Abstract
We have developed a software analysis package, HapScope, which includes a comprehensive analysis pipeline and a sophisticated visualization tool for analyzing functionally annotated haplotypes. The HapScope analysis pipeline supports: (i) computational haplotype construction with an expectation-maximization or Bayesian statistical algorithm; (ii) SNP classification by protein coding change, homology to model organisms or putative regulatory regions; and (iii) minimum SNP subset selection by either a Brute Force Algorithm or a Greedy Partition Algorithm. The HapScope viewer displays genomic structure with haplotype information in an integrated environment, providing eight alternative views for assessing genetic and functional correlation. It has a user-friendly interface for: (i) haplotype block visualization; (ii) SNP subset selection; (iii) haplotype consolidation with subset SNP markers; (iv) incorporation of both experimentally determined haplotypes and computational results; and (v) data export for additional analysis. Comparison of haplotypes constructed by the statistical algorithms with those determined experimentally shows variation in haplotype prediction accuracies in genomic regions with different levels of nucleotide diversity. We have applied HapScope in analyzing haplotypes for candidate genes and genomic regions with extensive SNP and genotype data. We envision that the systematic approach of integrating functional genomic analysis with population haplotypes, supported by HapScope, will greatly facilitate current genetic disease research.
INTRODUCTION
Advances in the understanding of the biological mechanisms of complex diseases require a knowledge of human population history as well as of gene function. A classic example is the parallel study in genetics and immunochemistry in which the association between ApoE4 haplotype and Alzheimer’s disease was discovered. Extensive association studies revealed a higher frequency of ApoE4 haplotypes in Alzheimer patients compared with the control population (1), and functional assays showed that ApoE plays a critical role in the pathogenesis of the lesions of Alzheimer’s disease (2).
Recent progress in human genomics and genetics research, highlighted by the completion of the human genome draft sequence (3,4) and the ongoing effort to develop a whole genome, high density haplotype map (5–9), has resulted in a wealth of information that can be employed to identify correlations between population genetics and functional genomics in a systematic approach. Improvements in statistical methods for constructing haplotypes with genotyping data (10–12) have the potential to significantly reduce the cost of haplotype mapping without compromising accuracy. In addition, computational tools for predicting deleterious effects of genetic variations (13) may reveal the significance of DNA variations on mRNA and protein expression and protein structure and function. Assembling and interpreting the data from the public domain and research laboratories in candidate regions or candidate genes can be difficult and challenging owing to the complex processes required for genetic and genomic data gathering, computational analysis and results integration. Many of the recently developed tools lack follow-up studies that analyze their relative strengths and weaknesses, which makes it difficult to apply the appropriate method to specific research and to interpret the results accurately. Furthermore, evaluation of the correlation between genetic variation and its potential functional impact requires parallel presentation of genetic and genomic data. Existing genome viewers, such as NCBI Map Viewer (http://www.ncbi.nlm.nih.gov) and the UCSC Genome Viewer (14) as well as static genotype or haplotype viewers such as Visual Genotype or Visual Haplotype (15,16) are capable of presenting donor genotype or haplotype information but lack more sophisticated features, such as SNP functional classification, haplotype phase probability assessment, display of population haplotype structure and haplotype blocks, and SNP subset selection, that are required to determine the functional significance of genetic variations and plan for follow-up genetic or functional experiments.
As a first step towards providing a systematic approach for establishing genetic and functional correlation, we have developed HapScope, a software tool for analyzing the genetic and functional correlation of haplotypes in a population of interest. Our system, which includes an automated analysis pipeline and a sophisticated visualization tool, is the first to combine genomic analysis and population haplotype analysis in an integrated environment. To enable users of the HapScope system to select the most appropriate algorithm for computational analysis of haplotypes, we have analyzed the performance and accuracy of two popular haplotype construction algorithms, PHASE (10) and SNPHAP (http://www-gene.cimr.cam.ac.uk/clayton/software/), the former based on Bayesian statistics and the latter based on the expectation-maximization (EM) algorithm. We have compared the computationally constructed haplotypes with those experimentally derived from allele-specific PCR in genomic regions encoding the ApoE (17) and LPL (18) genes, representing genomic regions with low and high nucleotide diversity, respectively. We have developed a Brute Force Algorithm (BFA) and a Greedy Partition Algorithm (GPA) for selection of the minimum (or near minimum in the case of the GPA) subset of SNPs that is able to represent the haplotype diversity observed in the population of interest, thus reducing the cost of genotyping without compromising the power of linkage disequilibrium (LD) mapping with haplotypes. The HapScope viewer is the first visualization tool to display haplotype data in parallel with genomic information such as coding region structure, repeats, SNPs, putative regulatory regions and conserved regions in model organisms. This will assist evaluation of potential susceptibility markers from both genetic and genomic perspectives. The software package was developed and tested with the genotype data for F2, ApoE and LPL, three candidate genes for cardiovascular disease, and for 5q31, a genomic region with susceptibility to Crohn’s disease (9). It has been successfully applied in a genotyping project (J.P.Struewing, unpublished data) to identify and assess SNPs in the BRIP1 (19) and ZBRK1 (20) genes in breast cancer studies.
MATERIALS AND METHODS
Sequence, SNP, genotype and haplotype data
We obtained genomic sequence, SNPs and genotype data for the F2 gene from the website of the UW-FHCRC Variation Discovery Resource (http://pga.mbt.washington.edu). The data set includes 103 SNPs discovered in 48 individuals. To analyze the success rate of haplotype construction algorithms and to test and validate the HapScope analysis pipeline, we obtained LPL and ApoE haplotype data (17,18) from Andy Clark and Charlie Sing. The LPL and ApoE haplotypes were originally determined by an iterative procedure involving application of Clark’s haplotype inference algorithm (21) and allele-specific PCR sequencing of multiple heterozygous individuals. The ApoE data set includes 23 SNPs discovered in 96 individuals; haplotype phases were determined experimentally for 22 SNPs. The LPL data set includes a total of 88 SNPs discovered in 71 individuals; 69 SNPs have experimentally determined haplotype phases. The SNP locations and their flanking sequence context for ApoE and LPL were derived from GenBank accession nos AF050163 and AF261279, respectively. To compare the effectiveness of our minimum SNP selection algorithms with manual htSNP selection (7), we retrieved haplotype data for eight genes (CTLA4, CASP10, CASP8, CFLAR, H19, INS, SDF1 and TCF8) from the original publication. The genomic sequence, SNPs, genotype data and haplotype block structure for 5q31 were obtained from the Whitehead Institute (http://www.genome.wi.mit.edu/humgen/IBD5/haplodata.html). The BRIP1 and ZBRK1 SNPs and genotypes were obtained from one of the authors (J.P.Struewing, unpublished data).
External haplotype construction programs
Two programs were used to construct haplotypes: SNPHAP and PHASE. SNPHAP was developed by David Clayton (http://www-gene.cimr.cam.ac.uk/clayton/software/) and uses the EM algorithm. PHASE was developed by Stephens et al. (10) and is based on Bayesian statistics. Both versions of the PHASE program, PHASE.big and PHASE.small, were incorporated into the pipeline.
Minimum SNP set selection algorithms
The minimum SNP set is the smallest possible subset of SNPs in a haplotype block that represents the diversity of haplotypes within the block; such SNPs have also been referred to as ‘haplotype tagging SNPs’ or ‘htSNPs’ (7). HapScope offers two minimum SNP set algorithms: a BFA that always finds the minimum SNP set and a GPA that finds either the minimum SNP set or one close to it in size.
The BFA iteratively generates all possible SNP combinations (i.e. putative minimum SNP sets) starting with the smallest possible until it finds a set of SNPs that represents all haplotype diversity within the haplotype block.
The GPA successively partitions the minimum SNP set discovery problem, finding a solution for the largest partition at each iteration. By combining intermediate solutions, a new set of smaller partitions is generated by each iteration. Eventually, all partitions are of size one, indicating that a minimum SNP set (or one close to it) has been found and the algorithm ends (an algorithm that, at each iteration, solves the biggest available sub-problem is referred to as a ‘greedy’ algorithm, hence the name).
Evaluation of haplotype construction algorithms
Donor haplotypes constructed with the PHASE.big, PHASE.small and SNPHAP programs were compared with those obtained experimentally from allele-specific PCR experiments for ApoE and LPL (17,18). The default parameters were used for all three programs. The programs were run on a Sparc II station with a 400 MHz CPU and 3.5 GB of RAM. The operating system was Solaris 2.6.
In the original allele-specific PCR experiments, haplotype phases for SNPs with multiple alleles and those identified in only one or two chromosomes were not determined experimentally. For this reason, 19 SNPs in LPL and one SNP in ApoE were excluded in our analysis of haplotype construction algorithms. We analyzed the accuracy of haplotype prediction by pre-filtering these SNPs from the input or post-filtering them from the output. In the post-filtering analysis, the multi-allele indel SNP at position 4827 of LPL was converted into a microsatellite marker in the input to PHASE. Since SNPHAP can handle only bi-allelic variations, alleles other than the two most frequent ones for this marker were treated as missing data in the input to SNPHAP. For each donor subject, we selected the matching haplotype pairs that represent minimum haplotype phase differences between computational prediction and experimental assay. We determined the proportion of subjects whose computationally constructed haplotypes are identical to experimental results to measure the accuracy of haplotype prediction. To take into account the differences in SNP density and nucleotide diversity in genomic regions encoding the ApoE and LPL genes, we also tabulated the proportion of individual heterozygous genotypes with correct phase assignment. Missing genotype data are excluded in all analyses.
A useful feature of the PHASE algorithm is its assignment of a probability score to the phase determination for each heterozygous genotype. To assess the accuracy of this probability value we compared the observed phase error rate with the predicted error rate. For each predicted error rate (= 1 – probability of phase being correct), the estimated number of incorrect calls and the actual number were tabulated. We plotted the percentage of all predicted and the percentage of all observed incorrect phase assignments in a moving window of size eight. For example, in ApoE, PHASE assigned an error probability in the range 41–50% to 20 heterozygous genotypes; the sum of its estimated inaccurate phase calls in this range was 9.08, giving an estimated incorrect call percentage of 45.40%. The percentage of observed inaccurate calls for the same range was 45.00%. Thus 45.50 and 45.00 are the first two data points on the chart. The window was then moved one position and new values of the percentages were computed. This proceeded until the entire table was covered. Error probability = 0 was plotted in its own interval.
Programming language, platform and availability
The HapScope analysis pipeline was written in C and Perl. Currently it runs on Solaris and can be re-compiled for other UNIX platforms, such as SGI and Sun. The HapScope viewer was written in C using NCBI’s Vibrant Software Toolkit. It runs on Windows 2000 and Windows NT as well as the UNIX platform.
The HapScope analysis pipeline, the viewer and the test data set are available free of charge and can be obtained by anonymous ftp (ftp://ftp1.nci.nih.gov/pub/HapScope).
RESULTS
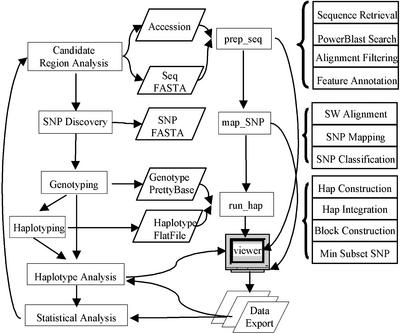
As shown in Figure 1, HapScope supports the iterative process of target region analysis, SNP discovery, genotyping, haplotype analysis and statistical analysis in genetics studies using SNPs or haplotypes. The analysis pipeline provides tools for both genomic and haplotype analysis and the viewer provides a graphical interface for expert review of the computational results. The analysis presented here of the accuracy of the computational haplotype algorithms used by the system will assist the user in selecting the method that is most appropriate to the genomic region of interest. The minimum SNP selection algorithms ensure the identification of the minimum or near minimum number of SNPs that are required to represent haplotype diversity in the population of interest within a user-defined frequency range.
Figure 1.
System design of the HapScope analysis pipeline. Rectangles indicate computational or experimental processes, parallelograms represent data files. The arrows indicate the process flow as well as input or output data generated from a process.
Analysis pipeline
The analysis pipeline is a flatfile-based system that consists of three modules, prep_seq for reference sequence annotation, map_SNP for SNP mapping and classification, and run_hap for haplotype construction and integration. The three modules can be run independently, in serial order or in a pipeline process, which enables the user to substitute results from computational predictions with experimental data and to perform manual data editing where applicable. Results from each of the modules can be viewed and edited with the HapScope viewer.
As outlined in Figure 1, the prep_seq module can perform de novo sequence annotation on raw sequence data or download an annotated sequence record from NCBI (http://www.ncbi.nlm.nih.gov) or combine de novo analysis with annotations on a GenBank record. The user can define a region of interest by specifying the start and stop positions and orientation of the region on the original record. For example, we used the sub-sequence between 790 and 1020 kb of NCBI’s contig NT_031907 to represent the genomic region encoding the BRIP1 gene. The sub-sequence was derived in reverse orientation to contig NT_031907 so that SNPs and haplotypes are presented in the same orientation as the coding sequence. A repeat database file and a collection of public or local databases can be specified for repeat masking and homology search using the program powerblast (23). Features such as coding regions, mRNAs, homologous regions to related organisms and repetitive regions are annotated automatically based on the database search results. Prep_seq was able to generate accurate coding region structures for the five genes analyzed in the study. For example, it was able to duplicate the 20 exon coding region structure for BRIP1 that was originally generated by manual analysis. Prep_seq can also be run as a stand-alone process to identify target regions such as coding or regulatory regions for SNP discovery projects.
SNPs of interest are discovered either by laboratory experiments or by mining the public databases. The map_SNP module first aligns the sequences representing the 5′ and 3′ flanking regions of SNPs to the reference sequence using a modified version of the sim (23) program. The SNPs are mapped to the reference sequence using these alignments and are subsequently classified into UTR, silent, missense, nonsense and splice site variations using the coding region and mRNA annotations on the reference sequence record. Starting with raw sequence data, prep_seq and map_SNP are able to accurately reproduce the SNP classification for ApoE and LPL in the original publication.
The run_hap module takes genotype data in Prettybase format and converts them into the input data files required for the PHASE or SNPHAP program. The computationally constructed haplotypes are stored as alignment data in the reference sequence record. Haplotype block information can be incorporated with user-supplied haplotype block boundaries. The minimum SNP subset required to represent haplotype diversity for all haplotypes (or a subset of haplotypes within a user-defined population frequency range) can be determined using the GPA or the BFA.
HapScope viewer
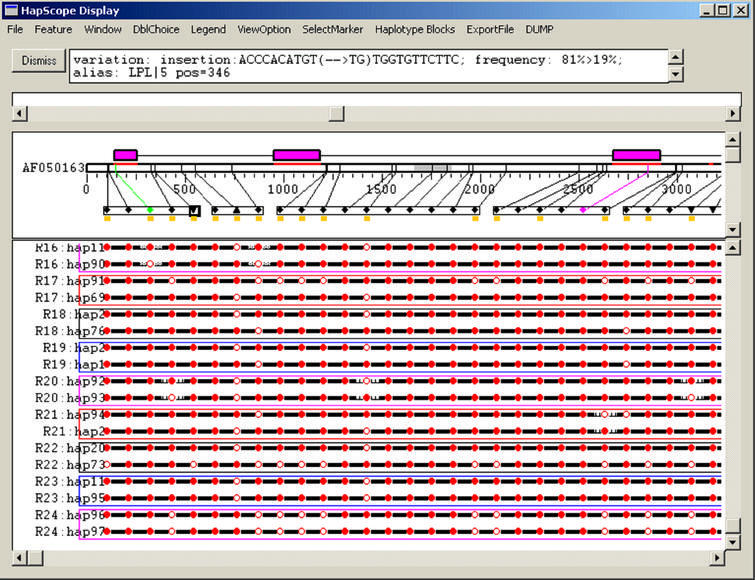
Figure 2 is a screenshot of the HapScope viewer displaying the LPL haplotypes computed by the PHASE program for 71 donors. The viewer divides the display window horizontally into two panels. The top panel is a text box containing a description of a user-selected object, which can be a SNP, a coding region, an mRNA transcript, a repeat or a region with conserved sequence homology across multiple species. In Figure 2, a 2 bp insertion SNP has been selected by mouse click and the top panel displays data associated with the SNP, including allelic variations with 10 bp flanking sequences, allele frequencies in the population, SNP position on the reference sequence and SNP type such as substitution, insertion or deletion, as well as user-supplied synonyms of the SNP. The bottom panel displays an integrated graphic view of genomic and genetic data. At the top of this panel, features annotated on the reference sequence are laid out in a compressed format to preserve the majority of the display space for genotype or haplotype data. The display panel can be split horizontally into two sub-panels with independent vertical scrolling so the user can navigate haplotype data at the bottom without losing connection to the genomic annotation displayed at the top. Features related to a SNP object are rendered graphically in detail. Different display symbols are used to differentiate substitution, insertion and deletion SNPs and different color schemes are applied to represent silent, missense and nonsense SNPs. SNPs belonging to the same haplotype block, determined either by an external process or manually constructed with the HapScope viewer interface, are grouped into a rectangle and subset SNPs from the user query are highlighted. For example, in Figure 2 SNPs with ≥5% minor allele frequency are highlighted in yellow; in Figure 3 the minimum SNP index for each haplotype block is highlighted
Figure 2.
A screen shot of the standard haplotype view for computed LPL haplotypes with the split display panel. The top panel displays the genome annotation computed with the prep_seq module. At the top, the magenta rectangles are used to represent the coding exons. The gray area inside the sequence represents the repeat region and the red lines on top of the sequence represent human–mouse conserved regions. The substitution SNPs are displayed as diamonds; the deletions and insertions are displayed as triangles or inverted triangles. Red, magenta or green are used to represent nonsense, missense and silent SNPs. Yellow squares at the bottom of this panel highlight SNPs selected by a user query (all SNPs with ≥5% allele frequency in this case). Donor haplotypes are displayed at the bottom panel. Major alleles are shown with filled red circles; minor alleles are shown with filled white circles; missing genotypes are not marked with circles. Gray and vertical hatched lines on either side of the circles present PHASE probability scores in the ranges 75–99% and 50–47%, while solid black lines indicate 100% probability.
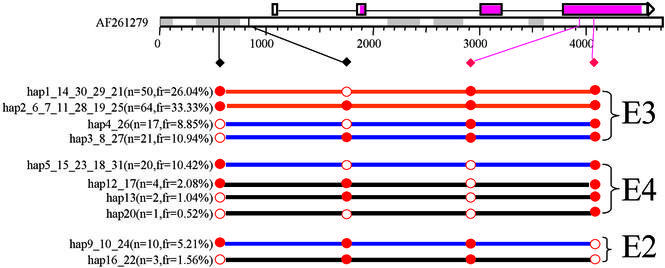
Figure 3.
Haplotype block view of the 5q31 region with haplotypes sorted in descending order of haplotype frequency within each block. The red, blue and black strings connecting the circles represent haplotypes with ≥15%, ≥5% and ≥1% population frequency.
Haplotype and/or genotype data are displayed in a ‘beads on a string’ fashion. The color of the beads indicates the major or minor allele for haplotype data, homozygous or heterozygous genotype for genotype data. The color of the strings illustrates population frequency of a haplotype or a haplotype in a block (Fig. 3). The probability score of haplotype phase determination is displayed with a gray or hatched line when the zoom feature is used to expand the image (Fig. 2). The user has the option of selecting from the eight alternative graphic views for visualizing haplotype or genotype data. With the exception of the male sex chromosomes, the standard view uses double strings to display the two haplotypes for each donor (Fig. 2). The genotype view uses a single string to display major allele homozygous, minor allele homozygous and heterozygous genotypes in a donor. The haplotype frequency view displays the haplotypes in descending order of population frequency (Fig. 3). The haplotype similarity view displays the haplotypes by their similarity to the most common haplotype. If haplotype blocks are defined by the user, the viewer can also display the haplotype frequency or haplotype similarity view for SNPs within each block (Fig. 3). If a selected subset of SNPs is displayed, the viewer has the option of consolidating identical haplotypes and recalculating population frequency. Figure 4 shows a consolidated view for molecularly determined haplotypes for ApoE with SNPs selected from the promoter and the coding region. Four SNPs are selected and as a result the original 31 haplotypes have been consolidated into 10.
Figure 4.
A consolidated haplotype view for ApoE haplotypes constructed with two promoter SNPs and two coding SNPs. The labels for haplotype name, count and frequency have been recomputed after haplotype consolidation.
In addition to its visualization features, the HapScope viewer also provides a variety of analysis functions, including SNP subset selection, manual construction of haplotype blocks and data export. A user can select a subset of SNPs by specifying one or more functional classifications, such as silent, missense and nonsense, by defining a range of minor allele frequency, by defining the threshold for missing genotype data, by requesting the minimum SNP subset that represents haplotype diversity or by manually selecting SNPs with a mouse click. Haplotype blocks can be manually built by selecting SNPs representing the start and/or stop positions of the blocks. Selected SNPs can be exported in FASTA format for primer design. Genotype and haplotype data files can be exported for additional statistical analysis. HapScope supports haplotype clade analysis by exporting the data in the format required for the RM network software (http://www.fluxus-engineering.com). HapScope can export flatfiles representing genomic, mRNA and protein haplotypes, which can be used to investigate potential deleterious effects on coding regions structure, mRNA expression and protein functions. Figure 5 shows the protein haplotype export file listing the most common ApoE3 haplotype as well as a rare variant of the ApoE4 haplotype with a Leu28Pro change that has been previously reported to be more acidic than the common E4 isoform (24).
Figure 5.
ApoE protein haplotype file exported by the HapScope viewer. Upper case characters indicate protein variations. Underlined characters are amino acid residues that define the ApoE3 and ApoE4 isoforms. The bold and underlined format does not appear in the exported file; it is used here for illustration purposes.
Minimum SNP set selection
To compare the effectiveness of GPA and BFA in manual htSNP identification (7), we applied both algorithms to the eight genes for the subset of haplotypes at or above the 5% frequency level, the same threshold used in the publication. Results from both algorithms agreed with each other in every case; minimum SNP set size agreed with the size of the manually derived htSNP set given in the paper, except for genes CASP10 and SDF1, where GPA and BFA found minimum SNP sets smaller by one SNP in each case.
A more detailed test was carried out against molecularly determined haplotypes for LPL. The results obtained from haplotypes filtered with four population frequency thresholds are summarized in Table 1. In all cases, GPA generates a minimum SNP set of the same size as BFA. In an additional test, unfiltered haplotypes from the htSNP publication (7) were run through both algorithms and again the minimum SNP set sizes were the same.
Table 1. Comparison of the GPA and BFA using LPL haplotypesa.
| Haplotype frequency (%) | Data | GPA | BFA | |||
|---|---|---|---|---|---|---|
| No. of haplotypes | No. of unique SNPsb | Size of minimum index | Run time (s) | Size of minimum index | Run time (s) | |
| All | 88 | 59 | 22 | 103 | ?c | ?c |
| >1 | 8 | 11 | 5 | 0.01 | 5 | 0.01 |
| >2 | 6 | 7 | 4 | <0.01 | 4 | <0.01 |
| >3 | 5 | 4 | 3 | <0.01 | 3 | <0.01 |
aThe input data included 88 haplotypes and 69 SNPs from Clark et al. (18).
bThe value in this column represents the number of SNPs that are polymorphic after: (i) removal of haplotypes with population frequencies lower than the threshold in the first column; (ii) consolidation of SNPs in LD. For example, the original 69 SNPs for all 88 haplotypes are consolidated into 59 unique SNPs.
cWe were unable to obtain the minimum SNP index for all haplotypes using BFA since the computing time exceeded the time required to prepare the manuscript.
For small inputs with fewer than 20 SNPs, the run times of the algorithms were approximately the same. For large inputs with more than 40 SNPs, the run time of the BFA was measured in days, versus minutes for the GPA.
Evaluation of computational haplotypes
The results of the three haplotype construction programs, PHASE.big, PHASE.small and SNPHAP, are summarized in Table 2. Pre-filtering low frequency SNPs produced a minor improvement in the accuracy of haplotype prediction (from 1 to 7%) in all programs. As a result, we implemented an option for pre-filtering input data with a user-defined SNP allele frequency in run_hap.
Table 2. Comparison of the PHASE.big, PHASE.small and SNPHAP programs using phase known data sets.
| Gene name | ApoE | LPL | |||||
| Target region (kb) | 5.5 | 9.7 | |||||
| No. of subjects | 96 | 71 | |||||
| No. of SNPs | 22 | 69 | |||||
| No. of heterozygous genotypes | 283 | 1154 | |||||
| Average no. of heterozygous genotypes per subject | 3 | 16 | |||||
| Nucleotide diversitya | 0.0005 ± 0.0003 | 0.002 ± 0.001 | |||||
| Algorithm | PHASE.big | PHASE.small | SNPHAP | PHASE.big | PHASE.small | SNPHAP | |
| Run time | Pre-filterb | 2 h | 6 h | 1 min | 3 h | 9 days | 90 s |
| Post-filterc | 2 h | 6 h | 1 min | 3 h | 7 days | 1 min | |
| Accuracy of full haplotyped | Pre-filter | 74 (77%) | 74 (77%) | 79 (82%) | 37 (52%) | 38 (53%) | 30 (42%) |
| Post-filter | 73 (76%) | 73 (76%) | 77 (80%) | 32 (45%) | 38 (53%) | 27 (39%) | |
| Accuracy of heterozygous genotypese | Pre-filter | 253 (91%) | 253 (91%) | 258 (93%) | 1051 (91%) | 1058 (92%) | 1033 (89%) |
| Post-filter | 252 (91%) | 252 (91%) | 256 (92%) | 1035 (90%) | 1053 (91%) | 1022 (88%) |
bPre-filter, haplotypes predicted after excluding SNPs with no experimentally determined haplotype phase.
cPost-filter, all SNPs were included in haplotype prediction, but SNPs with no experimentally determined haplotype phase were excluded in the comparison analysis.
dThe number of subjects (or percentage of donors) whose complete haplotypes are correctly constructed.
eThe number of heterozygous genotypes (or percentage of heterozygous genotypes) with correct haplotype phase assignment.
In general, both SNPHAP and PHASE performed well on the ApoE data set, with ≥75% of complete haplotypes accurately predicted. The accuracy of complete haplotype prediction was 4–5% higher for SNPHAP than PHASE and the CPU-intensive PHASE.small generated the same results as PHASE.big, the faster version of the two PHASE programs. The accuracy of complete haplotype prediction for the 69 SNP LPL data is significantly lower, ranging from a maximum of 53% using PHASE.small to a minimum of 39% using SNPHAP. Of the two PHASE programs, there was a 1–5% performance improvement when the CPU-intensive PHASE.small was used. SNPHAP had lower haplotype accuracy than PHASE and it failed to determine haplotypes for three donor subjects in the pre-filter analysis and one donor subject in the post-filter analysis. Despite the differences in their success rates for predicting complete haplotypes, the accuracy of individual heterozygous genotypes was high and consistent (88–92%) in ApoE and LPL, suggesting that the low success rate of LPL complete haplotype prediction is related to its high nucleotide diversity.
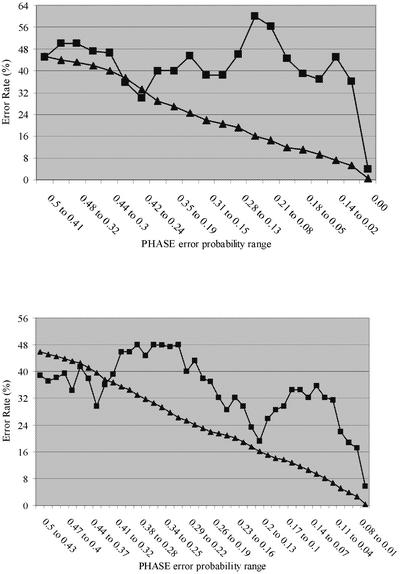
The results of the comparison of the predicted errors calculated from the probability scores with the observed heterozygous genotypes assigned with incorrect phase are shown in Figure 6. In general, the accuracy of prediction deteriorated as the PHASE error probability decreased, but improved again for error probability = 0; this was true for both the ApoE and LPL genes. In ApoE, of the 222 heterozygous genotypes predicted by PHASE to have an error probability of 0, two (0.01) were found to be different from the experimental data. In LPL, 39 (0.04) of 896 heterozygous genotypes predicted with 0 error probability were assigned with incorrect haplotype phase. Though there is a substantial difference between observation and prediction for lower error probability values, the correlation coefficient for these ranges was high (e.g. for LPL in the range 0.87–0.94 the correlation coefficient was 0.84, in ApoE in the range 0.87–0.99 it was 0.67). This suggests that the inaccuracy of the PHASE probability is a scaling problem.
Figure 6.
Graph representation of observed versus expected errors calculated by the PHASE program. The expected errors are shown in triangles and the observed errors are shown in squares. (Top) Results for ApoE. (Bottom) Results for LPL. Most of the heterozygous genotypes are assigned with 0% error probability, representing 78% and 77% of the heterozygous genotypes in ApoE and LPL.
DISCUSSION
Success in applying haplotypes in the identification of genes associated with Mendelian diseases such as cystic fibrosis (25), as well as complex diseases (9,26,27), and ongoing active research in the haplotype structures of the human genome (5,6,8) have led to increasing interest in the use of computational tools for haplotype analysis. HapScope is the first software package in the public domain to offer automated analysis and sophisticated graphical presentation for haplotypes and functional annotations. The design of the analysis pipeline follows the workflow of the SNP/haplotype-based genetic study, integrating genomic and genetic data for SNP and haplotype analysis. It is straightforward to incorporate additional computational tools into the analysis pipeline; for example, it took 2 days to implement the code changes required to add the SNPHAP program as an alternative to the PHASE program for computational haplotype construction. In addition, experimental data can be used in place of data derived by computational prediction; for example, we used the system to analyze molecularly derived haplotypes for ApoE and LPL. In theory, computing a high density haplotype map for an entire chromosome with the HapScope pipeline is possible with a ‘divide and conquer’ approach. It requires defining the chromosomal haplotype block structure and then computing haplotypes within each block. The challenge in this approach is the lack of consensus on the definition of a haplotype block and the variation in haplotype block structure across different populations (6).
The HapScope viewer can be considered a ‘polymorphism/population centric’ viewer that specializes in parallel analysis of information that will support the assessment of the possible functional relevance of polymorphisms. Polymorphism data are shown in great detail, with graphic features that indicate whether a polymorphism occurs in a mRNA encoding or regulatory region, the nature of the amino acid change, if applicable, and the type of variation, such as substitution, insertion or deletion. In contrast, presentation of genomic features is concise and compressed, with related computational results and annotated features superimposed on each other and details displayed only when the user manually selects an object of interest. This compact display, though rarely found in sequence-centric genomic viewers such as the UCSC genome viewer, is commonly used in figures manually drawn for journal publication and is exportable in HapScope. In addition to saving space for population data, it can also facilitate the detection of relationships among different types of features. For example, the 5′- and 3′-UTRs can be identified in the HapScope viewer since the coding region features are overlaid on the related mRNA features (Fig. 4). Similarly, conserved regions between human and mouse can be correlated with the predicted coding exons. To explore haplotype structure at the genome scale, HapScope provides an interface for querying and browsing chromosomal region or candidate genes of interest.
Unlike ViewGene (28), another recently developed SNP viewer, the HapScope viewer focuses on presenting results from haplotype analysis, while ViewGene’s main utility is for analyzing sequence data for SNP discovery. Many of HapScope’s novel display features, such as graphical presentation and manipulation of haplotype block structures, are specifically designed to support current active research in human haplotype structure analysis (5,6,8). In the donor haplotype view, HapScope displays haplotype probability scores, a novel feature that can assist interpretation of computational haplotype construction and aid laboratory scientists in the design of experiments for resolving ambiguities in computational prediction. For example, in ApoE >50% of the haplotype phase discrepancies computed by the PHASE program involve the SNP in the promoter region (also known as –491AT) of ApoE. Detection of such sites with the HapScope viewer can assist targeted experimental verification of computational haplotype prediction. The dynamic haplotype view with the option for selecting a subset of SNPs and consolidating redundant haplotypes in the subset gives more flexibility for haplotype analysis than the static Visual Haplotype or Visual Genotype view (15,16). Of the four haplotypes representing ApoE4 constructed with two promoter SNPs, –491AT and Th1/E47cs, the protective effect of –491AT and the deleterious effect of Th1/E47cs reported previously (29) suggest that the subtype of ApoE4 consolidated from haplotypes 5, 15, 23, 18 and 31 has the most severe adverse impact on Alzheimer’s disease patients as it lacks the protective allele of –491AT and harbors the deleterious allele of Th1/E47 (Fig. 4).
Our analysis of the two haplotype construction programs, PHASE and SNPHAP, showed variations in their success rates in genomic regions with different degrees of nucleotide diversity. The PHASE program, based on Bayesian statistics, is ∼10% more accurate than SNPHAP in analyzing the LPL gene, a region with high nucleotide diversity. In contrast, the SNPHAP program, based on the EM algorithm, is ∼5% more accurate than PHASE in analyzing the ApoE gene, a region with low nucleotide diversity. In a later analysis involving 17 SNPs in the ZBRK1 gene (19; J.P.Struewing, unpublished data) and 109 donors with an average of three heterozygous genotypes per donor, SNPHAP and PHASE generated identical results. Because haplotypes were not molecularly determined, we can conclude from this analysis that the two programs had equal, but unknown, accuracy. Our experience suggests that a prior analysis of regional nucleotide diversity for the human genome may assist in the selection of the best algorithm for generating an accurate human haplotype map.
The results of the comparison of predicted versus observed errors in haplotype phase cast doubt upon PHASE probability scores in the range 0.7–0.99. In ApoE, PHASE assigned a probability value of 0.57 to five heterozygous genotypes; of these, 43%, i.e. about two, should have been incorrect. In fact, two discrepancies were observed, a good match. In the same gene, PHASE assigned a probability value of 0.98 to four heterozygous genotypes, predicting effectively 0 incorrect calls. In fact, PHASE made two incorrect calls at this probability level, a poor match. We decided to use the moving window plot to display the relationship between predicted and expected errors for haplotype phase probability because this presentation shows most clearly the discrepancy between the two sets of values.
Minimum SNP set selection enables the user to obtain the minimum subset of SNPs required to represent haplotype diversity, thus reducing the cost of genotyping by assaying the minimum number of SNPs required. BFA is a robust and accurate method of obtaining a minimum SNP set, practicable for common haplotypes that only include a small number of unique SNPs. In comparison with the previously published htSNP identification result (7), BFA was able to identify smaller SNP set sizes for two of the eight genes. This suggests that manual derivation of a minimum SNP set is problematic even for small haplotype blocks (≤6 haplotypes). Though current haplotype mapping focuses on haplotypes with ≥5% population frequency, HapScope offers minimum SNP set solutions for less common haplotypes that may involve many SNPs. We developed the GPA partly because the exhaustive search for all possible combinations of SNPs, implemented in BFA, is an NP-complete problem and the run time becomes intolerably long when the number of unique SNPs exceeds 40. GPA is an approximation algorithm; however, in all the data we analyzed we have so far not found a real case in which the algorithm-generated minimum SNP set differs in size from that generated by BFA. This is an unusual result and cannot be expected to hold for all genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Andy Clark and Charlie Sing for providing molecularly determined haplotype data for ApoE and LPL genes and Dr John Rioux for providing the 5q31 genotype data. We thank Mr Richard Finney for implementing the SNPHAP parser code.
REFERENCES
- 1.Corder E.H., Saunders,A.M., Strittmatter,W.J., Schmechel,D.E., Gaskell,P.C., Small,G.W., Roses,A.D., Haines,J.L. and Pericak-Vance,M.A. (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science, 261, 921–923. [DOI] [PubMed] [Google Scholar]
- 2.Strittmatter W.J., Weisgraber,K.H., Huang,D.Y., Dong,L.M., Salvesen,G.S., Pericak-Vance,M., Schmechel,D., Saunders,A.M., Goldgaber,D. and Roses,A.D. (1993) Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc. Natl Acad. Sci. USA, 90, 8098–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 4.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 5.Daly M.J., Rioux,J.D., Schaffner,S.F., Hudson,T.J. and Lander,E.S. (2001) High-resolution haplotype structure in the human genome. Nature Genet., 29, 229–232. [DOI] [PubMed] [Google Scholar]
- 6.Gabriel S.B., Schaffner,S.F., Nguyen,H., Moore,J.M., Roy,J., Blumenstiel,B., Higgins,J., DeFelice,M., Lochner,A., Faggart,M. et al. (2002) The structure of haplotype blocks in the human genome. Science, 296, 2225–2229. [DOI] [PubMed] [Google Scholar]
- 7.Johnson G.C., Esposito,L., Barratt,B.J., Smith,A.N., Heward,J., Di Genova,G., Ueda,H., Cordell,H.J., Eaves,I.A., Dudbridge,F. et al. (2001) Haplotype tagging for the identification of common disease genes. Nature Genet., 29, 233–237. [DOI] [PubMed] [Google Scholar]
- 8.Patil N., Berno,A.J., Hinds,D.A., Barrett,W.A., Doshi,J.M., Hacker,C.R., Kautzer,C.R., Lee,D.H., Marjoribanks,C., McDonough,D.P. et al. (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science, 294, 1719–1723. [DOI] [PubMed] [Google Scholar]
- 9.Rioux J.D., Daly,M.J., Silverberg,M.S., Lindblad,K., Steinhart,H., Cohen,Z., Delmonte,T., Kocher,K., Miller,K., Guschwan,S. et al. (2001) Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nature Genet., 29, 223–228. [DOI] [PubMed] [Google Scholar]
- 10.Stephens M., Smith,N.J. and Donnelly,P. (2001) A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet., 68, 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J.S., Sabatti,C., Teng,J., Keats,B.J. and Risch,N. (2001) Bayesian analysis of haplotypes for linkage disequilibrium mapping. Genome Res., 11, 1716–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abecasis G.R., Cherny,S.S., Cookson,W.O. and Cardon,L.R. (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nature Genet., 30, 97–101. [DOI] [PubMed] [Google Scholar]
- 13.Sunyaev S., Ramensky,V., Koch,I., Lathe,W.,III, Kondrashov,A.S. and Bork,P. (2001) Prediction of deleterious human alleles. Hum. Mol. Genet., 10, 591–597. [DOI] [PubMed] [Google Scholar]
- 14.Kent W.J., Sugnet,C.W., Furey,T.S., Roskin,K.M., Pringle,T.H., Zahler,A.M. and Haussler,D. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nickerson D.A., Taylor,S.L., Weiss,K.M., Clark,A.G., Hutchinson,R.G., Stengard,J., Salomaa,V., Vartiainen,E., Boerwinkle,E. and Sing,C.F. (1998) DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nature Genet., 19, 233–240. [DOI] [PubMed] [Google Scholar]
- 16.Rieder M.J., Taylor,S.L., Clark,A.G. and Nickerson,D.A. (1999) Sequence variation in the human angiotensin converting enzyme. Nature Genet., 22, 59–62. [DOI] [PubMed] [Google Scholar]
- 17.Fullerton S.M., Clark,A.G., Weiss,K.M., Nickerson,D.A., Taylor,S.L., Stengard,J.H., Salomaa,V., Vartiainen,E., Perola,M., Boerwinkle,E. et al. (2000) Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am. J. Hum. Genet., 67, 881–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark A.G., Weiss,K.M., Nickerson,D.A., Taylor,S.L., Buchanan,A., Stengard,J., Salomaa,V., Vartiainen,E., Perola,M., Boerwinkle,E. et al. (1998) Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am. J. Hum. Genet., 63, 595–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cantor S.B., Bell,D.W., Ganesan,S., Kass,E.M., Drapkin,R., Grossman,S., Wahrer,D.C., Sgroi,D.C., Lane,W.S., Haber,D.A. et al. (2001) BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell, 105, 149–160. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L., Pan,H., Li,S., Flesken-Nikitin,A., Chen,P.L., Boyer,T.G. and Lee,W.H. (2000) Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol. Cell, 6, 757–768. [DOI] [PubMed] [Google Scholar]
- 21.Clark A.G. (1990) Inference of haplotypes from PCR-amplified samples of diploid populations. Mol. Biol. Evol., 7, 111–122. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J. and Madden,T.L. (1997) PowerBLAST: a new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res., 7, 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X.Q., Hardison,R.C. and Miller,W. (1990) A space-efficient algorithm for local similarities. Comput. Appl. Biosci., 6, 373–381. [DOI] [PubMed] [Google Scholar]
- 24.Wieland H., Funke,H., Krieg,J. and Luley,C. (1991) Abstract Book of the Ninth International Symposium on Atherosclerosis. Rosemont, IL, p. 164.
- 25.Kerem B., Rommens,J.M., Buchanan,J.A., Markiewicz,D., Cox,T.K., Chakravarti,A., Buchwald,M. and Tsui,L.C. (1989) Identification of the cystic fibrosis gene: genetic analysis. Science, 245, 1073–1080. [DOI] [PubMed] [Google Scholar]
- 26.Hugot J.P., Chamaillard,M., Zouali,H., Lesage,S., Cezard,J.P., Belaiche,J., Almer,S., Tysk,C., O’Morain,C.A., Gassull,M. et al. (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature, 411, 599–603. [DOI] [PubMed] [Google Scholar]
- 27.Ogura Y., Bonen,D.K., Inohara,N., Nicolae,D.L., Chen,F.F., Ramos,R., Britton,H., Moran,T., Karaliuskas,R., Duerr,R.H. et al. (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature, 411, 603–606. [DOI] [PubMed] [Google Scholar]
- 28.Kashuk C., SenGupta,S., Eichler,E. and Chakravarti,A. (2002) ViewGene: a graphical tool for polymorphism visualization and characterization. Genome Res., 12, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambert J.C., Berr,C., Pasquier,F., Delacourte,A., Frigard,B., Cottel,D., Perez-Tur,J., Mouroux,V., Mohr,M., Cecyre,D. et al. (1998) Pronounced impact of Th1/E47cs mutation compared with –491 AT mutation on neural APOE gene expression and risk of developing Alzheimer’s disease. Hum. Mol. Genet., 7, 1511–1516. [DOI] [PubMed] [Google Scholar]