Abstract
RNA molecules fold into characteristic secondary and tertiary structures that account for their diverse functional activities. Many of these RNA structures, or certain structural motifs within them, are thought to recur in multiple genes within a single organism or across the same gene in several organisms and provide a common regulatory mechanism. Search algorithms, such as RNAMotif, can be used to mine nucleotide sequence databases for these repeating motifs. RNAMotif allows users to capture essential features of known structures in detailed descriptors and can be used to identify, with high specificity, other similar motifs within the nucleotide database. However, when the descriptor constraints are relaxed to provide more flexibility, or when there is very little a priori information about hypothesized RNA structures, the number of motif ‘hits’ may become very large. Exhaustive methods to search for similar RNA structures over these large search spaces are likely to be computationally intractable. Here we describe a powerful new algorithm based on evolutionary computation to solve this problem. A series of experiments using ferritin IRE and SRP RNA stem–loop motifs were used to verify the method. We demonstrate that even when searching extremely large search spaces, of the order of 1023 potential solutions, we could find the correct solution in a fraction of the time it would have taken for exhaustive comparisons.
INTRODUCTION
RNA secondary structures have been described in several important classes of RNAs, including non-coding RNAs such as rRNAs, tRNAs, RNase P and SRP, as well as cellular and viral mRNAs where these structures are known to be important regulators of translation and stability. Some examples of the mRNA structures include the iron-responsive element (IRE) located in the 5′- or 3′-untranslated regions (UTRs) of mRNAs involved in iron metabolism and transport (1,2), stem–loops in the 3′-UTRs of histones and vimentin (3,4) and IRES elements in the 5′-UTRs of picornaviruses, pestiviruses and flaviviruses (5). In all of the known RNA structures, secondary structure is conserved during evolution, despite substantial sequence variation. While a number of tools exist for performing sequence similarity searches, currently there are no useful techniques for performing RNA structure similarity searches. A computational tool to explore nucleotide sequence space for conserved but unknown RNA structures might lead to the discovery of new structures and improve our understanding of functional and regulatory relationships amongst related RNAs.
One way to approach the task of de novo identification of conserved structural elements is to define the space of all structures that match a particular hypothesized motif and evaluate the presence or absence of these motifs in the sequences under consideration. Computational tools such as RNAMotif have previously been developed to define and search for RNA secondary structure motifs (6–10). These tools allow abstraction of the structural pattern into a ‘descriptor’ with a pattern language that gives details regarding pairing information, length and sequence. A list of all possible structures that match any given descriptor within a set of sequences can be easily generated. Depending on the specificity of the descriptor and the number of nucleotides in the sequence database, this can result in a few hits or a very large number of hits (i.e. of the order of 105 hits, or more, for a given bacterial genome). When the number of hits is large, an exhaustive search for a set of maximally similar structures can be computationally intractable. Here we describe methods based on evolutionary computation (EC) to search possible RNA secondary structures for common elements across multiple sequences without requiring pre-alignment or sequence constraints in the descriptor.
All evolutionary algorithms (EAs) require a population of contending solutions to be generated (11–13). Each solution in the population is then scored with respect to a measure of its worth or ‘fitness’. Solutions of low fitness are more likely to be removed from the population than solutions of higher fitness during a process of selection. Following selection, the surviving solutions are used to generate new contending solutions with random variations until the population size is re-established. This process of variation and selection is iterated for a specified number of generations or until the population has discovered a solution of adequate worth.
Previous attempts at RNA structure prediction using EC have focused on genetic algorithms, but alternative representations and methods exist and have yet to be explored, not only for structure prediction but also for calculation of RNA structure similarity (14). Here, we focus on an alternative representation and set of operators to search via evolution. The procedure seeks a set of similar structures in a top-down fashion rather than focusing mainly on a bottom-up combination of useful ‘building blocks’, as is common to genetic algorithm approaches (for additional information on these differences see 12). These representations and evolutionary techniques are described in Materials and Methods. Details of the algorithm and implementation are provided as Supplementary Material online.
MATERIALS AND METHODS
Evolutionary computation
RNAMotif produces a list of structures (or ‘hits’) that conform to a particular structure descriptor. The RNAMotif output file contains the following information: structure pairing information, a sequence identifier (ID), the position of a hit relative to the start of the sequence, the number of nucleotides in the structure, the strand (sense or antisense) and the nucleotide sequence associated with the RNA structure. The information contained in the RNAMotif output serves as input to the EA.
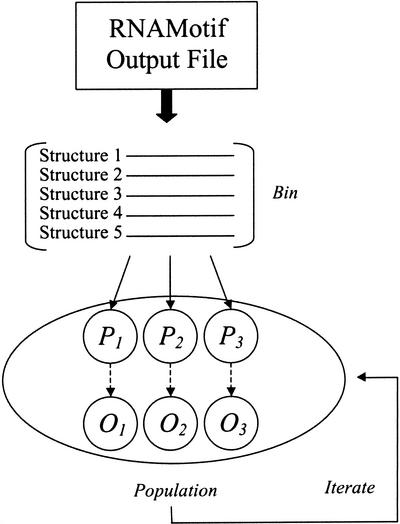
Population initialization. A collection or ‘bin’ of structures is chosen at random without replacement from structures represented in the RNAMotif output file. Each bin represents one contending solution in the population and is referred to as a ‘parent bin’ for the initial generation of evolution. The initialization process is repeated until P parent bins are created. The number of structures contained in each bin is referred to as the ‘bin size’ (B) (Fig. 1). Both B and P are user defined and are fixed throughout one run of evolution. During initialization, each of P bins is constructed by selecting B structures at random from the RNAMotif output file, where 1 < B ≤ Bmax (Bmax = the total number of structures in the RNAMotif file). When B is larger than the number of organisms represented in the RNAMotif file, multiple structures for a given sequence ID will occur. A user-defined parameter can be used to force only one structure to be drawn at random from each sequence ID.
Figure 1.
A schematic of the initialization process. Hits from an RNAMotif file are placed randomly into bins, where the bin size (B) is determined by the user (here B = 5). For initialization, the randomly generated bins are considered ‘parent’ bins (P). Each P is then used to generate O offspring bins with variation (see text). After initialization, the P + O bins are considered as one evolving population.
Variation. For the initial generation, each P is copied to form O ‘offspring bins’, where O is a user-defined parameter. Once O offspring bins have been generated, the parent and offspring bins are treated as one evolving population. During the copying process, variation operators are applied so that each offspring will have some difference relative to the parent. A first random variable is drawn from a probability distribution (e.g. Poisson or Gaussian) to determine which of the variation operators are chosen. A second random variable is drawn from a probability distribution to determine the number of times a particular variation operator is applied. The possible variation operators are: (i) structure replacement within a specified sequence ID; (ii) structure replacement from a different sequence ID; (iii) random single-point bin recombination; (iv) random multi-point bin recombination.
For the structure replacement within a specified sequence ID operator, structures in a bin are replaced at random with new structures from the same organism in the RNAMotif file. With this operator, a new set of random variables is required. A first random variable is chosen to determine which structure(s) to replace with a minimum number of replacements 1 and maximum B – 1. A second random variable is selected to determine between two choices of range (local or global) for the difference in structural similarity between the old and new structures. RNAMotif output files contain structures that are listed in order of position relative to the 5′ end of the target sequence. Therefore, within the file, neighboring RNAMotif structure hits have a high probability of structural similarity. The local version of the structure replacement within a specified sequence ID operator chooses a replacement structure from the RNAMotif file that neighbors the original structure in the file. The global version of the structure replacement within a specified sequence ID operator chooses a replacement structure at random from the RNAMotif file without replacement. The global version of this operator allows for the possibility of large variation whereas the local variation operator has a higher probability of small variation.
The structure replacement from a different sequence ID variation operator is used to randomly replace a structure in a bin with a new structure from a different organism in the RNAMotif file. Assume hits from 10 different organisms in the RNAMotif file and B = 5. A random number is drawn for the number of structures to be replaced in the bin, with a minimum and maximum number of replacements of 1 and B – 1, respectively. When a structure is chosen for replacement, a new structure is chosen at random from the set of structure hits from a different sequence ID in the RNAMotif file.
The random single-point bin recombination operator makes use of the information in two parent bins to generate two new offspring bins via single-point recombination. When using the random single-point bin recombination operator, one parent bin (P1) is selected at random from the population whereas the other parent bin (P2) is a newly constructed random draw of structures from the RNAMotif file. For example, assume B = 5. Within P1, a random variable is used to select a structure to serve as a position of single-point recombination between P1 and P2 and generate two new offspring bins, O1 and O2. One of the offspring is selected at random as a new member of the population. During the evolutionary process, this operator therefore combines a parent bin containing implicit evolutionary history (P1) with a new parent bin (P2) constructed completely at random in order to allow for very large jumps across the search space. The random multi-point bin recombination operator makes use of the same procedure except with multiple points of recombination. Methods of self-adaptation can be incorporated concurrent with the process of evolution (see Supplementary Material).
Fitness. The fitness function is an aggregate of components that measure RNA structure similarity. These measures are applied pairwise (15) by each structural component and then summed into a final bin fitness score. The scoring components are: (i) nucleotide sequence similarity within a structural component; (ii) structure component length similarity; (iii) structure thermodynamic stability similarity (see Supplementary Material for details).
Selection. Based on these scores, a mechanism of selection determines which bins will be removed from the current population. Under a tournament selection approach (12), a bin from the current population is chosen at random and is ‘competed’ with a set of R randomly chosen bins in the same population, where R is user defined. Each time the first bin’s fitness score is higher than (or ties) the opponent’s score, the first bin scores a ‘win’. The number of wins is recorded for all competitions and this process is iterated over all members of the population. All bins are then ranked with respect to the number of wins scored during the competition. Selection is used to remove the lower O bins on this ranked list. In the case of a tie in the number of wins, those specific bins are re-ranked by fitness prior to selection. After selection, the P remaining bins are saved to serve as parents for the next generation.
Program implementation
Evolution was performed in parallel on four, dual processor Intel Pentium III, 450 MHz, 256 MB RAM computers, running Linux O/S using server/client architecture. A ‘master’ server was used as the user interface, reading parameters, and RNAMotif data files. This program then spawned one or more clients that performed the evolution. Each client was initialized with a random number seed, periodically transmitting its best solution set back to the master. Although the clients acted as parallel evolutionary ‘islands’, data were also communicated between clients. This sharing of information between clients is known to facilitate escape from local optima and improve the rate of convergence.
For all the experiments presented here, tournament selection, Poisson distributions for the number of mutations and Gaussian distribution for self-adaptation were used with varying population sizes for 1000 generations of evolution to find similar structures in the sense strand. The time taken to converge on the known solutions for these RNA structures was measured and the remainder of evolution was monitored to ensure that ‘better’ solutions were not generated.
RESULTS
Motif search examples
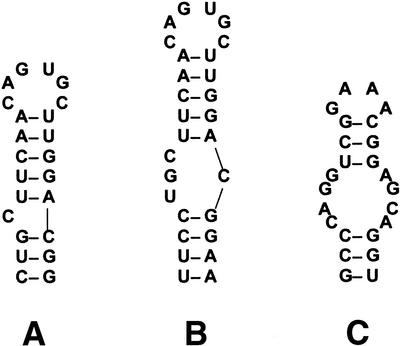
Iron-responsive element (IRE). IREs have been described in the 5′- and 3′-UTRs of several mRNAs (1,16–20). IREs bind iron-regulatory proteins (IRPs) and regulate iron homeostasis in eukaryotes. Two forms of the RNA secondary structure for IRE have been proposed in the literature (20). The stem–loop structure proposed differs in the structure of the internal loop disrupting the helix. The IRE secondary structure is most frequently shown with a C bulge on the 5′ side of the helix (Fig. 2A). An alternate structure has an asymmetrical internal loop at this same position with three unpaired bases on the 5′ side of the helix and a single C on the 3′ side (Fig. 2B). A single, highly specific RNAMotif descriptor can be written to capture both of these structural elements and identifies IREs in a number of iron-regulated transporters (10). A less specific descriptor for this same structure element increases the number of false positives significantly but may also allow discovery of distantly related IREs over many species. We used a series of three descriptors of increasing generality over four experiments to test the ability of the EA to discover common IRE structures in ferritin mRNA sequences from a number of orthologous sequences.
Figure 2.
Structures of the human ferritin IRE (A and B) and structure of human SRP domain IV (C) found in the literature (20,23).
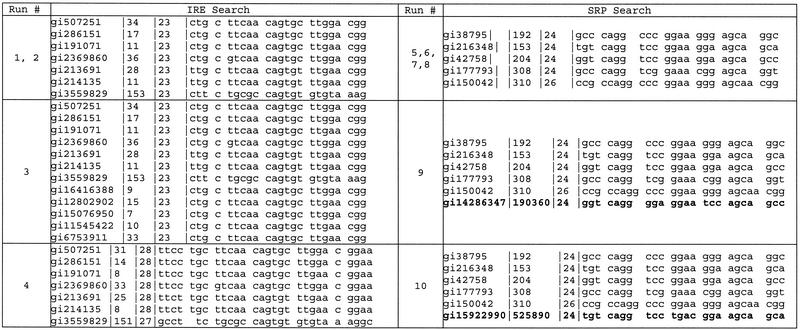
For the first experiment, seven full-length ferritin mRNA sequences (Homo sapiens, gi|507251; Sus scrofa, gi|286151; Cricetulus griseus, gi|191071; Gallus gallus, gi|2369860; Rana catesbeiana, gi|213691; Xenopus laevis, gi|214135; Drosophila melanogaster, gi|3559829) were obtained from GenBank. The descriptor shown in Figure 3A was used to generate structure hits using RNAMotif. The number of hits for each experiment is given in Table 1. Statistics regarding the number of possible bins, evolution parameters and time to completion for all experiments are provided in Table 2. When each bin is allowed to contain one structure from each of the seven organisms and all possible combinations are allowed, there are 7.6 × 108 possible bins in the search space (Table 2). The evolutionary search examined only a fraction of the possible bins (1.4 × 10–5) before converging on a solution, which contained a set of structures that exactly matched the proposed IRE structure (Table 3). This was achieved by the 13th generation in <3 min. Exhaustive evaluation of all possible bin combinations for this experiment at the same rate of calculation would have required 125 days.
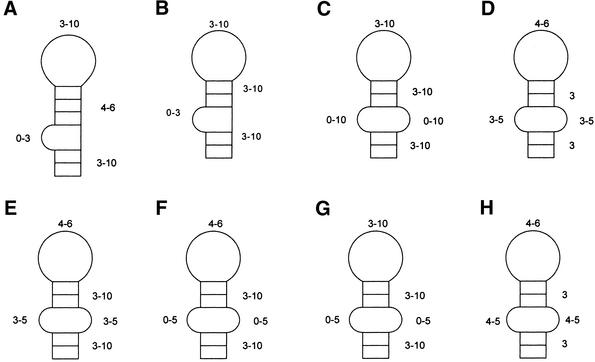
Figure 3.
RNAMotif descriptors used IRE experiments 1–4 (A–C) and SRP experiments 5–10 (D–H). Descriptors (A–C) include the possibility for one potential mispair at the base of the upper stem (not shown). Descriptor (C) has unpaired nucleotides on the 3′ side of the stem in opposition to the original bulge providing the possibility for internal loops in the final product.
Table 1. Number of RNAMotif hits for experiments 1–10 using descriptors for the ferritin IRE or the SRP listed by organism.
| Organism | Ferritin IRE | SRP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exp. 1 | Exp. 2 | Exp. 3 | Exp. 4 | Exp. 5 | Exp. 6 | Exp. 7 | Exp. 8 | Exp. 9 | Exp. 10 | |
| H.sapiens | 45 | 154 | 154 | 785 | 20 | 136 | 418 | 903 | 903 | 903 |
| S.scrofa | 25 | 122 | 122 | 570 | ||||||
| C.griseus | 15 | 67 | 67 | 260 | ||||||
| G.gallus | 37 | 91 | 91 | 1228 | ||||||
| R.catesbeiana | 9 | 100 | 100 | 142 | ||||||
| X.laevis | 9 | 62 | 62 | 148 | ||||||
| D.melanogaster | 15 | 137 | 137 | 554 | ||||||
| C.porcellus | 128 | |||||||||
| O.nerka | 57 | |||||||||
| C.familiaris | 59 | |||||||||
| D.rerio | 116 | |||||||||
| M.musculus | 71 | |||||||||
| A.fulgidus | 15 | 69 | 200 | 724 | 724 | 724 | ||||
| B.subtilis | 12 | 62 | 374 | 520 | 520 | 520 | ||||
| E.coli | 14 | 45 | 258 | 523 | 523 | 523 | ||||
| M.voltae | 11 | 30 | 121 | 315 | 315 | 315 | ||||
| S.pyogenes | 57295 | |||||||||
| S.aureus | 13591 | |||||||||
| Total | 155 | 733 | 1164 | 3687 | 72 | 342 | 1371 | 2985 | 60280 | 16576 |
The total number of RNAMotif hits is also provided.
Table 2. Results of experiments with IRE and SRP using RNAMotif coupled with an evolutionary search.
| Experiment | Possible bins | P | O | G | Time (min) | FE |
|---|---|---|---|---|---|---|
| 1 | 7.6 × 108 | 40 | 20 | 13 | 3 | 1.4 × 10–5 |
| 2 | 9.7 × 1013 | 40 | 20 | 33 | 6 | 2.7 × 10–19 |
| 3 | 3.5 × 1023 | 100 | 50 | 21 | 66 | 3.0 × 10–19 |
| 4 | 1.7 × 1018 | 100 | 50 | 115 | 180 | 3.4 × 10–13 |
| 5 | 5.5 × 105 | 80 | 40 | 3 | 2 | 1.7 × 10–2 |
| 6 | 7.9 × 108 | 80 | 40 | 7 | 4 | 2.8 × 10–5 |
| 7 | 9.8 × 1011 | 80 | 40 | 27 | 13 | 8.8 × 10–8 |
| 8 | 5.6 × 1013 | 80 | 40 | 27 | 12 | 5.9 × 10–11 |
| 9 | 3.2 × 1018 | 200 | 100 | 25 | 90 | 1.5 × 10–13 |
| 10 | 7.6 × 1017 | 200 | 100 | 13 | 41 | 3.4 × 10–13 |
For each of the eight experiments, the number of possible bins in the search space, the number of parents (P), offspring (O) and generations (G) to the correct solution are provided as well as the time taken to arrive at the correct solution in minutes, and the fraction of the total bin space evaluated (FE) during evolution (the result of equation 24 in Supplementary Material divided by the number of possible bins).
Table 3. Top bin structures in each experiment with the IRE and SRP motifs.
New results (the structures for S.pyogenes and S.aureus) are shown in bold in experiments 9 and 10.
For the second experiment, the descriptor was altered to provide additional variation in the length of the upper stem (Fig. 3B). This resulted in an increased number of hits and possible bin combinations (Table 2). By generation 33, the best bin in the population contained a set of structures identical to the IRE (Table 3). This calculation took 6 min and an even smaller fraction of the possible search space was evaluated, demonstrating the efficiency of the approach. In a third experiment, five additional ferritin mRNA sequences (Cavia porcellus, gi|16416388; Oncorhynchus nerka, gi|12802902; Canis familiaris, gi|15076950; Danio rerio, gi|11545422; Mus musculus, gi|6753911) were added to increase the size of the search space when using the descriptor from experiment 2. The results of this experiment are shown in Table 3. The size of this space was larger than that of experiment 1 by 15 orders of magnitude. A larger population size of 100 parents and 50 offspring was used to converge on the correct solution in 21 generations (1.1 h) (Table 2).
A fourth experiment was designed with a very generic stem–loop descriptor with an optional internal loop (symmetric or asymmetric) on either side of a helical region (Fig. 3C). This descriptor, while capturing both known variants of the IRE element, could represent any short RNA helical motif, and generated a very large search space (1.7 × 1018 bins). Using a population size of 100 parents and 50 offspring, our algorithm converged on the correct IRE structure in 115 generations (<3 h), once again having sampled only a small fraction of the search space (Table 2). The best solution (Table 3) matches the known variant of the ferritin IRE structure, with the exception of the D.melanogaster sequence that does not fit the 3:1 internal loop structure hypothesis. This is consistent with previous reports.
SRP RNA domain IV stem–loop descriptor. The signal recognition particle (SRP) targets signal peptide-containing proteins to plasma membranes (prokaryotes) or the endoplasmic reticulum (eukaryotes) (21–23). The SRP RNA (4.5S RNA in prokaryotes and 7S RNA in eukaryotes) is an essential component of the particle. A key portion of SRP is the domain IV stem–loop, which has been conserved from bacteria to mammals. Domain IV is the binding site for the protein component of the particle (23). Key features of the domain IV stem–loop have been identified. These include two internal loops, a symmetrical loop near the top of the stem and a variable asymmetric loop closer to the base of the stem (10). The helices are of varying length and the loop is typically one of two predominant types, either a tetraloop or hexaloop (Fig. 2C). Previous experimentation demonstrated that a single, highly specific RNAMotif descriptor is capable of finding SRP RNA domain IV structures in a wide range of bacterial genomes (10). Here we tested the hypothesis that a more general descriptor with appropriate fitness functions would be able to find the motif from a much larger search space.
Experiment 5 (Table 1) used five full-length sequences for 4.5S/7S rRNA (Archaeoglobus fulgidus, gi|38795; Bacillus subtilis, gi|216348; Escherichia coli, gi|42758; H.sapiens, gi|177793; Methanococcus voltae, gi|150042) obtained from GenBank. The descriptor shown in Figure 3D was used to screen these sequences for structures using RNAMotif. The resulting number of hits for each organism within the sense strand of these sequences is listed in Table 1. When each bin contains one structure from each of the five organisms and all combinations are allowed, there are 5.5 × 105 possible bins (Table 2). Only three generations of evolution (2.4 min) were required to generate a set of structures that matched the known SRP domain IV structure (Table 3).
For the sixth experiment, the descriptor was modified to allow greater length variation in the stems (Fig. 3E), resulting in a search space of 7.9 × 108 possible bins. A population of 80 parents and 40 offspring arrived at the correct solution in seven generations (4 min) (Table 3). In experiment 7, the internal loops of the descriptor were allowed to have additional length variation (Fig. 3F). With this change, the number of possible bins increased significantly (Table 2); however, evolution arrived at the correct SRP solution (Table 3) in 27 generations (13 min). An eighth experiment adding variability to the hairpin loop (Fig. 3G) further increased the number of hits per organism (5.6 × 1013 possible bins). The known SRP structure was identified in 27 generations (12 min) (Table 3).
To test the utility of the system for discovery, we reviewed GenBank and the SRPDB (24) (http://psyche.uthct.edu/dbs/SRPDB/SRPDB.html) for bacterial genomes that had recently been fully sequenced but did not have their SRP RNA identified. The SRPDB website has a useful alignment for this region for over 100 organisms and was last updated in August 2001. Streptococcus pyogenes M1 (gi|14286347) and Staphylococcus aureus Mu50 (gi|15922990) are two of 33 recently sequenced bacteria not represented in SRPDB. We searched the S.pyogenes genome with the descriptor shown in Figure 3H and generated over 58 000 matches scattered over the entire genome. In order to see if the SRP domain IV stem–loop was represented in this set, we added the hits from this search to the hits used in the previous experiments and used evolutionary computation to search for similarity (experiment 9). A population of 200 parents and 100 offspring was used with one structure for each gi in the RNAMotif output file. After 25 generations (90 min), the best solution contained SRP structures identical to experiment 8 with the addition of a very similar sequence and structure from S.pyogenes (Table 3). We used the genomic coordinates of this hit to extract flanking sequences from the S.pyogenes genomic sequence. This entire region compares favorably to the structure-based alignment of the larger SRP RNA from SRPDB (Fig. 4).
Figure 4.
Alignment of the S.pyogenes and S.aureus SRP RNA domain IV structures discovered, relative to the four closest organisms found in the SRPDB. The top line of the alignment provides information on proposed base pairing for the S.pyogenes structure. The symbols ( and ) are used to denote the 5′ and 3′ sides of the helix, respectively.
Using a similar approach with the S.aureus Mu50 genome (Table 3), we obtained a best-match solution within 13 generations (40 min). This best solution compared favorably with the SRPDB alignment (Fig. 4) but had an atypical tetraloop sequence UGAC instead of GGAA commonly observed in the rest of the bacterial SRPs.
DISCUSSION
We have coupled EC with RNAMotif to discover similar RNA structure motifs over a wide phylogenetic range of organisms. Our hypothesis is that by identifying regions of structural similarity over a number of orthologs, one can find kernels that could lead to the discovery of larger structures through downstream investigation. The current tool described above provides a method for discovering these kernels. By itself, the RNAMotif algorithm can be used to specify RNA secondary structure in the form of a descriptor that contains both sequence and structure context with varying complexity. This approach is more robust than previous motif searching tools and can be used to find structures that match a particular descriptor in a sequence database. However, development of an appropriate descriptor for RNAMotif is typically problem dependent and presumes prior knowledge of the structure, which may or may not be available for a target of interest.
An alternative approach might be to ‘loosen’ the descriptor by allowing for mispairing in helices, variation in length for structural elements, variation in nucleotide sequence, etc. This method gives greater flexibility to the descriptor, allows the possibility for the detection of more distantly related structure elements, but at the same time increases the size of the space that must be searched for similarity. The number of hits can be too large to be searched with exhaustive calculation, but can be efficiently searched with an EA. For the experiments above, no information regarding correct base sequence or structure location was provided to the algorithm and the approach was able, in all cases, to converge on the solution known to be correct even when the descriptors were very generic.
Comparison of results from the IRE (Fig. 3C) and SRP RNA (Fig. 3G) experiments demonstrates that in both cases generic stem–loop descriptors with very little sequence constraints could find the known, correct structural elements in a large set of RNA structures. Further, as demonstrated in SRP experiments 9 and 10, we were able to discover SRP RNA-containing regions in newly sequenced genomes without prior knowledge of the SRP sequences specific to these genomes. We validated our findings by matching a larger region flanking our motif hits with the global features of the SRP RNA seen in the SRPDB.
As we were finishing our analysis, a method for predicting SRP RNA was published (25) and included the same results from the S.pyogenes and S.aureus genomes. In addition, they also showed the presence of the unusual UGAC tetraloop in the Lactococcus lactis genome. We verified this using our technique (data not shown). It is noteworthy that in order for Regalia et al. (25) to find these atypical SRPs, a number of additional bioinformatic steps had to be undertaken, such as BLAST or FASTA searches with closely related organism genomes, as well as modification of their original search to specifically include the tetraloop nucleotide sequences. Further, their technique depended on using a specific search on the target organism class, archaea, eubacteria, plant, yeast and metazoans. We have previously shown similar results using RNAMotif (10), where, with the exception of Buchnera sp., we could identify SRPs in all branches of life with a single descriptor. The current method overcomes all of the limitations of these specific motif-based descriptors that require some prior knowledge of expected structures in the target genome, and allows the overall similarity of the structures in the search results to identify novel variants of structural motifs such as those described above. Further, the methods described here using EAs can search very large spaces (∼1023 structures) in a fraction of the time it would require to search this space exhaustively, making this a convenient method for motif discovery.
Comparative analysis has been used extensively in determining RNA structure (26–28). This is a very reliable method for inferring RNA secondary structure but requires multiple sequences and alignment. Unfortunately, for many molecules (especially mRNAs), very few related sequences are known. Previous computational approaches for the task of discovering common stem–loop structures have relied on software that first builds an alignment to then build a model representing the sequences and structures found in the alignment (29). Here, we present a method to efficiently discover common structures in even just a few homologous sequences without the requirement for sequence alignment. The utility of this method does increase with an increasing number of orthologous sequences in a manner similar to comparative sequence analysis.
Many measures of RNA structure similarity can easily be incorporated into the fitness function for further refinement (30,31). For instance, energy minimization can be used to discover potential lowest energy structures for a single sequence (31). Previous methods using EC for RNA structure folding (32–42) and for discovering common structures (43) used free energy calculations to define RNA structure. Rather than exclusively relying on thermodynamic calculations (efn), we use this as one of many optional metrics that can be computed in arriving at the final solution. In all the examples shown here, we did not use any thermodynamic calculations in the fitness function, and showed that successful results can still be achieved. In the rare case where two solutions in the final generation might share the same fitness value, efn could be used to resolve these ties. Information regarding relative motif position can also be used as a measure of similarity. Following evolution, similar hits are mapped back to true coordinates based on the original sequence. Should the structures lie in similar locations with respect to a gene of interest, the results are considered more valuable. Position similarity can also be included as an additional fitness function term during evolution.
Taking a broad view, one might consider generating a space of all possible stem structures for each of a number of orthologous mRNA sequences, and then comparing folded structures from this space to arrive at the set of structures that is most similar across all species. An exhaustive search through this stem space may be plausible if the sequence lengths for each mRNA and/or number of orthologous sequences are small. Algorithms have been developed previously to generate all potential helices of a specific length given a sequence of RNA and base pairing rules (44,45). However, it is known that the number of potential RNA structures in a single sequence of n nucleotides increases on the order of 2n (46). Therefore, an exhaustive search of this space for similar helices across two or more mRNAs of any biological relevance is likely to be computationally intractable. We have initiated a series of experiments to examine the potential of EC for searching these spaces. Such an approach obviates the requirement for RNAMotif but increases the complexity of the search space dramatically.
PROGRAM AVAILABILITY
Please contact the authors for information regarding program implementation and/or motif searches using the code.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Harold Levene, Dr Robin Gutell, Dr Daniel Gautheret, Tim Henderson, Tom Macke and the reviewers for their valuable input. The ideas for this application were formalized in the spring of 1999 at the 2nd RNA Summit held at Ibis Therapeutics, Carlsbad, CA.
REFERENCES
- 1.Theil E.C. (1998) The iron responsive element (IRE) family of mRNA regulators. Regulation of iron transport and uptake compared in animals, plants and microorganisms. Met. Ions Biol. Syst., 35, 403–434. [PubMed] [Google Scholar]
- 2.Kim H.Y., LaVaute,T., Iwai,K., Klausner,R.D. and Rouault,T.A. (1996) Identification of a conserved and functional iron-responsive element in the 5′-untranslated region of mammalian mitochondrial aconitase. J. Biol. Chem., 271, 24226–24230. [DOI] [PubMed] [Google Scholar]
- 3.Son S.Y. (1993) The structure and regulation of histone genes. Saenghwahak Nyusu, 13, 64–70. [Google Scholar]
- 4.Zehner Z.E., Shepherd,R.K., Gabryszuk,J., Fu,T.-F., Al-Ali,M. and Holmes,W.M. (1997) RNA–protein interactions within the 3′ untranslated region of vimentin mRNA. Nucleic Acids Res., 25, 3362–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le S.Y., Siddiqui,A. and Maizel,J.V.,Jr (1996) A common structural core in the internal ribosome entry sites of picornavirus, hepatitis C virus and pestivirus. Virus Genes, 12, 135–147. [DOI] [PubMed] [Google Scholar]
- 6.Gautheret D., Major,F. and Cedergren,R. (1990) Pattern searching/alignment with RNA primary and secondary structures: an effective descriptor for tRNA. Comput. Appl. Biosci., 6, 325–331. [DOI] [PubMed] [Google Scholar]
- 7.Laferriere A., Gautheret,D. and Cedergren,R. (1994) An RNA pattern matching program with enhanced performance and portability. Comput. Appl. Biosci., 10, 211–212. [DOI] [PubMed] [Google Scholar]
- 8.Billoud B., Kontic,M. and Viari,A. (1996) Palingol: a declarative programming language to describe nucleic acids’ secondary structures and to scan sequence database. Nucleic Acids Res., 24, 1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pesole G., Liuni,S. and D’Souza,M. (2000) PatSearch: a pattern matcher software that finds functional elements in nucleotide and protein sequences and assesses their statistical significance. Bioinformatics, 16, 439–450. [DOI] [PubMed] [Google Scholar]
- 10.Macke T.J., Ecker,D.J., Gutell,R.R., Gautheret,D., Case,D.A. and Sampath,R. (2001) RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res., 29, 4724–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fogel D.B. (ed.) (1998) Evolutionary Computation: The Fossil Record. IEEE Press, Piscataway, NJ.
- 12.Fogel D.B. (2000) Evolutionary Computation: Toward a New Philosophy of Machine Intelligence, 2nd Edn. IEEE Press, Piscataway, NJ.
- 13.Fogel G.B. (1997) The application of evolutionary computation to selected problems in molecular biology. In Angeline,P.J., Reynolds,R.G., McDonnell,J.R. and Eberhart,R. (eds), Evolutionary Programming VI: Sixth International Conference, EP97. Springer-Verlag, Berlin, Germany, pp. 23–33.
- 14.Matthews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence on thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 15.Myers E.W. and Miller,W. (1988) Optimal alignments in linear space. Comput. Appl. Biosci., 4, 11–17. [DOI] [PubMed] [Google Scholar]
- 16.Ke Y., Sierzputowska-Gracz,H., Gdaniec,Z. and Theil,E.C. (2000) Internal loop/bulge and hairpin loop of the iron-responsive element of ferritin mRNA contribute to maximal iron regulatory protein 2 binding and translational regulation in the iso-iron-responsive element/iso-iron regulatory protein family. Biochemistry, 39, 6235–6242. [DOI] [PubMed] [Google Scholar]
- 17.McKie A.T., Marciani,P., Rolfs,A., Brennan,K., Wehr,K., Barrow,D., Miret,S., Bomford,A., Peters,T.J., Farzaneh,F. et al. (2000) A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol. Cell, 5, 299–309. [DOI] [PubMed] [Google Scholar]
- 18.Thomson A.M., Rogers,J.T. and Leedman,P.J. (1999) Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int. J. Biochem. Cell Biol., 31, 1139–1152. [DOI] [PubMed] [Google Scholar]
- 19.Schlegl J., Gegout,V., Schlager,B., Hentze,M.W., Westhof,E., Ehresmann,C., Ehresmann,B. and Romby,P. (1997) Probing the structure of the regulatory region of human transferrin receptor messenger RNA and its interaction with iron regulatory protein-1. RNA, 3, 1159–1172. [PMC free article] [PubMed] [Google Scholar]
- 20.Gdaniec Z., Sierzputowska-Gracz,H. and Theil,E.C. (1998) Iron regulatory element and internal loop/bulge structure for ferritin mRNA studied by cobalt(III) hexamine binding, molecular modeling and NMR spectroscopy. Biochemistry, 37, 1505–1512. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz U., Behrens,S., Freymann,D.M., Keenan,R.J., Lukavsky,P., Walter,P. and James,T.L. (1999) Structure of the phylogenetically most conserved domain of SRP RNA. RNA, 5, 1419–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz U., James,T.L., Lukavsky,P. and Walter,P. (1999) Structure of the most conserved internal loop in SRP RNA. Nature Struct. Biol., 6, 634–638. [DOI] [PubMed] [Google Scholar]
- 23.Batey R.T., Rambo,R.P., Lucast,L., Rha,B. and Doudna,J.A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science, 287, 1232–1239. [DOI] [PubMed] [Google Scholar]
- 24.Gorodkin J., Knudsen,B., Zwieb,C. and Samuelson,T. (2001) SRPDB (Signal recognition particle database). Nucleic Acids Res., 29, 169–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regalia M., Rosenblad,M.A. and Samuelson,T. (2002) Prediction of signal recognition particle RNA genes. Nucleic Acids Res., 30, 3368–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutell R.R., Cannone,J.J., Shang,Z., Du,Y. and Serra,M.J. (2000) A story: unpaired adenosine bases in ribosomal RNAs. J. Mol. Biol., 304, 335–354. [DOI] [PubMed] [Google Scholar]
- 27.Gutell R.R., Cannone,J.J., Konings,D. and Gautheret,D. (2000) Predicting U-turns in ribosomal RNA with comparative sequence analysis. J. Mol. Biol., 300, 791–803. [DOI] [PubMed] [Google Scholar]
- 28.Cannone J.J., Subashchandran,S., Schnare,M.N., Collett,J., D’Souza,L.M., Du,Y., Feng,B., Lin,N., Madabusi,L.V., Muller,K.M. et al. (2002) The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron and other RNAs. BMC Bioinformatics, 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins G.D., Le,S. and Zhang,K. (2001) A new algorithm for computing similarity between RNA structures. Inf. Sci., 139, 59–77. [Google Scholar]
- 30.Gorodkin J., Sticklin,S.L. and Stormo,G.D. (2001) Discovering common stem–loop motifs in unaligned RNA sequences. Nucleic Acids Res., 29, 2135–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 32.Van Batenburg F.H.D., Gultyaev,A.P. and Pleij,C.W.A. (1995) An APL-programmed genetic algorithm for the prediction of RNA secondary structure. J. Theor. Biol., 174, 269–280. [DOI] [PubMed] [Google Scholar]
- 33.Gultyaev A.P., van Batenburg,F.H.D. and Pleij,C.W.A. (1995) The computer simulation of RNA folding pathways using a genetic algorithm. J. Mol. Biol., 250, 37–51. [DOI] [PubMed] [Google Scholar]
- 34.Gultyaev A.P., van Batenburg,F.H.D. and Pleij,C.W.A. (1998) Dynamic competition between alternative structures in viroid RNAs simulated by an RNA folding algorithm. J. Mol. Biol., 276, 43–55. [DOI] [PubMed] [Google Scholar]
- 35.Benedetti G. and Morosetti,S. (1995) A genetic algorithm to search for optimal and suboptimal RNA secondary structures. Biophys. Chem., 55, 253–259. [DOI] [PubMed] [Google Scholar]
- 36.Shapiro B.A., Bengali,D., Kasprzak,W. and Wu,J.C. (2001) RNA folding pathway functional intermediates: their prediction and analysis. J. Mol. Biol., 312, 27–44. [DOI] [PubMed] [Google Scholar]
- 37.Shapiro B.A. and Navetta,J. (1994) A massively parallel genetic algorithm for RNA secondary structure prediction. J. Supercomput., 8, 195–207. [Google Scholar]
- 38.Shapiro B.A. and Wu,J.C. (1996) An annealing mutation operator in the genetic algorithm for RNA folding. Comput. Appl. Biosci., 12, 171–180. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro B.A., Wu,J.C., Bengali,D. and Potts,M.J. (2001) The massively parallel genetic algorithm for RNA folding: MIMD implementation and population variation. Bioinformatics, 17, 137–148. [DOI] [PubMed] [Google Scholar]
- 40.Wu J.C. and Shapiro,B.A. (1999) A Boltzmann filter improves the prediction of RNA folding pathways in a massively parallel genetic algorithm. J. Biol. Struct. Dyn., 17, 581–595. [DOI] [PubMed] [Google Scholar]
- 41.Shapiro B.A., Bengali,D., Kasprzak,W. and Wu,J.C. (2001) Computational insights into RNA folding pathways: getting from here to there. In Proceedings of the Atlantic Symposium on Computational Biology and Genome Information Systems and Technology. ACM Inc., New York, NY, pp. 10–13.
- 42.Shapiro B.A. and Wu,J.C. (1997) Predicting RNA H-type pseudoknots with the massively parallel genetic algorithm. Comput. Appl. Biosci., 13, 459–471. [DOI] [PubMed] [Google Scholar]
- 43.Chen J.-H., Le,S.-Y. and Maizel,J.V. (2000) Prediction of common secondary structures of RNAs: a genetic algorithm approach. Nucleic Acids Res., 28, 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McMahon J.E. (1975) Computer method for predicting RNA secondary structure, PhD dissertation, Florida State University. [DOI] [PMC free article] [PubMed]
- 45.Pipas J.M. and McMahon,J.E. (1975) Method for predicting RNA secondary structure. Proc. Natl Acad. Sci. USA, 72, 2017–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waterman M.S. (1995) Introduction to Computational Biology. Maps, Sequences and Genomes. Chapman & Hall, London, UK.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.