNA pseudogene and an Alu repeat in the human alpha-1 globin gene. Nucleic Acids Res. 1988;16:766. doi: 10.1093/nar/16.2.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soares MB, et al. RNA-mediated gene duplication: The rat preproinsulin I gene is a functional retroposon. Mol Cell Biol. 1985;5:2090–2103. doi: 10.1128/mcb.5.8.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavlicek A, et al. Processed pseudogenes of human endogenous retroviruses generated by LINEs: their integration, stability, and distribution. Genome Res. 2002;12:391–399. doi: 10.1101/gr.216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- 17.Gentles AJ, Karlin S. Why are human G-protein-coupled receptors predominantly intronless? Trends Genet. 1999;15:47–49. doi: 10.1016/s0168-9525(98)01648-5. [DOI] [PubMed] [Google Scholar]
- 18.Brosius J. Many G-protein-coupled receptors are encoded by retrogenes. Trends Genet. 1999;15:304–305. doi: 10.1016/s0168-9525(99)01783-7. [DOI] [PubMed] [Google Scholar]
- 19.Dupuy D, et al. SCAN domain-containing 2 gene (SCAND2) is a novel nuclear protein derived from the zinc finger family by exon shuffling. Gene. 2002;289:1–6. doi: 10.1016/s0378-1119(02)00543-7. [DOI] [PubMed] [Google Scholar]
- 20.Goncalves I, et al. Nature and structure of human genes that generate retropseudogenes. Genome Res. 2000;10:672–678. doi: 10.1101/gr.10.5.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohshima K, et al. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torrents D, et al. A genome-wide survey of human pseudogenes. Genome Res. 2003;13:2559–2567. doi: 10.1101/gr.1455503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, et al. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adel K, et al. HOPPSIGEN: a database of human and mouse processed pseudogenes. Nucleic Acids Res. 2005;33:D59–66. doi: 10.1093/nar/gki084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleene KC, et al. The mouse gene encoding the testis-specific isoform of Poly(A) binding protein (Pabp2) is an expressed retroposon: intimations that gene expression in spermatogenic cells facilitates the creation of new genes. J Mol Evol. 1998;47:275–281. doi: 10.1007/pl00006385. [DOI] [PubMed] [Google Scholar]
- 26.Pain D, et al. Multiple retropseudogenes from pluripotent cell-specific gene expression indicates a potential signature for novel gene identification. J Biol Chem. 2005;280:6265–6268. doi: 10.1074/jbc.C400587200. [DOI] [PubMed] [Google Scholar]
- 27.Pavlicek A, et al. Similar integration but different stability of Alus and LINEs in the human genome. Gene. 2001;276:39–45. doi: 10.1016/s0378-1119(01)00645-x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, et al. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 2004;20:62–67. doi: 10.1016/j.tig.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Yang E, et al. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maquat LE, Carmichael GG. Quality control of mRNA function. Cell. 2001;104:173–176. doi: 10.1016/s0092-8674(01)00202-1. [DOI] [PubMed] [Google Scholar]
- 31.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 32.Lejeune F, et al. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 33.Mendell JT, et al. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 34.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 35.Lingappa VR, et al. Chicken ovalbumin contains an internal signal sequence. Nature. 1979;281:114–121. doi: 10.1038/281117a0. [DOI] [PubMed] [Google Scholar]
- 36.Imanishi T, et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavlicek A, et al. Length distribution of long interspersed nucleotide elements (LINEs) and processed pseudogenes of human endogenous retroviruses: implications for retrotransposition and pseudogene detection. Gene. 2002;300:189–194. doi: 10.1016/s0378-1119(02)01047-8. [DOI] [PubMed] [Google Scholar]
- 38.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 39.Sassaman DM, et al. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 40.Wei W, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boeke JD. LINEs and Alus--the polyA connection. Nat Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- 42.Hirokawa T, et al. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


