Abstract
Expansion of a CAG tract within the huntingtin gene, leading to the production of a protein with an expanded polyglutamine tract, is responsible for Huntington’s disease. We show here that the 5′ untranslated region (UTR) of the huntingtin gene plays an important role in controlling the synthesis of huntingtin. In particular, the 5′ UTR contains an upstream open reading frame (uORF) encoding a 21 amino acid peptide. We demonstrate that the presence of this uORF negatively influences expression from the huntingtin mRNA. Our results suggest a role for the uORF in limiting ribosomal access to downstream initiation sites. Mechanisms involving the post-transcriptional regulation of huntingtin are not well understood, and this may be an important way of regulating huntingtin protein levels.
INTRODUCTION
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder caused by the expansion of a (CAG)n repeat in a gene of unknown function (1). The CAG repeats are within the HD coding region and are successfully transcribed and translated—producing a protein, called huntingtin, containing a polyglutamine tract (2,3). The mutated protein is thought to acquire a novel deleterious function (4), a process involving the formation of ubiquitinated neuronal intranuclear inclusions (5,6). Despite the selective cell death, the HD transcript is ubiquitously expressed (7). Huntingtin is extremely conserved across species and its normal function appears critical for fetal development (8–10).
Normal chromosomes are polymorphic and contain less than (CAG)36 repeats in the huntingtin gene, while chromosomes with (CAG)36 or more repeats are associated with the disease (11). In general, early manifestation is caused by repeat lengths of more than (CAG)50 and in patients with late manifestations one finds less than (CAG)43 repeats. However, there is significant variation of the age of onset in the group of patients with a specific repeat length. This is well illustrated by patients with repeat lengths in the range of (CAG)36–39, where some remain unaffected in later age (70–95 years), whereas some develop symptoms as early as their third decade of life (11). The underlying reason for this variable penetrance in HD is not understood, but may reflect polymorphisms of interacting proteins or of proteins in a biochemical pathway that ties into the HD pathway. Polymorphisms of the HD gene, either as insertion/deletion polymorphisms at position 2642 or within the HD promoter, do not appear to affect the age of onset (12,13), although it is unclear if previously reported promoter polymorphisms affect transcription rates. There are developmentally related differences in levels of the rat HD mRNA in non-neuronal tissues (14), suggesting possible transcriptional or mRNA turnover regulation of the HD gene. Alternatively, variable expression of the huntingtin protein could be responsible, given that variation in HD protein levels occurs within the human striatum (15) and cell types displaying increased expression correspond to affected sites (16). A transgene dosage-dependent phenotype is exhibited by HD transgenic mice consistent with differences in protein levels being responsible for the variable penetrance (17). It is therefore important to understand transcriptional and post-transcriptional mechanisms that regulate HD expression levels.
We have examined the effect on translation of an upstream open reading frame (uORF) present within the 5′ untranslated region (UTR) of the HD mRNA. Previous studies have demonstrated that the presence of uORFs in the 5′ UTR can affect translation of a downstream major ORF (18–20). This can happen by a variety of mechanisms, depending on the configuration of the uORF relative to the downstream major ORF and the primary sequence of the uORF. In cases where the uORF does not overlap with the downstream ORF, translation at the downstream ORF can occur due to one of three events (none of which need to be exclusive): reinitiation, leaky scanning or internal initiation. Reinitiation can occur after translation of a uORF when the 40S subunit remains bound to the mRNA, resumes scanning and initiates at a downstream ORF. It is thought that reinitiation is favored by long intercistronic distances—allowing ribosomes to reacquire initiation factors that would enable them to initiate at the downstream ORF—although direct evidence for this hypothesis is lacking (18). In some cases the frequency of initiation at the downstream ORF depends on the coding content of the uORF [e.g. the Neurospora crassa arg-2 gene (21)], the sequence context of the termination codon (22) or the ability of the uORF to induce shunting (23).
In the current report, we demonstrate that an uORF within the HD 5′ UTR inhibits expression of the huntingtin gene product. Mutational analysis indicates that the majority of ribosomes initiate translation at the uORF, with only a small fraction initiating at the downstream start site. These observations suggest a mechanism by which HD expression might be targeted and regulated.
MATERIALS AND METHODS
Materials and general methods
Restriction endonucleases, calf intestinal alkaline phosphatase, the Klenow fragment of DNA polymerase I, T4 DNA ligase and T4 DNA polymerase were purchased from New England Biolabs. [γ-32P]ATP (6000 Ci/mmol), 35S-methionine (>1000 Ci/mmol), d-threo-[dichloroacetyl-1-14C]-chloroamphenicol (54.0 Ci/mmol), and [3H]CTP (21.8 Ci/mmol) were purchased from New England Nuclear.
Preparation of plasmid DNA, restriction enzyme digestion, PCR amplifications, agarose gel electrophoresis of DNA, DNA ligation and bacterial transformations were accomplished using standard methods (24). DNA clones of PCR-amplification products were always sequenced by the chain termination method using double-stranded DNA templates to ensure the absence of mutations.
5′ Rapid amplification of cDNA ends (RACE) and S1 nuclease mapping
5′ RACE reactions were performed using a commercially available SMART kit from Clontech following the manufacturer’s recommendations. Human fetal brain RNA was purchased from Clontech and used in the 5′ RACE reactions. The oligonucleotide used to prime the cDNA reactions and in the PCR amplification was HD-5′ UTR (5′-CACGGCAGT CCCCGGAGGCCTCGGGCCGACTCG-3′). Clones obtained from seven different 5′ RACE reactions were directly analyzed by sequencing (368 clones).
The probe used in the S1 analysis was HD-S1(Rev) (5′-CTGAGCGGCCGTCCATCTTGGACCCGTCCCGGCA GCCCCCACGGCGCCTTGCGTCCCAGACGTGCGCCG G-3′), a 70-nt oligomer containing 33 nt at its 5′-terminus complementary to the human HD promoter (Fig. 1A, nucleotides –110 to –139). The oligonucleotide probe was radiolabeled with [γ-32P]ATP (6000 Ci/mmol) and T4 polynucleotide kinase. S1 nuclease analyses with gel purified probes were performed as described (24). Briefly, RNA was ethanol precipitated with ∼50 000 c.p.m. of radiolabeled probe, resuspended in 20 µl of S1 hybridization solution [80% deionized formamide, 40 mM PIPES (pH 6.4) 400 mM NaCl, 1 mM EDTA (pH 8.0)], denatured at 90°C for 10 min and hybridized at 30°C overnight. The samples were treated with 250 U of S1 nuclease (Boehinger Mannheim, Germany) at 60°C for 30 min, ethanol precipitated and anlayzed on an 8 M urea–8% polyacrylamide gel (acrylamide:bisacrylamide ratio 18:1).
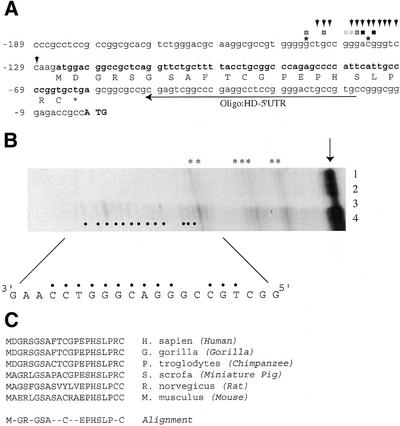
Figure 1.
Schematic representation of the HD promoter region and mapping of the HD 5′ end. (A) The nucleotide sequence of the human HD promoter and 5′ UTR are shown. The initiator ATG codon of the human HD gene is shown in bold uppercase letters and the numbering scheme is relative to the adenosine of the ATG, which is set as position +1. The uORF present in the human 5′ UTR is shown in bold and the encoded amino acids are depicted below the ORF. Two transcription start sites identified by Lin et al. (31) are denoted by asterisks at positions –145 and –135. The 5′ end positions defined by 5′ RACE analysis (seven independent assays) are shown by squares above the appropriate nucleotide positions between –145 and –134. The light grey squares indicate that 5% of the clones analyzed terminated at that position. The darker grey squares indicate that 7–10% of the clones analyzed terminated at the nucleotide. The two blackened squares indicate that 16% (nucleotide position –136) and 25% (nucleotide position –134) of the clones analyzed terminated at that position. The location of the oligonucleotide primer (HD-5′ UTR) used in the 5′ RACE reactions is shown. The 5′ ends identified by S1 nuclease analysis (in B) are shown by downward arrowheads. (B) S1 nuclease mapping of the HD 5′ UTR. S1 nuclease analysis was performed as described in the Materials and Methods and the products fractionated on an 8 M urea–8% polyacrylamide gel. The gel was dried and exposed to X-Omat film (Kodak) for 3 weeks at –70°C with an intensifying screen. The input RNA used in each experiment was: lane 1, 10 µg yeast tRNA; lane 2, 50 µg yeast tRNA; lane 3, 50 µg human fetal brain poly(A–) RNA; lane 4, 50 µg human fetal brain total RNA. The arrow denotes the position of migration of undigested probe. The size of the products was determined in reference to a sequencing ladder. Radiolabeled fragments only present in total RNA from human fetal brain (lane 4) are denoted by blackened circles. The asterisks denote non-specific S1 products obtained with yeast tRNA. (C) Comparison of the polypeptide sequence encoded by the HD uORF found in humans (GenBank accession no. L27350), gorilla (GenBank accession no. Y07988), chimpanzee (GenBank accession no. Y07990), miniature pig (GenBank accession no. AB016793), rat (GenBank accession no. AJ224997) and mouse (GenBank accession no. L34008). Alignment of conserved amino acids among the HD uORFs from different mammals is shown.
Plasmid construction
The generation of the CAT reporter, pSP/CAT, has been previously described (25). For translation studies, pSP/CAT was linearized with HindIII. For generation of HD–145/CAT, the HD 5′ UTR was amplified from genomic DNA using oligonucleotide HD-rev(BamHI) (5′-TCCCGCGGATCCG GCGGTCTCCCGCCCGGCACGGC-3′; BamHI site underlined) and HD-forward (5′-CCTTCGAATTCGCTGCCGGG ACGGGTCCAAGATGGACGGCCGCTCAGGTTCTGC-3′; EcoRI site underlined). The product was cloned into the EcoRI and BamHI site of pSP/CAT, and sequenced. For HD–145(A-125T)/CAT, the HD 5′ UTR was amplified with HD-rev(BamHI) and HD–145(A-125T)-forward (5′-CCTTC GAATTCGCTGCCGGGACGGGTCCAAGTTGGACGGC CGCTCAGGTTCTGC-3′; EcoRI site underlined, altered nucleotide in bold). The product was cloned into the EcoRI and BamHI site of pSP/CAT and sequenced. For HD–145(insG-121)/CAT, the HD 5′ UTR was amplified with HD-rev(BamHI) and HD–145(insG-121)/CAT (5′-CCTTC GAATTCGCTGCCGGGACGGGTCCAAGATGGGACGG CCGCTCAGGTTCTGC-3′; EcoRI site underlined, extra G in bold).
For HD–142/CAT, the HD 5′ UTR was amplified with HD-rev(BamHI) and SP6-HD–142 (5′-ATTTAGGTGACACTAT AGAAGCCGGGACGGGTCCAAGATGGAC-3′) and transferred into pSKII/CAT which had been cleaved with Ecl136B and BamHI. For HD–134/CAT, the HD 5′ UTR was amplified with HD-rev(BamHI) and SP6-HD–134 (5′-ATTTAGGT GACACTATAGAAGGGTCCAAGATGGACGGCCGCTC- 3′) and transferred into pSKII/CAT digested with Ecl136B and BamHI. For HD–134(A-125T)/CAT, the HD 5′ UTR was amplified with HD-rev(BamHI) and SP6-HD–134(A-125T) (5′- ATTTAGGTGACACTATAGAAGGGTCCAAGTTGGAC GGCCGCTC-3′; altered nucleotide in bold) and transferred into pSKII/CAT which had been cleaved with Ecl136B and BamHI.
The bicistronic plasmid, pGEMCAT/Luc, has been previously described (26). CAT/HDLuc and CAT/HD(as)Luc were generated by inserting the BamHI fragment from HD–142/CAT into the BamHI site of pGEMCAT/Luc, and selecting for recombinants that had inserts in the sense and antisense orientation.
For expression studies, a modified pcDNA3 vector was generated in which the polylinker and T7 promoter were replaced by an EcoRI site positioned 10 nt downstream of the CMV promoter initiation site, utilizing site-directed mutagenesis. Expression vectors pcDNA/HD–142 CAT and HD–142(A-125T)/CAT were generated by digesting pSP/HD–142 CAT and pSP/HD–142(A-125T)/CAT with EcoRI. The EcoRI fragments containing the 5′ UTRs and part of the CAT coding region were transferred into pcDNA/CAT that had been linearized with EcoRI. Recombinant clones where verified by sequencing.
In vitro translations and ribosome bindings
In vitro transcriptions, in the presence of m7GpppG, were performed as previously described (27). RNA transcripts were quantitated by monitoring incorporation of [3H]CTP (20 Ci/mmol; New England Nuclear) and the quality of each preparation assessed by SYBR gold staining of RNA that had been fractionated on formaldehyde/1.2% agarose gels. In vitro translations in wheat germ and rabbit reticulocyte lysates were performed using 35S-methionine as directed by the manufacturer (Promega). Krebs translation extracts were prepared and used in in vitro translation reactions, as previously described (28). The final KOAc concentration in the wheat germ, rabbit reticulocyte lysate and Krebs translations was 130 mM. Translations were performed at mRNA concentrations of 20 µg/ml. TCA precipitation of translation products was used to compare the relative translation efficiencies.
Ribosome binding assays were performed essentially as previously described (27). Briefly, 32P-labeled mRNA was incubated in 25 µl of wheat germ extract in the presence of 0.6 mM cyclohexamide (to monitor 80S assembly) at 20°C for 10 min. The final KOAc concentration was adjusted to be 150 mM. Initiation complexes formed were analyzed by sedimentation through 10–30% glycerol gradients. Centrifugation was for 4 h at 39 000 r.p.m. at 4°C in an SW40 rotor. Fractions of ∼500 µl were collected and radioactivity was determined by scintillation counting.
Translations in Xenopus oocytes
Translations in Xenopus oocytes consisted of microinjecting 10 oocytes with 50 nl containing 3H-labeled mRNA (1.75 ng). Given an average oocyte diameter of 1.2 mm (∼0.7 µl) (29), we aimed to achieve a final concentration of 2.5 µg/ml mRNA. Oocytes were incubated at 20°C for 2 h, homogenized in lysis buffer [20 mM Tris–HCl (pH 7.6), 0.1 M NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride], centrifuged in a microfuge for 5 min at 14 000 g and the supernatant used to measure CAT activity (30). Quantitation was accomplished using a Fuji BAS2000 with a Fuji imaging screen. Since each injection was spiked with a known amount of [3H]CTP, portions of the homogenate were counted in a scintillation counter to normalize for variations in microinjection and sample preparation.
Cell culture, transfection, and CAT assays
HeLa and COS-7 cell lines were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal calf serum (Gibco-BRL), penicillin and streptomycin. For transient transfections, cells were plated at a density of 2 × 105 to 5 × 105 cells per 100 mm diameter dish 24 h prior to transfection. Cells were transfected by the calcium phosphate precipitation method (24). The total DNA per plate of cells was 3 µg of pRSV/β-gal vector and 5 µg of expression vector. Cells were washed and re-fed after an overnight incubation and harvested ∼48 h post-transfection. CAT activity was determined as previously described (30). CAT activity values were normalized to β-galactosidase values, which served as internal controls for the transfections.
RESULTS
Mapping of the HD transcription initiation sites
Lin et al. (31) reported mapping the transcription initiation sites of the human and murine HD genes (denoted by asterisks in Fig. 1A). However, the data that led to this conclusion are not presented in their study and are only mentioned in the Discussion of their report. We therefore undertook to independently confirm their results and to also determine whether additional transcription initiation sites are present within the HD promoter. In our hands, primer extension utilizing an oligonucleotide positioned upstream of the initiator ATG codon and priming from poly(A+) RNA did not yield signals that could be interpreted as specific products (data not shown). We therefore performed 5′ RACE analysis since this approach is more sensitive than primer extension. From direct sequencing of 368 clones, we identified that the majority of clones had 5′ ends that terminated between nucleotides –134 and –145 (Fig. 1A). This places the transcription start site of the human HD gene in the same region as defined by Lin et al. (31) but indicates that the start of transcription is more heterogenous than previously reported. To confirm these results, we performed S1 nuclease protection experiments using a radiolabeled oligonucleotide that was complementary to nucleotides –110 to –139 of the HD gene. S1 analysis with this oligonucleotide and yeast tRNA produced few non-specific cleaved products (Fig. 1B, lanes 1 and 2; indicated by asterisks). No additional products were present when the S1 analysis was performed with poly(A–) RNA (lane 3). However, when the S1 protection was performed with total RNA, a set of discrete bands was observed which mapped to nucleotides –129 to –143 (lane 4, denoted by arrowheads). The products of the S1 analysis had approximately the same intensity indicating that these sites are used to the same extent. The S1 nuclease analysis is consistent with the 5′ RACE results and taken together indicate that the HD gene contains a set of heterogenous transcription sites spanning nucleotides –129 to –145.
Presence of an uORF within the HD 5′ UTR
Mapping the 5′ ends of the HD transcript enabled us to define the 5′ UTR of the HD mRNA. We noted the presence of a small uORF within the human HD mRNA 5′ UTR that has the potential to encode a polypeptide of 21 amino acids (Fig. 1A). The uORF is present in the HD genes from human, gorilla, chimpanzee, pig, rat and mouse (Fig. 1C) and there is a conserved motif (EPHSLP) at the C-terminus of the encoded polypeptide of the uORF that is present among the mammalian HD genes analyzed. The initiator codon of the uORF is in a favorable context (5′-AxxATGG-3′) suggesting that it should be recognized by the 43S pre-initiation complex—if the 43S pre-initiation complex binds upstream of the uORF and scans in a linear fashion on the mRNA template (32). Clearly, if initiation of translation occurs by internal binding of the 43S pre-initiation complex on the HD mRNA template (33), then the peptide encoded by the uORF would not be synthesized. It was therefore important to determine whether the HD 5′ UTR could support internal binding of ribosomes.
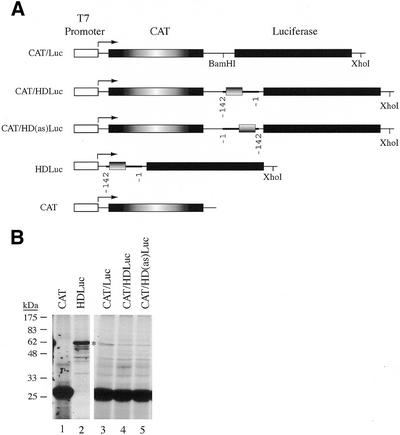
We placed the HD 5′ UTR (nucleotides –1 to –142) as the intercistronic spacer between the CAT and luciferase genes (Fig. 2A) and asked whether this bicistronic transcript could support translation of the second cistron, independent of the first cistron. We found no evidence that the HD 5′ UTR could support internal initiation of translation in either wheat germ extracts (data not shown) or rabbit reticulocyte extracts (Fig. 2B). Translation of CAT mRNA produced a polypeptide of expected molecular mass ∼26 kDa (Fig. 2B, lane 1). Translation of HDLuc mRNA in rabbit reticulocyte lysate produced a polypeptide of ∼61 kDa—the expected molecular mass for Luciferase (Fig. 2B, lane 2). Expression of CAT/Luc mRNA in rabbit reticulocyte lysates produced CAT protein and a small amount of ∼61 kDa product with the same molecular mass as Luciferase (compare lanes 2 and 3) which was not present when CAT mRNA was translated (compare lanes 3 and 1). This product could reflect a small amount of Luciferase produced as a result of either (i) reinitiation of ribosomes from the upstream CAT ORF, (ii) inefficient internal initiation or (iii) initiation on truncated CAT/Luc mRNAs that still maintain an intact luciferase coding region. We have not further characterized the responsible mechanism. Translation of CAT/HDLuc mRNA (containing 142 nt from the HD 5′ UTR as an intercistronic spacer) produced CAT protein when translated (lane 4) and a small amount of ∼61 kDa product with the same molecular mass as Luciferase. Translation of CAT/HD(as)Luc (containing the reverse complement sequence of the HD 5′ UTR) produced the same protein profile as obtained with translation of CAT/HDLuc mRNA (lane 5). If the HD 5′ UTR could initiate translation by internally recruiting ribosomes, one would expect to see levels of Luciferase protein produced from CAT/HDLuc mRNA comparable to those observed with HDLuc (compare lane 4 with 2). Our results indicate that under the conditions tested, the HD 5′ UTR is unable to efficiently internally recruit ribosomes. However, we cannot eliminate the possibility that the HD 5′ UTR can behave as a weak IRES or functions as an IRES in specific cell types.
Figure 2.
(A) Schematic representation of bicistronic expression constructs. The T7 promoter is shown as a white box, the CAT coding region is shown as a box with radial shading, the luciferase coding region is represented by a blackened box, the HD 5′ UTR is shown as a thickened line with the HD uORF denoted as a box with a horizontally shaded gradient. Nucleotide positions of the HD 5′ UTR are denoted. (B) In vitro translation of bicistronic mRNAs in rabbit reticulocyte lysates. Following in vitro translations, samples were treated in SDS sample buffer and electrophoresed into a 12% SDS–polyacrylamide gel. The gel was treated with EN3Hance, dried and exposed to X-Omat (Kodak) film. The mRNA species used to program the lysate are indicated above the panel. Molecular mass markers (NEB) are shown to the left.
Inhibition of translation by the HD uORF
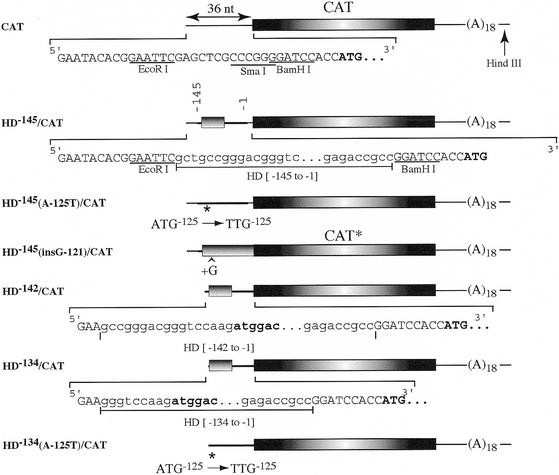
To assess whether the presence of the uORF influenced downstream expression, we built a series of reporters driving CAT expression and containing the HD 5′ UTR. HD–145/CAT contains 145 nt of the HD 5′ UTR placed in the EcoRI site of pSP/CAT. The first 15 nt of this transcript are derived from the pSP65 polylinker. HD–145(A-125T)/CAT harbors a mutation in the initiator codon of the uORF, thus removing this coding region from the 5′ UTR. HD–145(InsG-121)/CAT inserts a guanine residue immediately downstream of the uORF initiation codon, causing a frameshift and placing the coding region of the uORF in-frame with that of the downstream CAT ORF. Thus, the production of a slightly larger protein product (CAT*) from this transcript would suggest that the initiation codon of the uORF is utilized. HD–142/CAT and HD–134/CAT contain the 5′ UTR of HD extending to nucleotides –142 or –134, respectively (Fig. 3).
Figure 3.
Schematic representation of expression constructs. The sequence within the 5′ UTR is presented and the CAT initiation codon is shown in bold. The 5′ UTR derived from plasmid pSP65(A)18 is shown as a thin line whereas the 5′ UTR derived from the HD gene is shown as a thick line. The CAT ORF is denoted by a box with radial shading and the HD uORF is denoted by a box with horizontal shading. The mutation that converts the ATG codon of the uORF into a TTG codon is denoted by an asterisk [HD–145(A-125T)/CAT], whereas the insertion of a guanine residue immediately after the ATG codon is shown by a caret [HD–145(insG-121)/CAT]. Partial nucleotide sequence of the HD 5′ UTR is shown in lowercase letters, whereas sequence derived from the vector polylinker is shown in uppercase letters. CAT* refers to the predicted product obtained from HD(InsG–121)/CAT, where the CAT ORF is fused in-frame with sequences derived from the HD 5′ UTR.
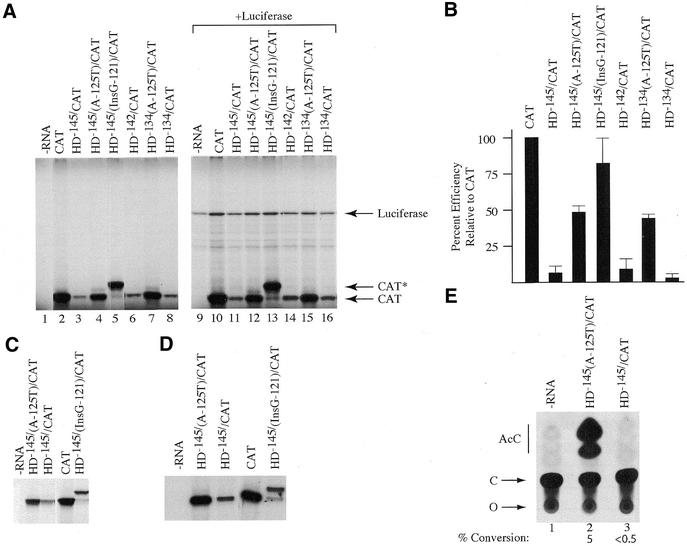
Translation of the HD/CAT reporters was first assessed in wheat germ extracts (Fig. 4A). CAT was efficiently translated in this system and produced a single product (compare lane 2 with 1). Translation of HD–145/CAT was reduced ∼20-fold compared with CAT mRNA, as was the translation of HD–142/CAT and HD–134/CAT (Fig. 4A, compare lanes 3, 6 and 8 with lane 2; quantitation summarized in Fig. 4B). The translation of HD–145(A-125T)/CAT and HD–134(A-125T)/CAT, in which the initiation codon of the uORF is abolished, was 50% that of CAT mRNA and 10-fold higher than HD–145/CAT or HD–134/CAT (Fig. 4A and B). To assess whether the uORF was being translated, we translated HD–145(InsG-121)/CAT, which contains an additional nucleotide within the uORF and puts it in-frame with the downstream CAT ORF. Analysis of the product revealed a protein of higher molecular mass (CAT*) (Fig. 4A, lane 5) consistent with initiation starting at the ATG codon of the uORF. The efficiency of translation of HD–145(InsG-121)/CAT indicated that this transcript expressed CAT* at levels comparable to translation of CAT mRNA (Fig. 4B). In addition, very little CAT product was generated from HD–145(InsG-121)/CAT (Fig. 4A, lane 5; <5%), suggesting that the majority of the 43S initiation complexes (>95%) initiate translation at the first encountered ATG codon. These results suggest that 43S ribosomes efficiently recognize the HD uORF initiation codon and that its presence inhibits translation of a downstream ORF.
Figure 4.
In vitro and in vivo translation of HD expression constructs. (A) Translation of HD/CAT reporter constructs in wheat germ extracts. Following in vitro translations, samples were treated in SDS sample buffer and electrophoresed into a 12% SDS–polyacrylamide gel. The gel was treated with EN3Hance, dried and exposed to X-Omat (Kodak) film. The mRNA species used to program the lysate are indicated above the panel. The position of migration of Luciferase, CAT* and CAT protein is indicated to the right of the panel. Lanes 1 and 9, control translation performed without endogenously added mRNA template; lanes 2–8, translation of CAT and HD/CAT mRNAs; lane 10–16, co-translation of luciferase mRNA and either CAT or HD/CAT mRNAs. (B) Bar graph representing the average translation efficiencies of HD/CAT reporters. TCA precipitation of translated products was used to quantitate levels of protein product. Values are standardized to the efficiency obtained with CAT mRNA. All values represent the average of at least four experiments and include the experiment presented in (A). Standard deviations are presented. (C) Translation of HD/CAT reporter constructs in rabbit reticulocyte lysates. Following in vitro translations, samples were treated in SDS sample buffer and electrophoresed into a 12% SDS–polyacrylamide gel. The gel was treated with EN3Hance, dried and exposed to X-Omat (Kodak) film. The mRNA species used to program the lysate are indicated above the panel. (D) Translation of HD/CAT reporter constructs in Krebs extracts. The mRNA species used to program the lysate are indicated above the panel. (E) In vivo translation in X.laevis oocytes. An autoradiograph of a representative TLC for the CAT assays performed is shown. The percent conversion shown below the panel is the average obtained from four independent experiments. AcC, acetylated forms of chloramphenicol; C, chloramphenicol; O, origin.
To eliminate the possibility that some of the transcript preparations used in this study harbored trans-acting inhibitors of translation, we co-translated the HD/CAT reporter transcripts in the presence of luciferase transcripts (Fig. 4A). Whereas the same relative levels of CAT produced from the various reporter transcripts was observed (compare lanes 10–16 with lanes 2–8, respectively), only slight variations (2-fold) in the amount of luciferase product synthesized was noted (lanes 9–16), consistent with the absence of any trans-acting inhibitor in the CAT-based mRNA reporter preparations.
When CAT, HD–145/CAT, HD–145/(A-125T)/CAT and HD–145/(InsG-121)/CAT mRNA were translated in rabbit reticulocyte lysates (Fig. 4C) or Krebs extracts (Fig. 4D), similar relative translational efficiencies were observed as in the wheat germ extracts. In addition, HD–145/CAT mRNA was not expressed in injected Xenopus laevis oocytes, whereas HD–145(A-125T)/CAT mRNA was expressed (Fig. 4E). To assess the relative expression levels of HD–142/CAT and HD–142/(A-125T)/CAT in cell lines, we transfected pcDNA3 based expression vectors into COS-7 and HeLa cells. In duplicate experiments, the percentage of CAT activity of extracts from pcDNA/HD–142/(A-125T)/CAT transfected cells varied from 37 to 54% conversion, whereas those of extracts from pcDNA/HD–142/CAT were 13–20%. Thus, transfection of pcDNA/HD–142/CAT into COS-7 or HeLa cells yielded 2.5–3 times lower CAT activity than did pcDNA/HD–142/(A-125T)/CAT. These results are consistent with the idea that the HD uORF is inhibitory to translation of downstream ORFs, although the inhibitory effect was less in the transfection experiments than was observed in cell-free systems.
Stability of HD/CAT reporter transcripts in wheat germ extracts
One explanation that could account for the observed difference in translation between HD–145/CAT and HD–145(A-125T)/CAT (Fig. 4) would be if there was a significant difference in stability between these transcripts. To directly address this, we performed a kinetic analysis of HD–145/CAT and HD–145(A-125T)/CAT mRNA stability when translated in wheat germ extracts (Fig. 5). Re-isolation of radiolabeled transcripts from programmed translation extracts, followed by fractionation on formaldehyde agarose gels, revealed that the stability of HD–145/CAT and HD–145(A-125T)/CAT mRNA are similar (Fig. 5). Thus, the observed differences in translation between HD–145/CAT and HD–145(A-125T)/CAT cannot be attributed to differences in mRNA stability.
Figure 5.
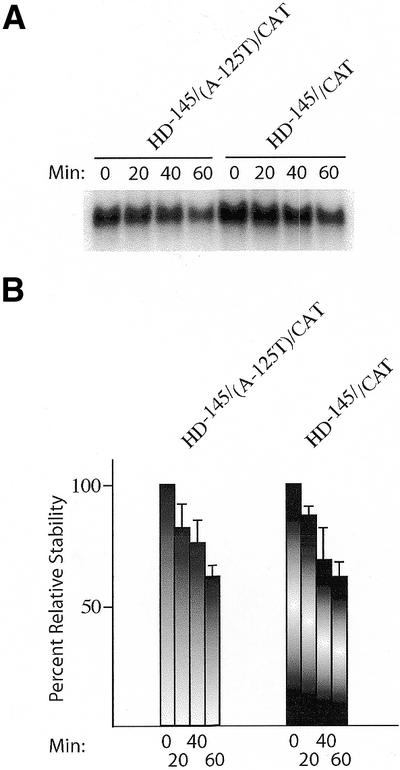
Stability of HD–145/CAT and HD–145(A-125T)/CAT mRNA in wheat germ translation extracts. (A) 32P-labeled HD–145/CAT and HD–145(A-125T)/CAT mRNA were translated in wheat germ extracts and, at the indicated time points, an aliquot was removed (10 µl), incubated with 50 µg of Proteinase K at 37°C for 15 min, after which the sample was phenol/chloroform extracted. Following ethanol precipitation, RNA samples were fractionated on a 1.4% agarose/formaldehyde gel. The gel was stained with SYBR gold (Molecular Probes) to ensure equal recovery of rRNA from the translation extracts (data not shown), dried and exposed to X-Omat film (Kodak) at –70°C with an intensifying screen. The time at which mRNA was extracted is shown above the panel. (B) Summary of three experiments measuring the relative stability of HD–145/CAT and HD–145(A-125T)/CAT in wheat germ extracts. Following fractionation on 1.0% formaldehyde agarose gels, the radioactivity associated with each mRNA was quantitated on a Fujix BAS2000 with a Fuji imaging screen. For each mRNA species, the amount of radiolabel present at the 0 min time point is taken as 100%. All gels were stained with SYBR gold (Molecular Probes) to ensure equal recovery of rRNA from the translation extracts (data not shown). The nature of the mRNA species being assayed is indicated above the panel.
Ribosome binding is not affected by the uORF
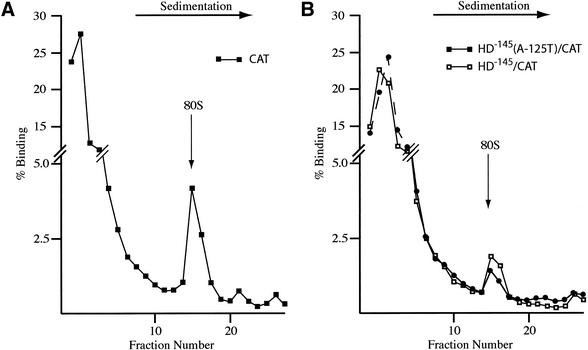
One way to explain the reduced expression of HD–145/CAT versus HD–145(A-125T)/CAT would be if HD–145/CAT was less efficient at loading ribosomes than HD–145(A-125T)/CAT. To directly assess this, we performed ribosome binding assays in the presence of cycloheximide to trap 80S ribosomes on the mRNA templates (Fig. 6). We used CAT mRNA as a positive control, and observed that ∼6% of the mRNA was bound to 80S ribosomes (Fig. 6A). We observed similar ribosome binding profiles between HD–145/CAT and HD–145(A-125T)/CAT (Fig. 6B). Both mRNA templates were less efficient than CAT mRNA at recruiting 80S ribosomes, with ∼1.5–2% of the input mRNA binding to ribosomes (Fig. 6B). We have not toe-printed the position of the initiation complexes to determine where the 80S ribosomes are positioned on the two templates. However, our results indicate that differences in ribosome recruitment cannot account for the observed differences in translation between HD–145/CAT and HD–145(A-125T)/CAT.
Figure 6.
Effect of the HD uORF on 80S assembly in wheat germ extracts. 32P-labeled CAT (A) or HD–145/CAT (B) and HD–145(A-125T)/CAT (B) were incubated with wheat germ extracts as described in the Materials and Methods. 80S complexes were visualized by treatment of the extracts with cycloheximide. Fractions from each sucrose gradient were collected using a Brandel Tube Piercer connected to an ISCO fraction collector and individually counted. The total counts recovered from each gradient and percentage mRNA bound to 80S ribosomes were: (A) CAT mRNA (52 314 c.p.m., 6% binding), (B) HD–145/CAT mRNA (83 043 c.p.m., 1.5% binding); HD–145(A-125T)/CAT mRNA (85 986 c.p.m., 2% binding).
DISCUSSION
The HD promoter has been previously studied in order to better understand cell-specific differences in HD mRNA expression and functional effects of HD promoter polymorphisms. Both the human and rodent upstream regions have been sequenced and functionally characterized (31,34–36). Lin et al. (31) reported mapping the transcription initiation sites of the murine and human HD genes but did not present any supporting data. Their report suggests the presence of two initiation sites within both species, placing the two human sites at nucleotides –135 and –145 (relative to the adenosine nucleotide of the ATG codon) (see Fig. 1A of the current study). Holzman et al. (35) cloned the rat promoter and indicated that they could not define the start sites of the rat transcript by using primer-extension or nuclease protection (S1 and mung bean) assays. Functional analysis of the human promoter suggested that the main regulatory elements responsible for transcription lie within a fragment extending from position –324 to +20 (34), with binding sites for Sp1, AP-2, AP-4 and NF-AT transcription factors (34,36). Neither the human or rodent promoters have TATA or CCAAT elements (31). Importantly, a 1.9 kb genomic fragment, including ∼1 kb of sequence upstream of the initiator AUG codon, is capable of driving expression of HD genes carrying CAG expansions in mice and reproducing many of the features of HD (17). These latter results indicate that all of the elements required for HD gene expression lie within 1 kb of the initiation codon.
In the current report we have mapped the 5′ end of the human HD gene by using a combination of 5′ RACE and S1 nuclease mapping (Fig. 1). Our results indicate that the start site of the HD gene is more heterogenous than previously suspected. Both 5′ RACE and S1 nuclease mapping revealed the presence of multiple start sites spanning nucleotides –145 to –129. Multiple transcription initiation sites and GC-rich domains (as found within the HD promoter) are common features of many housekeeping genes (37), of which the HD gene has been postulated to be a member (31). Our study does not exclude the possibility that the HD transcript contains alternative 5′ ends in other tissues or cell types than examined herein (fetal brain).
Mapping of the HD 5′ end was a prerequisite to allow us to confidently place the uORF within the 5′ UTR of the HD gene. uORFs regulate translation in eukaryotes (19,20,38). There are several mechanisms by which this can be achieved. uORFs can encode peptides that control reinitiation at the downstream ORF [e.g. the GCN4 system (39)] or that stall ribosomes and block scanning [e.g. the arginine attenuator peptide (39)]. They can have a passive effect and influence reinitation efficiency as a consequence of the distance between their termination codon and the initiation codon of the downstream ORF (18), given that the longer intercistronic region allows more time for a scanning ribosome to reacquire factors necessary for initiation at the downstream ORF, and that the type of termination event at the uORF puts the ribosome in a position to resume scanning. There have also been reports for additional roles for uORFs such as stimulation of translation and effects on mRNA stability (reviewed in 19).
We document that the huntingtin uORF significantly impairs translation of a downstream reporter ORF (Fig. 4). The presence and length (21 amino acids) of the uORF in the HD 5′ UTR is conserved in a number of species, but only the C-terminal domain of the peptide is conserved (Fig. 1C). Translation of HD–145/CAT is reduced ∼5-fold in wheat germ extracts compared with translation of HD–145(A-125T)/CAT, where the initiation codon of the uORF has been mutated (Fig. 4B). A similar effect is observed in translations performed with rabbit reticulocyte lysates, Kreb-2 cell extracts and in injected frog oocytes (Fig. 2C–E), indicating that the results are not extract- or cell-specific. Expression is also reduced ∼3-fold in COS-7 and HeLa cells when the uORF is present in the 5′ UTR of a pcDNA/CAT expression vector.
Even without the uORF, the HD 5′ UTR appeared inhibitory to translation. The 5′ UTR of the huntingtin mRNA can be folded into stable stem–loop structures with a free energy value of –71 kcal/mol (data not shown). Such structures are known to be inhibitory to translation (27) and may explain the reduced translation of HD–145(A-125T)/CAT compared with CAT mRNA (Fig. 4A and B). However, differences in translation efficiency between HD–145/CAT and HD–145(A-125T)/CAT cannot be attributed to differences in overall mRNA stability, since both showed equivalent stabilities when used to program a wheat germ translation system (Fig. 5).
The initiation codon of the HD uORF is close to the 5′ ends of the huntingtin mRNAs (Fig. 1A). It has been proposed that ribosomes may have a ‘blind spot’ where initiation codons positioned close to the 5′ cap structure are not recognized by the 43S preinitiation complex (40). We assessed whether there was a difference in the inhibitory potential of transcripts that had the uORF initiation codon 12 nt (HD–134/CAT) or 20 nt (HD–142/CAT) distal to the cap structure and found little difference in the inhibition conferred by the uORF from either HD–134/CAT or HD–142/CAT, compared with HD–145/CAT (Fig. 4B).
To gauge the fraction of ribosomes initiating at the uORF ATG codon versus those displaying leaky scanning and initiating at the downstream ORF ATG codon, we compared the translational efficiency of HD–145/(InsG-121)/CAT with HD–145/CAT mRNA (Fig. 4B–D). Since this mutation fuses the coding region of the uORF in-frame with the downstream ORF producing a larger protein product (called CAT*), we were able to monitor the production of the two proteins from both ATG codons (Fig. 4). The results indicate that >95% of the product obtained from HD–145/(InsG-121)/CAT is an extended version of CAT (CAT*), suggesting that the majority of ribosomes efficiently recognize the initiation codon of the uORF. Both HD–145/CAT and HD–145(A-125T)/CAT mRNAs are able to form similar amounts of 80S initiation complexes (Fig. 5), indicating that loading of ribosomes onto HD–145/CAT mRNA is not impaired. Finally, the HD 5′ UTR does not efficiently recruit ribosomes by internal initiation in vitro (Fig. 2).
Many mRNAs whose major ORF initiates downstream of one or several uORF(s) encode regulatory proteins whose expression has to be finely regulated. In support of this, the oncogenic potential of c-mos and c-lck are increased by rearrangements that remove the uORFs present within their 5′ UTRs (41–43). Four uORFs regulate translation of GCN4 in response to amino acid availability by controlling which downstream AUG codons are utilized by the re-initiating ribosomes (39,44). The uORF of the S-adenosylmethioine decarboxylase gene can confer polyamide regulation of translation on a heterologous downstream ORF. The arginine attenuator peptide is encoded by a small ORF upstream of the N.crassa arg-2 gene and confers negative regulation in response to arginine by causing ribosomes to stall at the termination codon of the uORF and reducing loading at the downstream start codon (39,45). Given the importance of huntingtin in normal development (8), requirement for precise control of its expression may be necessary. We have described a new feature of huntingtin expression that involves translational repression by a conserved uORF. Better understanding this regulation may lead to ways by which HD expression can be manipulated
Acknowledgments
ACKNOWLEDGEMENTS
We thank Matthew Donlan for generating pcDNA/HD–142/CAT and pcDNA/HD–142/(A-125T)/CAT expression vectors. E.H.P. was supported by a McGill University Cancer Center Canderel Studentship. J.P. is a Canadian Institutes of Health Research (CIHR) Senior Investigator. This work was supported by grants from the Canadian Institutes of Health Research and from the Hereditary Disease Foundation Cure HD Initiative (CHDI) to J.P.
REFERENCES
- 1.The Huntington’s Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 2.Jou Y.S. and Myers,R.M. (1995) Evidence from antibody studies that the CAG repeat in the Huntington disease gene is expressed in the protein. Hum. Mol. Genet., 4, 465–469. [DOI] [PubMed] [Google Scholar]
- 3.Trottier Y., Devys,D., Imbert,G., Saudou,F., An,I., Lutz,Y., Weber,C., Agid,Y., Hirsch,E.C. and Mandel,J.L. (1995) Cellular localization of the Huntington’s disease protein and discrimination of the normal and mutated form. Nature Genet., 10, 104–110. [DOI] [PubMed] [Google Scholar]
- 4.Nasir J., Goldberg,Y.P. and Hayden,M.R. (1996) Huntington disease: new insights into the relationship between CAG expansion and disease. Hum. Mol. Genet., 5, 1431–1435. [DOI] [PubMed] [Google Scholar]
- 5.Davies S.W., Turmaine,M., Cozens,B.A., DiFiglia,M., Sharp,A.H., Ross,C.A., Scherzinger,E., Wanker,E.E., Mangiarini,L. and Bates,G.P. (1997) Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell, 90, 537–548. [DOI] [PubMed] [Google Scholar]
- 6.DiFiglia M., Sapp,E., Chase,K.O., Davies,S.W., Bates,G.P., Vonsattel,J.P. and Aronin,N. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science, 277, 1990–1993. [DOI] [PubMed] [Google Scholar]
- 7.Strong T.V., Tagle,D.A., Valdes,J.M., Elmer,L.W., Boehm,K., Swaroop,M., Kaatz,K.W., Collins,F.S. and Albin,R.L. (1993) Widespread expression of the human and rat Huntington’s disease gene in brain and nonneural tissues. Nature Genet., 5, 259–265. [DOI] [PubMed] [Google Scholar]
- 8.Duyao M.P., Auerbach,A.B., Ryan,A., Persichetti,F., Barnes,G.T., McNeil,S.M., Ge,P., Vonsattel,J.P., Gusella,J.F., Joyner,A.L. et al. (1995) Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science, 269, 407–410. [DOI] [PubMed] [Google Scholar]
- 9.Nasir J., Floresco,S.B., O’Kusky,J.R., Diewert,V.M., Richman,J.M., Zeisler,J., Borowski,A., Marth,J.D., Phillips,A.G. and Hayden,M.R. (1995) Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell, 81, 811–823. [DOI] [PubMed] [Google Scholar]
- 10.Zeitlin S., Liu,J.P., Chapman,D.L., Papaioannou,V.E. and Efstratiadis,A. (1995) Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nature Genet., 11, 155–163. [DOI] [PubMed] [Google Scholar]
- 11.Rubinsztein D.C., Leggo,J., Coles,R., Almqvist,E., Biancalana,V., Cassiman,J.J., Chotai,K., Connarty,M., Crauford,D., Curtis,A. et al. (1996) Phenotypic characterization of individuals with 30–40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36–39 repeats. Am. J. Hum. Genet., 59, 16–22. [PMC free article] [PubMed] [Google Scholar]
- 12.Novelletto A., Persichetti,F., Sabbadini,G., Mandich,P., Bellone,E., Ajmar,F., Squitieri,F., Campanella,G., Bozza,A., MacDonald,M.E. et al. (1994) Polymorphism analysis of the huntingtin gene in Italian families affected with Huntington disease. Hum. Mol. Genet., 3, 1129–1132. [DOI] [PubMed] [Google Scholar]
- 13.Coles R., Leggo,J. and Rubinsztein,D.C. (1997) Analysis of the 5′ upstream sequence of the Huntington’s disease (HD) gene shows six new rare alleles which are unrelated to the age at onset of HD. J. Med. Genet., 34, 371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitt I., Bachner,D., Megow,D., Henklein,P., Hameister,H., Epplen,J.T. and Riess,O. (1995) Expression of the Huntington disease gene in rodents: cloning the rat homologue and evidence for downregulation in non-neuronal tissues during development. Hum. Mol. Genet., 4, 1173–1182. [DOI] [PubMed] [Google Scholar]
- 15.Gutekunst C.A., Levey,A.I., Heilman,C.J., Whaley,W.L., Yi,H., Nash,N.R., Rees,H.D., Madden,J.J. and Hersch,S.M. (1995) Identification and localization of huntingtin in brain and human lymphoblastoid cell lines with anti-fusion protein antibodies. Proc. Natl Acad. Sci. USA, 92, 8710–8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrante R.J., Gutekunst,C.A., Persichetti,F., McNeil,S.M., Kowall,N.W., Gusella,J.F., MacDonald,M.E., Beal,M.F. and Hersch,S.M. (1997) Heterogeneous topographic and cellular distribution of huntingtin expression in the normal human neostriatum. J. Neurosci., 17, 3052–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangiarini L., Sathasivam,K., Seller,M., Cozens,B., Harper,A., Hetherington,C., Lawton,M., Trottier,Y., Lehrach,H., Davies,S.W. and Bates,G.P. (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell, 87, 493–506. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. (1987) Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol., 7, 3438–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geballe A. and Sachs,M. (2000) Translational control by upstream open reading frames. In Sonenberg,N., Hershey,J. and Matthews,M. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 595–614.
- 20.Morris D.R. and Geballe,A.P. (2000) Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol., 20, 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z. and Sachs,M.S. (1997) Arginine-specific regulation mediated by the Neurospora crassa arg-2 upstream open reading frame in a homologous, cell-free in vitro translation system. J. Biol. Chem., 272, 255–261. [DOI] [PubMed] [Google Scholar]
- 22.McCarthy J.E. (1998) Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev., 62, 1492–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemmings-Mieszczak M., Hohn,T. and Preiss,T. (2000) Termination and peptide release at the upstream open reading frame are required for downstream translation on synthetic shunt-competent mRNA leaders. Mol. Cell. Biol., 20, 6212–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J. and Russell,D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Harvey I., Garneau,P. and Pelletier,J. (2002) Forced engagement of a RNA/protein complex by a chemical inducer of dimerization to modulate gene expression. Proc. Natl Acad. Sci. USA, 99, 1882–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pause A., Belsham,G.J., Gingras,A.C., Donze,O., Lin,T.A., Lawrence,J.C.,Jr and Sonenberg,N. (1994) Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature, 371, 762–767. [DOI] [PubMed] [Google Scholar]
- 27.Pelletier J. and Sonenberg,N. (1985) Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell, 40, 515–526. [DOI] [PubMed] [Google Scholar]
- 28.Svitkin Y.V. and Agol,V.I. (1978) Complete translation of encephalomyocarditis virus RNA and faithful cleavage of virus-specific proteins in a cell-free system from Krebs-2 cells. FEBS Lett., 87, 7–11. [DOI] [PubMed] [Google Scholar]
- 29.Colman A. (1984) Translation of eukaryotic messenger RNA in Xenopus oocytes. In Hames,B.D. and Higgins,S.J. (eds), Transcription and Translation. A Practical Approach. IRL Press Ltd, Oxford, UK, pp. 271–302.
- 30.Gorman C. (1985) High efficiency gene transfer into mammalian cells. In Glover,D.M. (ed.), DNA Cloning II. A Practical Approach. IRL Press Ltd, Oxford, UK, pp. 143–190.
- 31.Lin B., Nasir,J., Kalchman,M.A., McDonald,H., Zeisler,J., Goldberg,Y.P. and Hayden,M.R. (1995) Structural analysis of the 5′ region of mouse and human Huntington disease genes reveals conservation of putative promoter region and di- and trinucleotide polymorphisms. Genomics, 25, 707–715. [DOI] [PubMed] [Google Scholar]
- 32.Kozak M. (1981) Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res., 9, 5233–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelletier J. and Sonenberg,N. (1988) Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature, 334, 320–325. [DOI] [PubMed] [Google Scholar]
- 34.Coles R., Caswell,R. and Rubinsztein,D.C. (1998) Functional analysis of the Huntington’s disease (HD) gene promoter. Hum. Mol. Genet., 7, 791–800. [DOI] [PubMed] [Google Scholar]
- 35.Holzmann C., Maueler,W., Petersohn,D., Schmidt,T., Thiel,G., Epplen,J.T. and Riess,O. (1998) Isolation and characterization of the rat huntingtin promoter. Biochem. J., 336, 227–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holzmann C., Schmidt,T., Thiel,G., Epplen,J.T. and Riess,O. (2001) Functional characterization of the human Huntington’s disease gene promoter. Brain Res. Mol. Brain Res., 92, 85–97. [DOI] [PubMed] [Google Scholar]
- 37.Gardiner-Garden M. and Frommer,M. (1987) CpG islands in vertebrate genomes. J. Mol. Biol., 196, 261–282. [DOI] [PubMed] [Google Scholar]
- 38.Lovett P.S. and Rogers,E.J. (1996) Ribosome regulation by the nascent peptide. Microbiol Rev., 60, 366–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaba A., Wang,Z., Krishnamoorthy,T., Hinnebusch,A.G. and Sachs,M.S. (2001) Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J., 20, 6453–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak M. (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res., 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rechavi G., Givol,D. and Canaani,E. (1982) Activation of a cellular oncogene by DNA rearrangement: possible involvement of an IS-like element. Nature, 300, 607–611. [DOI] [PubMed] [Google Scholar]
- 42.Steel L.F., Telly,D.L., Leonard,J., Rice,B.A., Monks,B. and Sawicki,J.A. (1996) Elements in the murine c-mos messenger RNA 5′-untranslated region repress translation of downstream coding sequences. Cell Growth Differ., 7, 1415–1424. [PubMed] [Google Scholar]
- 43.Marth J.D., Overell,R.W., Meier,K.E., Krebs,E.G. and Perlmutter,R.M. (1988) Translational activation of the lck proto-oncogene. Nature, 332, 171–173. [DOI] [PubMed] [Google Scholar]
- 44.Hinnebusch A.G. (1997) Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem., 272, 21661–21664. [DOI] [PubMed] [Google Scholar]
- 45.Fang P., Wang,Z. and Sachs,M.S. (2000) Evolutionarily conserved features of the arginine attenuator peptide provide the necessary requirements for its function in translational regulation. J. Biol. Chem., 275, 26710–26719. [DOI] [PubMed] [Google Scholar]