Abstract
The essential Aurora B kinase is a chromosomal passenger protein that is required for mitotic chromosome alignment and segregation. Aurora B function is dependent on the chromosome passenger, INCENP. INCENP, in turn, requires sister chromatid cohesion for its appropriate behaviour. Relatively few substrates have been identified for Aurora B, so that the precise role it plays in controlling mitosis remains to be elucidated. To identify potential novel mitotic substrates of Aurora B, extracted chromosomes were prepared from mitotically-arrested HeLa S3 cells and incubated with recombinant human Aurora B in the presence of radioactive ATP. Immunoblot analysis confirmed the HeLa scaffold fraction to be enriched for known chromosomal proteins including CENP-A, CENP-B, CENP-C, ScII and INCENP. Mass spectrometry of bands excised from one-dimensional polyacrylamide gels further defined the protein composition of the extracted chromosome fraction. Cloning, fluorescent tagging and expression in HeLa cells of the putative GTP-binding protein NGB/CRFG demonstrated it to be a novel mitotic chromosome protein, with a perichromosomal localisation. Identi fication of the protein bands corresponding to those phosphorylated by Aurora B revealed topoisomerase II alpha (topo IIα) as a potential Aurora B substrate. Purified recombinant human topo IIα was phosphorylated by Aurora B in vitro, confirming this proteomic approach as a valid method for the initial definition of candidate substrates of key mitotic kinases.
INTRODUCTION
Progression through mitosis is regulated by a number of protein kinases (1). One of these is Aurora B, an essential kinase that is required for chromosome alignment at the metaphase plate and for completion of cytokinesis in metazoan cells (2–7; reviewed in 8). Aurora B may also be involved in mitotic chromosome condensation, but the extent of this involvement is unclear (3,6,9,10). Aurora B is a chromosomal passenger protein. Chromosomal passengers are a class of proteins that show a complex and defined localisation during mitosis; associating along the chromosome during prophase, concentrating at the centromere at metaphase and moving from the centromere to the central region of the mitotic spindle at anaphase (11). Other chromosome passengers include INCENP and survivin, both of which physically interact with Aurora B. INCENP or survivin deficiency causes Aurora B mislocalisation and cellular defects indistinguishable from those occasioned by Aurora B deficiency, while the loss of Aurora B prevents both INCENP and survivin from behaving normally (2,4,6,12–15). The appropriate localisation of INCENP and survivin also requires the cohesin subunit, Scc1 [(16) and C.Morrison, P.Vagnarelli and W.C.Earnshaw, unpublished data], implying that the activities of the chromosome passengers are closely linked to the other key processes occurring during mitosis.
Histone H3 is serine-phosphorylated by Aurora B (2,3,6,14,17), as are CENP-A (18) and myosin II regulatory light chain (19), although the effects of these modifications are not yet clear. The in vitro activity of the Caenorhabditis elegans Aurora B kinase, AIR-2, is enhanced by the presence of ICP-1, the C.elegans INCENP, which is a target for serine phosphorylation by the kinase (20). The budding yeast homologue of Aurora B, Ipl-1p, which is necessary for symmetric chromosome segregation, phosphorylates the kinetochore component Ndc10p (21,22), and a recent model for Ipl-1p function has suggested that an unknown kinetochore Ipl-1p target may be the mediator of Ipl-1p control of the attachments between the spindle pole bodies and the kinetochores that result in bipolar kinetochore attachment (23). Clearly, the identification of further substrates of Aurora B kinase is an important issue in defining the mechanism by which the passengers control mitotic events.
Recent advances in the ability to identify peptides present in mixed protein samples using mass spectrometry have provided an invaluable tool for the analysis of cellular subfractions (24–26). One such fraction is the metaphase chromosome, which can be prepared from mitotically-blocked tissue culture cells (27). The chromosome scaffold fraction comprises the residual insoluble, non-histone proteins that remain after extraction of nuclease-digested, isolated metaphase chromosomes by high salt, low ionic strength or chaotropic buffers (27). Proteins that have been identified in this fraction include CENP-E, DNA topoisomerase II and the condensin subunit, ScII (28–31) and kinetochores are also components of the scaffold (32). Candidate Aurora B substrates might be anticipated in metaphase chromosomes, or in the chromosome scaffold fraction. Here we describe experiments in which we characterise the components of partially-extracted metaphase chromosomes using mass spectrometry in order to identify potential Aurora B substrates.
MATERIALS AND METHODS
Cloning, protein overexpression and purification and antibody preparation
An EST containing a cDNA for human Aurora B was obtained from the UK HGMP Resource Centre (Babraham, UK) and subcloned into pGEX4T (Amersham Biosciences UK, Little Chalfont, UK). For site-directed mutagenesis of the pGEX4T-Aurora B construct, the QuikChange kit (Stratagene, La Jolla, CA) was used, following the manufacturer’s instructions, with the primer 5′-CAT TTC ATC GTG GCG CTC CGA GTC CTC TTC AAG TCC-3′. Bacterial overexpression and chromatography over glutathione-sepharose was performed as previously described (2). For polyclonal antibody generation, rabbits were immunised with gel-purified GST-Aurora B: this yielded anti-Aurora B antiserum R902. Human topoisomerase II alpha (topo IIα) was expressed and purified from Saccharomyces cerevisiae as described previously (33,34).
Total RNA was extracted from HeLa S3 cells using Trizol (Invitrogen, Carlsbad, CA) and reverse transcription using oligo(dT) primers performed using the Superscript system (Invitrogen). Amplification of the CRFG sequence was performed with LA-Taq (Takara Bio, Shiga, Japan) and the product cloned into pEGFP-N1 and pEGFP-C1 (BD Clontech UK, Cowley, UK) using the XhoI and HindIII sites contained in the primers 5′-ACT CGA GAA ATG GCA CAT TAC AAC TTC AAG-3′ and 5′-AAA GCT TTC TCC TGT CCT TTT TAC CAG C-3′, respectively.
Cell culture, transfection and microscopy
HeLa JW cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% foetal bovine serum (FBS) and antibiotics, while HeLa S3 cells were grown in the same medium supplemented with only 5% FBS and antibiotics. HeLa S3 cells were treated with colcemid (0.1 µg/ml) for 2 h before harvest and were then hyptotonically swollen in 75 mM KCl for 10 min before being dropped on to polylysine coated slides. Transfection of HeLa JW cells was performed using Fugene 6 reagent (Roche, Basel, Switzerland). For microscopy, cells were fixed with 4% paraformaldehyde and permeabilised with 0.15% Triton in cytoskeleton buffer (137 mM NaCl, 5 mM KCl, 1.1 mM Na2HPO4, 0.4 mM KH2PO4,, 2 mM MgCl2, 2 mM EGTA, 5 mM PIPES, 5.5 mM glucose). Monoclonal anti-α-tubulin B512 was from Sigma (St Louis, MO) and was used in immunofluorescence applications at 1:2000 dilution. The monoclonal AIM-1, which recognises Aurora B, was used at 1:200 dilution and was obtained from BD Transduction Laboratories (Lexington, KY). Rabbit polyclonal anti-topoisomerase II (29) was used at 1:50. Fluorescently labelled secondary antibodies were from Jackson Immunoresearch Laboratories (West Grove, PA) and were used at 1:200 dilution. Micrographs consist of single plane projections of deconvolved, three-dimensional data sets taken with a DeltaVision microscope (Applied Precision, Issaquah, WA) or of images captured with a CCD camera (Princeton Instruments, Trenton, NJ) from an Axioplan 2 microscope (Zeiss, Jena, Germany).
Chromosome preparation and immunoblotting
Chromosomes were prepared by sequential sucrose and Percoll gradients from colcemid-treated HeLa S3 cells according to Lewis and Laemmli (27). Unless otherwise stated, chromosomes were resuspended in 5 mM Tris–Cl pH 7.4, 2 mM KCl, 375 µM spermidine (Sigma, St Louis MO), 0.01% Ammonyx Lo, 1 mM CaCl2 plus protease inhibitors (aprotinin, PMSF and chymostatin, leupeptin, antipain and pepstatin) and digested for 1 h at 4°C with 30 µg/ml micrococcal nuclease (Sigma). For partial extraction (27,35,36), chromosome suspensions were diluted 1:1 with 2× dextran sulphate/heparin lysis buffer [20 mM Tris–Cl pH 8.8, 20 mM Na-EDTA, 0.2% Ammonyx Lo, 0.4 mg/ml dextran sulphate (Amersham), 0.04 mg/ml heparin (Sigma)], incubated for 5 min on ice and spun for 10 min at 10 000 g. More extensive extractions were performed as previously described (27,36). Pellets were resuspended in SDS– polyacrylamide gel electrophoresis (SDS–PAGE) sample loading buffer and chromosome protein samples were resolved on SDS–PAGE gels and transferred to nitrocellulose membranes by standard means. Membrane strips were incubated with antibodies to ScII (37) at 1:2500, human anti-centromere antiserum GS (38) at 1:2500, anti-CENP-C (39) at 1:2500, anti-INCENP ra2 (40) at 1:1000 and anti-Aurora B (R902), prepared as described above, at 1:500. Antibody binding was visualised by incubation with 125I-Protein A (Amersham). Immunblotting with the monoclonal MPM-2 (Upstate Biotechnology, Lake Placid, NY) was performed at 1:1000 dilution and signal was visualised by enhanced chemiluminesence (Amersham).
Kinase assays
Chromosomes were resuspended in kinase buffer 1 (10 mM HEPES pH 7.5, 100 mM KCl), kinase buffer 2 (10 mM HEPES pH 7.5, 20 mM KCl), kinase buffer 3 (50 mM Tris–Cl pH 7.5, 100 mM KCl) or kinase buffer 4 (50 mM Tris–Cl pH 7.5, 20 mM KCl), containing 10 mM MgCl2 and 1 mM CaCl2, plus a protease inhibitor cocktail. Following micrococcal nuclease digestion as described above, heat treatment, if indicated, consisted of 10 min incubation at 65°C followed by at least 5 min incubation on ice. Topo IIα was heat-treated prior to its being incubated with or without Aurora B kinase in kinase buffer. Kinase assays were carried out in the presence of 1 mM ATP, 1 mM dithiothreitol, 20 mM EGTA and 2.5–5 µCi [γ-32P]ATP per 100 µl, along with fresh protease inhibitors, at 37°C for 20 min, at which point reactions were extracted as described above and prepared for SDS–PAGE.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS)
Protein identification was accomplished by peptide mass fingerprinting and sequence database searching (41). Proteins were separated by SDS–PAGE and relevant protein bands were cut from the gel by using a scalpel. In-gel digestion of protein by trypsin was performed as previously described (42), followed by sample preparation using miniaturised sample concentration/desalting techniques (43). Two separate mass spectrometry research laboratories were involved in these experiments. However, several proteins were identified in both sets of experiments, thereby confirming both protocols. For one preparation of proteins, the mass spectrometry system was Reflex IV (Bruker-Daltonics, Bremen, Germany), for another we used a PerSeptive Biosystems Voyager DE™STR MALDI-TOF mass spectrometer (Applied Biosystems, Foster City, CA). Peptide ion signals were assigned with a mass error <50 p.p.m. Lists of tryptic peptide masses were used to search protein sequence databases using the ProFound internet-based protein identification tool (44) available at http://prowl.rockefeller.edu/cgi-bin/ProFound or the MS-Fit tool (http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm). For positive identification, a MOWSE (45) score of 104 and at least 20% coverage of the protein by the peptide fragments was required for ‘small’ proteins (<100 kDa), but 5–10% coverage was allowed for high molecular weight proteins (>100 kDa). Where indicated, we used an ‘Exp value’, which is a probability based expectation value which in all cases provided a significant score for a positive protein identification.
RESULTS
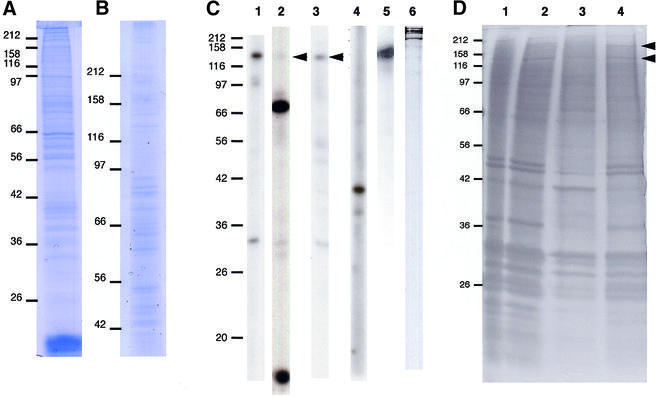
A metaphase chromosome fraction can be prepared from colcemid-treated tissue culture cells by Dounce homogenisation and sequential density gradients (27). The metaphase chromosome scaffold generated by stringent extraction of histones from this fraction includes the kinetochore and contains several prominent protein bands upon SDS–PAGE resolution, notably topoisomerase II and ScII (27,29,32, 35,36). A less stringent procedure was followed here, to retain more chromosome-associated material in the search for candidate Aurora B substrates. To confirm that the resultant fraction, shown in Figure 1A and B, indeed contained the chromosome scaffold proteins, we performed immunoblot analysis with antibodies specific for various proteins expected to be in the scaffold. As shown in Figure 1C, this partially-extracted scaffold contained CENP-A, CENP-B, CENP-C, INCENP, Aurora B and ScII. The monoclonal antibody MPM-2, which recognises a mitotic phosphoepitope, also detected a number of bands. To confirm the reproducibility of this extraction protocol, 11 separate chromosome preparations were run on the same one-dimensional gel. Visual inspection of the band patterns of these different preparations revealed them to have essentially the same composition (data not shown). To examine how this extraction procedure compared with the more stringent protocols previously published, we performed gel electrophoresis on equivalent amounts of chromosomal protein extracted by differing methods. As shown in Figure 1D, there exist clear differences between the protein scaffold fractions prepared by the different extraction methods, but further analysis will be necessary to determine to what extent the content of minor, possibly unknown, components of the scaffolds is increased under the different conditions.
Figure 1.
Preparation of chromosome scaffold fractions from colcemid-treated HeLa S3 cells. Proteins were separated by 12.5% (A) and 7.5% (B) SDS–PAGE and gels stained with Coomassie Blue. Apparent molecular masses are shown in kDa on the left. (C) Immunoblot characterisation of the partially-extracted chromosome scaffold fraction. After SDS–PAGE, proteins were transferred to a nitrocellulose membrane and strips of the same membrane were hybridised with antibodies as follows: lane 1, anti-INCENP; lane 2, anti-centromere antibodies (ACA); lane 3, anti-CENP-C; lane 4, anti-Aurora B; lane 5, anti-ScII; lane 6, MPM-2 monoclonal. Arrowheads indicate the CENP-C polypeptide recognised by the ACA and the CENP-C-specific antibody. Apparent molecular masses are shown in kDa on the left. (D) Comparison of scaffold fractions resulting from extraction under different conditions. A 12.5% polyacrylamide gel was silver stained after separation of proteins by electrophoresis. To control for protein loading, different cell equivalents were used as follows: lane 1, total chromosomes from 106 cells; lane 2, partially-extracted chromosome scaffold prepared as described in this paper from 5 × 106 cells; lane 3, chromosome scaffolds extracted by dextran sulphate-heparin as described (36) from 12.5 × 106 cells; lane 4, chromosome scaffolds extracted by NaCl as described (36) from 12.5 × 106 cells. Arrowheads indicate the two abundant protein bands typically found in chromosome scaffolds, ScI and ScII. Apparent molecular masses are shown in kDa on the left.
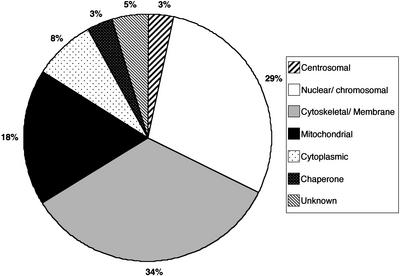
To define the components of this chromosome subfraction, we subjected the preparations to one-dimensional SDS– PAGE (Fig. 1A and B) and excised individual bands from Coomassie-stained gels. MALDI-TOF analysis of the peptide composition of these bands was performed and proteins in the fraction identified by database comparison. Table 1 lists the proteins identified in this initial analysis of the extracted chromosome fraction and Figure 2 summarises these results graphically. Of the 62 proteins identified in the fraction, 29% are nuclear or chromosome-associated. Some 34% are primarily associated with membranes or the cytoskeleton, 18% are mitochondrial, 3% centrosomal and the remainder are from the cytoplasm, act as chaperones or have unknown properties. Since the extent to which the cytoskeletal proteins interact with the chromosomes in forming a scaffold is not clear, some of the cytoskeletal proteins may actually represent chromosome-associated material, rather than artefactual or contaminant protein. We hypothesise that the mitochondria may be co-fractionating with the chromosomes in the Percoll density gradients, which would explain the relatively high proportion of mitochondrial proteins identified.
Table 1. Classification of proteins identified from partially-extracted metaphase chromosomes by intracellular localisation.
| Apparent mass (kDa) | Protein | MOWSE | Calculated mass | Coverage (%) | GenBank accession no. | Classification |
|---|---|---|---|---|---|---|
| >212 | A kinase anchor protein 9 (PRKA9, AKAP450, AKAP350) | 1.78E + 09 | 453671.0 | 8 | Q99996 | C |
| 43 | Actin | 1.76E + 08 | 40220.4 | 51 | BC012854 | S |
| >212 | Actin cross-linking family protein 7 | 2.80E + 05 | 620365.6 | 9 | Q9UPN3 | S |
| 29 | Adenine nucleotide translocator-2 | 1.94E + 04 | 32895.4 | 42 | M57424 | M |
| >212 | Ankyrin | 1.34E + 04 | 206278.5 | 8 | P16157 | S |
| 50 | APC2 protein | 5.80E + 04 | 80877.7 | 16 | AJ012652 | N |
| >212 | Apolipoprotein D-100 | 1.54E + 15 | 515566.9 | 11 | P04114 | Y |
| 50 | ATP synthase α subunit | 2.35E + 11, 8.3E – 5a | 59750.9 | 46, 31b | D14710 | M |
| 49 | ATP synthase β subunit | 1.54E + 11, 4.2E – 3a | 56560.2 | 44, 20b | BC010111 | M |
| 50 | β tubulin | 5.24E + 04 | 49907.3 | 17 | X79535 | S |
| 66 | BiP | 9.25E + 13, 0.039a | 72333.3 | 50, 17b | AJ271729 | S |
| >212 | Breast cancer type 2 susceptibility protein | 5.51E + 06 | 384228.5 | 6 | P51587 | N |
| >212 | Bullous pemphigoid antigen 1 | 8.43E + 07 | 313090.5 | 12 | Q03001 | S |
| >212 | Cadherin 23 precursor (Otocadherin) | 7.54E + 10 | 369527.8 | 9 | Q9H251 | S |
| 80 | Carnithine palmitoyltransferase I | 1.22E + 09 | 86239.6 | 30 | BC000185 | M |
| <20 | Cdc23 | 1.95E + 04 | 68285.9 | 17 | AB011472 | N |
| >212 | CENP-E | 3.77E + 07 | 312092.9 | 12 | Q02224 | N |
| ≥158 | Chromosome-associated polypeptide C | 0.016a | 147090 | 14b | NP_005487 | N |
| >212 | Chromodomain helicase-DNA-binding protein 4 | 7.89E + 05 | 217992.4 | 8 | Q14839 | N |
| >212 | Ciliary dynein | 1.14E + 07 | 511935.6 | 8 | Q9NYC9 | Y |
| >212 | Collagen α 3 (VI) chain precursor | 1.44E + 04 | 343555.2 | 4 | P12111 | S |
| 90 | Cullin 3 | 4.16E + 08 | 88930.7 | 29 | AF064087 | N |
| 62 | DEAD/H box polypeptide 30 | 2.1E – 3a | 124173 | 14b | XM_084133 | N |
| >212 | Desmoplakin | 3.36E + 10 | 331778.6 | 15 | P15924 | S |
| 170 | DNA topo II α | 1.95E + 04, 0.014a | 174386.6 | 10, 6b | P11388 | N |
| 170 | DNA topo II β | 3.17E + 04 | 183298.7 | 7 | Q02880 | N |
| >212 | DNA-dependent protein kinase | 9.94E + 17 | 469145.2 | 16 | P78527 | N |
| >212 | Dystrophin | 2.33E + 08 | 426680.1 | 9 | P11532 | S |
| 82 | Glutaminyl-tRNA synthetase | 3.43E + 05 | 87799.2 | 21 | X76013 | Y |
| <20 | Glycerol-3-phosphate dehydrogenase | 8.20E + 03 | 44734.8 | 23 | U79250 | M |
| 76 | GTP-binding protein NGB/CRFG | 0.020a | 73786 | 12b | AF120334 | N |
| 66 | Heat shock protein, 71 kDa | 2.27E + 09 | 70898.4 | 40 | Y00371 | H |
| 62 | Hypothetical protein FLJ10709 | 0.026a | 66177 | 15b | NP_060658 | U |
| 150 | Isoleucyl-tRNA synthetase | 1.58E + 07 | 144959.9 | 12 | P41252 | Y |
| >212 | Kendrin | 7.66E + 10 | 376355 | 12 | O95613 | C |
| 90 | Keratin 1 | 8.93E + 09 | 66018.0 | 35 | M98776 | S |
| 52 | Keratin 8 | 4.8E – 3a | 53671 | 25b | NP_002264 | S |
| 50 | Keratin 17 | 9.3E – 3a | 48076 | 34b | NP_000413 | S |
| 46 | KIAA1692 protein | 3.51E + 04 | 97050.6 | 20 | AB051479 | U |
| 95 | Ksam/FGFR2 | 1.15E + 07 | 92832.7 | 23 | AB030075 | S |
| 66 | Lamin A/C | 4.08E + 08 | 65135.1 | 21 | BC000511 | N |
| 95 | MAD1 | 8.16E + 07 | 83067.3 | 40 | BC009964 | N |
| 33 | Mitochondrial carrier homologue 2 | 1.29E + 04 | 33331.1 | 37 | BC000875 | M |
| 82 | Mitofilin, inner membrane protein mitochondrion | 2.24E + 10 | 83668.3 | 35 | D21092 | M |
| >212 | Monocytic leukemia zinc finger protein | 7.14E + 05 | 225057.8 | 10 | Q92794 | N |
| >212 | Myosin | 1.02E + 08 | 224037.7 | 15 | P11055 | S |
| >212 | Myosin XV | 7.92E + 10 | 395177.8 | 9 | Q9UKN7 | S |
| 66 | NADH dehydrogenase precursor, 75kDa subunit | 2.41E + 08, 9.8E – 3a | 79574.1 | 20, 20b | X61100 | M |
| >212 | Plectin1 | 6.83E + 17 | 531741.0 | 17 | Q15149 | Y |
| 29 | Prohibitin | 1.87E + 04 | 29804.2 | 34 | S85655 | M |
| >212 | Protein-tyrosine phosphatase delta precursor | 8.33E + 06 | 214761.7 | 13 | P23468 | N |
| >212 | Ryanodine receptor 2 | 9.68E + 09 | 564503.5 | 8 | Q92736 | N |
| >212 | Sacsin | 1.80E + 07 | 436980.0 | 6 | Q9NZJ4 | H |
| >212 | Spectrin α chain, fodrin | 6.61E + 15 | 284284.3 | 20 | Q13813 | S |
| >212 | Spectrin β chain | 1.64E + 05 | 288987.7 | 7 | Q9H254 | S |
| >212 | Talin | 4.99E + 07 | 269720.3 | 9 | Q9Y490 | S |
| 62 | TOB3 AAA ATPase | 3.96E + 09 | 66218.6 | 38 | AK001571 | M |
| 40 | Tubulin, β | 7.23E + 05 | 50433.0 | 25 | BC000748 | S |
| 95 | Un-named protein product | 7.56E + 06 | 69753.6 | 19 | AK023035 | U |
| >212 | Utrophin | 5.07E + 08 | 394497.6 | 11 | P46936 | S |
| 33 | VDAC (porin1) | 6.52E + 06 | 30722.7 | 50 | AJ250032 | M |
| >212 | Zinc finger protein HRX | 1.56E + 06 | 431895.2 | 6 | Q03164 | N |
Each protein identified was given only a single classification. Abbreviated classifications were: C, centrosomal; H, chaperone; M, mitochondrial; N, nuclear/chromosomal; S, cytoskeletal/membrane component; U, unknown; Y, cytoplasmic. Apparent masses were derived from the preparative gels from which MALDI-TOF identification was performed. Degradation and/or proteolytic cleavage is assumed to account for sizes significantly smaller than the published molecular weight. MOWSE scores were derived using the MS-Fit tool and ‘Exp values’a using the ProFound programme.
a‘Exp values’ obtained with ProFound analysis.
bCoverage in the experiments generating the Exp value.
Figure 2.
Putative subcellular distribution of 62 proteins identified by MALDI-TOF in the extracted chromosome fraction. Only one localisation was given for any one protein. Table 1 lists these proteins and their individual assignations to a subcellular localisation.
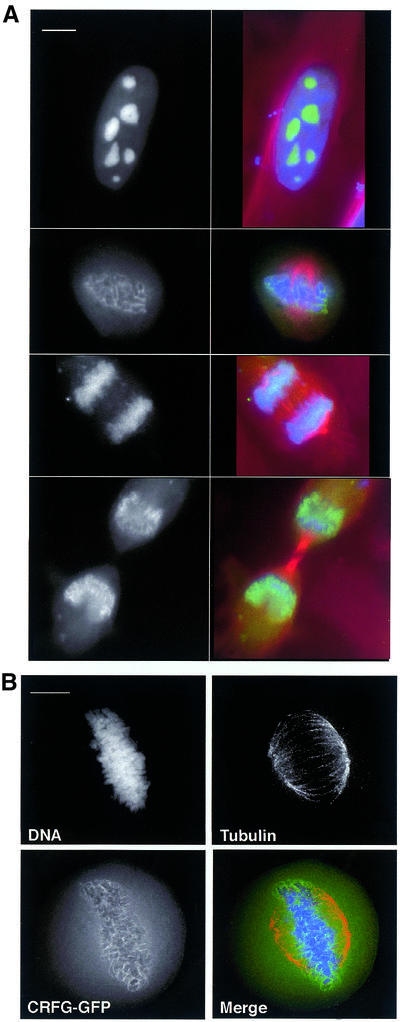
To test whether the identification of proteins by this method can define new chromosomal proteins, we examined the localisation of the putative GTP-binding protein NGB/CRFG during mitosis. Green fluorescent protein (GFP)-tagging in pEGFP-C1 and transfection of the gene encoding this protein into HeLa JW cells revealed that it forms subnuclear assemblies in interphase, as has been noted previously (46), but that it is associated with chromosomes throughout mitosis (Fig. 3). Transfection with pEGFP-N1-CRFG did not result in any detectable GFP signal and use of the vector alone gave rise to the typical diffuse staining of GFP throughout the cell cycle (data not shown). The perichromosomal localisation, seen clearly in metaphase (Fig. 3B), has been observed for a number of nucleolar proteins, but does not exclude a role for this protein in mitotic chromosomes, nuclear function or in cell cycle control (47,48). These findings confirm that the analysis of scaffolds in this manner is a useful means to discover new proteins associated with mitotic chromosomes.
Figure 3.
Localisation of NGB/CRFG as a chromosomal protein in mitosis. (A) 105 HeLa JW cells were transiently transfected with 2 µg pEGFP-C1-CRFG. Cells were fixed 24 h after transfection as described, and were immunostained for α-tubulin and stained with DAPI. Cells in interphase and in different stages of mitosis are shown. Black-and-white images show GFP-CRFG staining, which is shown in green in the merged images. Tubulin staining is shown in red and DNA in blue. Images are of single focal planes. Scale bar is 10 µm. (B) Deconvolved image of a transfected cell in metaphase to show the perichromosomal localisation of the GFP-CRFG signal more clearly. Separate channels are shown as indicated and the merged image uses the same colour scheme as in (A).
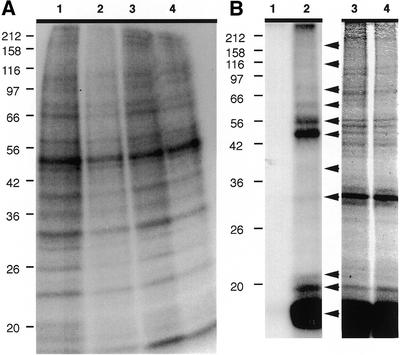
Since Aurora B itself was shown to be in the chromosome fraction, we tested whether there was a kinase activity associated with the preparation. A number of different buffer conditions were assayed and a reproducible pattern of phosphorylated bands was found to result upon incubation of the chromosomes with [γ-32P]ATP (Fig. 4A), irrespective of the buffer used. This activity was lost upon heat treatment (Fig. 4B). At present, the identities of the endogenous kinases that carry out this phosphorylation are unknown.
Figure 4.
(A) Assay for kinase activity in chromosome scaffold fraction. Lanes 1–4 show autoradiographic image of an SDS–polyacrylamide gel used to separate proteins after chromosomes in kinase buffers 1–4, respectively, were incubated with [γ-32P]dATP and then extracted. Apparent molecular masses are shown in kDa on the left. (B) Assay for Aurora B kinase substrates in chromosome scaffold fraction. Chromosomes were heat-treated and then incubated in the presence of [γ-32P]dATP without (lanes 1 and 3) or with (lanes 2 and 4) recombinant Aurora B and extracted before being subjected to SDS–PAGE. Lanes 1 and 2 show an autoradiographic image of a 12.5% gel and lanes 3 and 4 show the same gel stained with Coomassie Blue. Arrowheads indicate phosphorylated proteins and apparent molecular masses are shown in kDa on the left.
To identify new potential substrates of Aurora B, we next incubated the heat-inactivated, extracted chromosomes with recombinant human Aurora B kinase. As shown in Figure 4B, incubation of the chromosomes with the recombinant enzyme resulted in the phosphorylation of a number of proteins, notably giving rise to a strong band at the approximate size of the histones. Distinct bands were observed that indicated substrates with apparent molecular masses of approximately 16, 20, 22, 32, 39, 52, 56, 62, 76, 105 and 170 kDa (Fig. 4B). That these were not the same as those found following the incubation of the chromosomes with radiolabel but without recombinant enzyme indicates that Aurora B is not the only kinase in the fraction and that the recombinant enzyme we prepared has a restricted specificity for its activity. Next, to identify candidate substrates for Aurora B, we performed one-dimensional SDS–PAGE and excised bands that corresponded in size to those phosphorylated in parallel experiments (Fig. 4B). MALDI-TOF analysis of these bands identified the proteins listed in Table 2. MALDI-TOF analysis of a negative control region, which contained no distinct bands, gave a complex mixture of peptides and no protein was identified. This list includes a number of potentially relevant Aurora B substrates, based on intracellular distribution. It should be noted that the most common investigator-derived, artefactual keratin contaminants are keratins 1, 2 and 10, so that the keratins 8 and 17 found here are likely real components of the HeLa preparation. Three nuclear proteins—the GTP-binding protein NGB, topo IIα and CAP-C—were identified as candidate Aurora B substrates in this screen, with the DEAD/H box polypeptide 30 representing a possible fourth. It should be noted that the experiment performed to identify potential Aurora B substrates (using ProFound) was carried out at the beginning of our study, separately to the work used to generate the bulk of the data in Table 1 (using MS-Fit, generating MOWSE scores), so that the peptides identified in the screen in Table 2 are clearly not the only components of the bands we examined.
Table 2. Candidate Aurora B substrates identified by MALDI-TOF MS from partially-extracted metaphase chromosomes.
| Apparent mass (kDa) | Proteins identified |
|---|---|
| 52 | Keratin 17 |
| 56 | ATP synthase, H+ transporting, mitochondrial F1 complex α subunit; ATP synthase, β subunit; keratin 8 |
| 62 | DEAD/H box polypeptide 30; hypothetical protein FLJ10709 |
| 76 | GTP-binding protein NGB; NADH-ubiquinone oxidoreductase 75 kDa subunit precursor; BiP protein |
| 170 | Topo IIα; chromosome-associated polypeptide C (CAP-C/SMC-4) |
Note that additional data are provided for each of these identifications in Table 1; the ProFound analysis refers to these proteins.
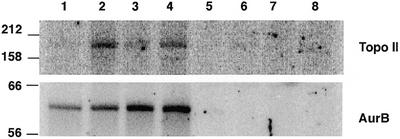
Since it is known that topo IIα is a phosphoprotein (49–54), we then tested whether it could serve as an in vitro substrate for Aurora B. Recombinant topo IIα and recombinant Aurora B were co-incubated in the presence of [γ-32P]ATP and we found that Aurora B did indeed phosphorylate the topoisomerase, as well as itself (Fig. 5). As a control for the specificity of this phosphorylation, we incubated topo IIα with a recombinant kinase-inactive Aurora B in which a key active site residue was mutated from lysine to arginine (7). This protein failed to phosphorylate topo IIα (Fig. 5), confirming that this is an activity derived only from functional Aurora B. It is noteworthy here that the Aurora B activity was very dependent on buffer choice, even though the kinase was active on chromosomes to essentially the same extent in either Tris- or HEPES-containing buffers (data not shown). This may reflect the likely presence of co-factors necessary for optimal activity of the enzyme, e.g. INCENP, in the chromosome preparation, which are not available to the reaction with recombinant protein. Attempts to localise phosphorylation sites using Fe(III)-IMAC and electrospray tandem mass spectrometry (55) revealed one endogenously phosphorylated residue in the recombinant topoisomerase II. No Aurora B phosphorylation sites in topo IIα were identified, due perhaps to the low efficiency of the phosphorylation activity and to the large size and complexity of the topoisomerase molecule (i.e., there are approximately 200 serines and threonines in the peptide sequence). Since there is, as yet, no clear consensus site for the Aurora B kinase, it has been difficult to further explore this observation.
Figure 5.
Confirmation of topo IIα as an Aurora B substrate in vitro, showing an autoradiograph of an SDS–polyacrylamide gel on which recombinant, heat-treated topo IIα was resolved following incubation with wild-type GST-Aurora B (lanes 1–4) or GST-Aurora B K106R (lanes 5–8) in kinase buffer 1 (lanes 1 and 5), kinase buffer 2 (lanes 2 and 6), kinase buffer 3 (lanes 3 and 7) or kinase buffer 4 (lanes 4 and 8). Apparent molecular masses are shown in kDa on the left.
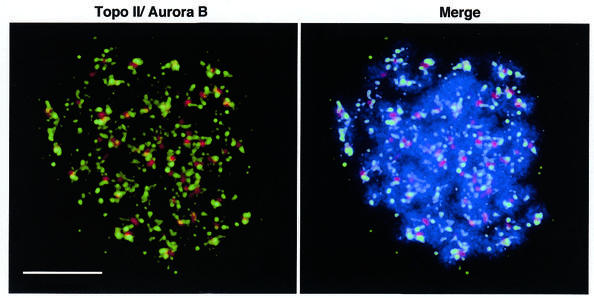
In order for the in vitro data to have any significance in vivo, topoisomerase II and Aurora B must be able to interact during mitosis. Both are known to be nuclear proteins during mitosis and to examine the relationship between them we performed immunofluorescence localisation experiments. As shown in Figure 6, topo IIα is located along the axis of the chromosomes, while Aurora B is centromeric, as expected. These findings show that at least a subfraction of the topo IIα population is available for phosphorylation by Aurora B and, given recent evidence for the mobility of the topoisomerase (56,57), suggests that Aurora B may be able to phosphorylate a significant amount of this substrate during mitosis.
Figure 6.
Immunofluorescence localisation of topo IIα and Aurora B. Colcemid-treated, hypotonically swollen HeLa S3 cells were spread and immunostained as described. Deconvolved images show staining for topoisomerase II (green) and Aurora B (red). DNA is shown in blue. Scale bar is 5 µm.
DISCUSSION
Here we present the first mass spectrometry-driven proteome analysis of metaphase chromosomes prepared from HeLa S3 cells. The preparative method used has been previously used successfully in the identification of CENP-E, DNA topoisomerase II and condensin (28–31), all of which are found in the preparation we describe here. Despite the levels of cytoskeletal and mitochondrial material, the useful percentage of chromosome-associated material found in this preparation makes it an attractive source for further analysis. It is important to note that the relative amount of any given protein in the preparation cannot be assessed by the techniques we used here, as the various peptides from tryptic digests may behave differently during mass spectrometry—indeed, the relatively low scores obtained for the known scaffold component, topoisomerase II, emphasise this point. We attribute the absence of many of the proteins known to be in the scaffold (e.g. the CENP proteins) from the MALDI-TOF profile to their relatively low abundance and to the possibility of their being poor subjects for mass spectrometric analysis. Our observations suggest that the further use of mass spectrometry against a more stringently-extracted scaffold fraction might reveal novel components of the chromosome scaffold that have not been identified by antibodies.
The further characterisation of the unknown and/or poorly-characterised proteins identified in the current screen will require their localisation. This was done for the putative GTP-binding protein NGB/CRFG by fluorescently tagging it and overexpressing it in tissue culture cells. The perichromosomal localisation, seen clearly in metaphase, does not clearly define the likely role of this protein (58–62). It may reflect a nucleolar function for the protein (47), as might be expected from its apparent localisation in interphase (46). However, it may also indicate some role in cell cycle control—a similar mitotic localisation has also been observed for the BCR oncogene product (47,48), which interacts with and activates GTP-binding proteins. The chromosomal localisation of NGB/CRFG we describe here may, therefore, be in some way related to its putative role in renal disease (46), but very little is known about this protein to date.
In the search for new Aurora B substrates, the most likely candidates are those known to have an association with chromosomes or the spindle, such as CAP-C/SMC-4 and topo IIα. Investigation of the Xenopus condensin complex showed no mitotic phosphorylation of CAP-C (63). Therefore, topo IIα, which has been well described as a phosphoprotein (49–54), was tested as an Aurora B substrate. Recombinant Aurora B indeed phosphorylated recombinant topo IIα in vitro. The functions of DNA topo IIα during mitosis have long been of interest and its phosphorylation, which varies throughout the cell cycle and is regulated by a number of kinases, appears to be of great importance in modulating its activities (64,65). While the localisation of topo IIα during mitosis is somewhat controversial (53,56,66–68), its locations are consistent with its being a potential Aurora B substrate. The localisation of topo IIα at the centromere (69) further enhances the likelihood of the Aurora B interaction being significant, but our inability to specify the target residue(s) by mass spectrometry has hampered our efforts to define this significance. To date, no consensus Aurora B phosphorylation signal has been described, so we cannot speculate on the potential target residues on topo IIα. While there remain further candidate substrates to be identified, the complexity of the metaphase chromosome fraction may necessitate the use of two-dimensional electrophoresis to resolve phosphorylated proteins sufficiently for their conclusive identification.
In conclusion, we present the proteomic analysis of partially-extracted metaphase chromosomes as a means by which novel mitotic chromosome components may be described and by which chromosomal kinase substrates may be identified. The ability to remove kinase activity endogenous to the preparation by heat treatment means that any kinase may be tested on chromosomes and the more abundant of its potential substrates described.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Rhys C. Roberts for assistance with phosphopeptide analysis by mass spectrometry and Sally P. Wheatley for help with microscopy. C.M. received an EMBO Long Term Postdoctoral Fellowship during this study. H.D. is the recipient of a Wellcome Trust Prize Studentship. Support was received from the Danish Natural Sciences Research Council to the Danish Biotechnology Instrument Center (O.N.J.). Work in N.O.’s laboratory was supported by NIH grant GM 33944. Work in W.C.E.’s laboratory is supported by the Wellcome Trust, of which he is a Principal Research Fellow.
REFERENCES
- 1.Nigg E.A. (2001) Mitotic kinases as regulators of cell division and its checkpoints. Nature Rev. Mol. Cell Biol., 2, 21–32. [DOI] [PubMed] [Google Scholar]
- 2.Adams R.R., Maiato,H., Earnshaw,W.C. and Carmena,M. (2001) Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction and chromosome segregation. J. Cell Biol., 153, 865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giet R. and Glover,D.M. (2001) Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol., 152, 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaitna S., Mendoza,M., Jantsch-Plunger,V. and Glotzer,M. (2000) Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol., 10, 1172–1181. [DOI] [PubMed] [Google Scholar]
- 5.Kallio M.J., McCleland,M.L., Stukenberg,P.T. and Gorbsky,G.J. (2002) Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol., 12, 900–905. [DOI] [PubMed] [Google Scholar]
- 6.Speliotes E.K., Uren,A., Vaux,D. and Horvitz,H.R. (2000) The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell, 6, 211–223. [DOI] [PubMed] [Google Scholar]
- 7.Terada Y., Tatsuka,M., Suzuki,F., Yasuda,Y., Fujita,S. and Otsu,M. (1998) AIM-1: a mammalian midbody associated protein required for cytokinesis. EMBO J., 17, 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams R.R., Carmena,M. and Earnshaw,W.C. (2001) Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell. Biol., 11, 49–54. [DOI] [PubMed] [Google Scholar]
- 9.MacCallum D.E., Losada,A., Kobayashi,R. and Hirano,T. (2002) ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol. Biol. Cell, 13, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumara I., Vorlaufer,E., Stukenberg,P.T., Kelm,O., Redemann,N., Nigg,E.A. and Peters,J.-M. (2002) The dissociation of cohesin from chromosomes in prophase is regulated by polo-like kinase. Mol. Cell, 9, 515–525. [DOI] [PubMed] [Google Scholar]
- 11.Earnshaw W.C. and Bernat,R.L. (1990) Chromosomal passengers: towards an integrated view of mitosis. Chromosoma, 100, 139–146. [DOI] [PubMed] [Google Scholar]
- 12.Adams R.R., Wheatley,S.P., Gouldsworthy,A.M., Kandels-Lewis,S.E., Carmena,M., Smythe,C., Gerloff,D.L. and Earnshaw,W.C. (2000) INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol., 10, 1075–1078. [DOI] [PubMed] [Google Scholar]
- 13.Oegema K., Desai,A., Rybina,S., Kirkham,M. and Hyman,A.A. (2001) Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol., 153, 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uren A.G., Wong,L., Pakusch,M., Fowler,K.J., Burrows,F.J., Vaux,D.L. and Choo,K.H. (2000) Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol., 10, 319–328. [DOI] [PubMed] [Google Scholar]
- 15.Wheatley S.P., Carvalho,A., Vagnarelli,P. and Earnshaw,W.C. (2001) INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol., 11, 886–890. [DOI] [PubMed] [Google Scholar]
- 16.Sonoda E., Matsusaka,T., Morrison,C., Vagnarelli,P., Hoshi,O., Ushiki,T., Nojima,K., Fukagawa,T., Waizenegger,I.C., Peters,J.-M. et al. (2001) Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell., 1, 759–770. [DOI] [PubMed] [Google Scholar]
- 17.Hsu J.-Y., Sun,Z.-W., Li,X., Reuben,M., Tatchell,K., Bishop,D.K., Grushcow,J.M., Brame,C.J., Caldwell,J.A., Hunt,D.F. et al. (2000) Mitotic phosphorylation of histone H3 is governed by Ipl/ aurora kinase and glc7/PP1 phosphatase in budding yeast and nematodes. Cell, 102, 272–291. [DOI] [PubMed] [Google Scholar]
- 18.Zeitlin S.G., Shelby,R.D. and Sullivan,K.F. (2001) CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol., 155, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murata-Hori M., Fumoto,K., Fukuta,Y., Iwasaki,T., Kikuchi,A., Tatsuka,M. and Hosoya,H. (2000) Myosin II regulatory light chain as a novel substrate for AIM-1, an aurora/Ipl1p-related kinase from rat. J. Biochem. (Tokyo), 128, 903–907. [DOI] [PubMed] [Google Scholar]
- 20.Bishop J.D. and Schumacher,J.M. (2002) Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B kinase stimulates Aurora B kinase activity. J. Biol. Chem., 277, 27577–27580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biggins S., Severin,F.F., Bhalla,N., Sassoon,I., Hyman,A.A. and Murray,A.W. (1999) The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev., 13, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sassoon I., Severin,F.F., Andrews,P.D., Taba,M.R., Kaplan,K.B., Ashford,A.J., Stark,M.J., Sorger,P.K. and Hyman,A.A. (1999) Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev., 13, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka T.U., Rachidi,N., Janke,C., Pereira,G., Galova,M., Sciebel,E., Stark,M.J.R. and Nasmyth,K. (2002) Evidence that the Ipl1-Sli15 (Aurora kinase- INCENP) complex promotes chromosome bi-orientation by latering kinetochore-spindle pole connections. Cell, 108, 317–329. [DOI] [PubMed] [Google Scholar]
- 24.Andersen J.S., Lyon,C.E., Fox,A.H., Leung,A.K., Lam,Y.W., Steen,H., Mann,M. and Lamond,A.I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol., 12, 1–11. [DOI] [PubMed] [Google Scholar]
- 25.Mann M., Hendrickson,R.C. and Pandey,A. (2001) Analysis of proteins and proteomes by mass spectrometry. Annu. Rev. Biochem., 70, 437–473. [DOI] [PubMed] [Google Scholar]
- 26.Mintz P.J., Patterson,S.D., Neuwald,A.F., Spahr,C.S. and Spector,D.L. (1999) Purification and biochemical characterization of interchromatin granule clusters. EMBO J., 18, 4308–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis C.D. and Laemmli,U.K. (1982) Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell, 29, 171–181. [DOI] [PubMed] [Google Scholar]
- 28.Compton D.A., Yen,T.J. and Cleveland,D.W. (1991) Identification of novel centromere/kinetochore-associated proteins using monoclonal antibodies generated against human mitotic chromosome scaffolds. J. Cell Biol., 112, 1083–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Earnshaw W.C., Halligan,B., Cooke,C.A., Heck,M.M. and Liu,L.F. (1985) Topoisomerase II is a structural component of mitotic chromosome scaffolds. J. Cell Biol., 100, 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fields A.P. and Shaper,J.H. (1988) A major 62-kD intranuclear matrix polypeptide is a component of metaphase chromosomes. J. Cell Biol., 107, 833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yen T.J., Compton,D.A., Wise,D., Zinkowski,R.P., Brinkley,B.R., Earnshaw,W.C. and Cleveland,D.W. (1991) CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J., 10, 1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Earnshaw W.C., Halligan,N., Cooke,C. and Rothfield,N. (1984) The kinetochore is part of the metaphase chromosome scaffold. J. Cell Biol., 98, 352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kingma P.S., Greider,C.A. and Osheroff,N. (1997) Spontaneous DNA lesions poison human topoisomerase IIalpha and stimulate cleavage proximal to leukemic 11q23 chromosomal breakpoints. Biochemistry, 36, 5934–5939. [DOI] [PubMed] [Google Scholar]
- 34.Wasserman R.A., Austin,C.A., Fisher,L.M. and Wang,J.C. (1993) Use of yeast in the study of anticancer drugs targeting DNA topoisomerases: expression of a functional recombinant human DNA topoisomerase II alpha in yeast. Cancer Res., 53, 3591–3596. [PubMed] [Google Scholar]
- 35.Adolph K.W., Cheng,S.M. and Laemmli,U.K. (1977) Role of nonhistone proteins in metaphase chromosome structure. Cell, 12, 805–816. [DOI] [PubMed] [Google Scholar]
- 36.Earnshaw W.C. and Laemmli,U.K. (1983) Architecture of metaphase chromosomes and chromosome scaffolds. J. Cell Biol., 96, 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitoh N., Goldberg,I.G., Wood,E.R. and Earnshaw,W.C. (1994) ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J. Cell Biol., 127, 303–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Earnshaw W.C. and Rothfield,N. (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma, 91, 313–321. [DOI] [PubMed] [Google Scholar]
- 39.Tomkiel J.E., Cooke,C.A., Saitoh,H., Bernat,R.L. and Earnshaw,W.C. (1994) CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J. Cell Biol., 125, 531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackay A.M., Eckley,D.M., Chue,C. and Earnshaw,W.C. (1993) Molecular analysis of the INCENPs (inner centromere proteins): separate domains are required for association with microtubules during interphase and with the central spindle during anaphase. J. Cell Biol., 123, 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen O.N., Larsen,M.R. and Roepstorff,P. (1998) Mass spectrometric identification and microcharacterization of proteins from electrophoretic gels: strategies and applications. Proteins Struct. Func. Genet., Suppl 2, 74–89. [DOI] [PubMed] [Google Scholar]
- 42.Shevchenko A., Wilm,M., Vorm,O. and Mann,M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- 43.Gobom J., Nordhoff,E., Mirgorodskaya,E., Ekman,R. and Roepstorff,P. (1999) Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom., 34, 105–116. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W. and Chait,B.T. (2000) ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem., 72, 2482–2489. [DOI] [PubMed] [Google Scholar]
- 45.Pappin D.J.C., Hojrup,P. and Bleasby,A.J. (1993) Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol., 3, 327–332. [DOI] [PubMed] [Google Scholar]
- 46.Laping N.J., Olson,B.A. and Zhu,Y. (2001) Identification of a novel guanosine triphosphate-binding protein differentially expressed in renal disease. J. Am. Soc. Nephrol., 12, 883–890. [DOI] [PubMed] [Google Scholar]
- 47.Hernandez-Verdun D. and Gautier,T. (1994) The chromosome periphery during mitosis. Bioessays, 16, 179–185. [DOI] [PubMed] [Google Scholar]
- 48.Wetzler M., Talpaz,M., Yee,G., Stass,S.A., Van Etten,R.A., Andreeff,M., Goodacre,A.M., Kleine,H.D., Mahadevia,R.K. and Kurzrock,R. (1995) Cell cycle-related shifts in subcellular localization of BCR: association with mitotic chromosomes and with heterochromatin. Proc. Natl Acad. Sci. USA, 92, 3488–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Escargueil A.E., Plisov,S.Y., Filhol,O., Cochet,C. and Larsen,A.K. (2000) Mitotic phosphorylation of DNA topoisomerase IIα by protein kinase CK2 creates the MPM-2 phosphoepitope on Ser-1469. J. Biol. Chem., 275, 34710–34718. [DOI] [PubMed] [Google Scholar]
- 50.Heck M.M., Hittelman,W.N. and Earnshaw,W.C. (1989) In vivo phosphorylation of the 170-kDa form of eukaryotic DNA topoisomerase II. Cell cycle analysis. J. Biol. Chem., 264, 15161–15164. [PubMed] [Google Scholar]
- 51.Ishida R., Iwai,M., Marsh,K.L., Austin,C.A., Yano,T., Shibata,M., Nozaki,N. and Hara,A. (1996) Threonine 1342 in human topoisomerase IIalpha is phosphorylated throughout the cell cycle. J. Biol. Chem., 271, 30077–30082. [DOI] [PubMed] [Google Scholar]
- 52.Kimura K., Nozaki,N., Enomoto,T., Tanaka,M. and Kikuchi,A. (1996) Analysis of M phase-specific phosphorylation of DNA topoisomerase II. J. Biol. Chem., 271, 21439–21445. [DOI] [PubMed] [Google Scholar]
- 53.Taagepera S., Rao,P.N., Drake,F.H. and Gorbsky,G.J. (1993) DNA topoisomerase IIα is the major chromosome protein recognized by the mitotic phosphoprotein antibody MPM-2. Proc. Natl Acad. Sci. USA, 90, 8407–8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wells N.J., Fry,A.M., Guano,F., Norbury,C. and Hickson,I.D. (1995) Cell cycle phase-specific phosphorylation of human topoisomerase II alpha. Evidence of a role for protein kinase C. J. Biol. Chem., 270, 28357–28363. [DOI] [PubMed] [Google Scholar]
- 55.Stensballe A. and Jensen,O.N. (2001) Simplified sample preparation method for protein identification by matrix-assisted laser desorption/ionization mass spectrometry: in-gel digestion on the probe surface. Proteomics, 1, 955–966. [DOI] [PubMed] [Google Scholar]
- 56.Christensen M.O., Larsen,M.K., Barthelmes,H.U., Hock,R., Andersen,C.L., Kjeldsen,E., Knudsen,B.R., Westergaard,O., Boege,F. and Mielke,C. (2002) Dynamics of human DNA topoisomerases IIα and IIβ in living cells. J. Cell Biol., 157, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tavormina P.A., Come,M.G., Hudson,J.R., Mo,Y.Y., Beck,W.T. and Gorbsky,G.J. (2002) Rapid exchange of mammalian topoisomerase II alpha at kinetochores and chromosome arms in mitosis. J. Cell Biol., 158, 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chaly N., Bladon,T., Setterfield,G., Little,J.E., Kaplan,J.G. and Brown,D.L. (1984) Changes in distribution of nuclear matrix antigens during the mitotic cell cycle. J. Cell Biol., 99, 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dilworth S.M. (1991) A perichromosomal region contains proteins phosphorylated during mitosis in Xenopus laevis cells. J. Cell Sci., 98, 309–315. [DOI] [PubMed] [Google Scholar]
- 60.McKeon F.D., Tuffanelli,D.L., Kobayashi,S. and Kirschner,M.W. (1984) The redistribution of a conserved nuclear envelope protein during the cell cycle suggests a pathway for chromosome condensation. Cell, 36, 83–92. [DOI] [PubMed] [Google Scholar]
- 61.Wataya-Kaneda M., Kaneda,Y., Sakurai,T., Sugawa,H. and Uchida,T. (1987) A monoclonal antibody against the nucleus reveals the presence of a common protein in the nuclear envelope, the perichromosomal region, and cytoplasmic vesicles. J. Cell Biol., 104, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yasuda Y. and Maul,G.G. (1990) A nucleolar auto-antigen is part of the major chromosomal surface component. Chromosoma, 99, 152–160. [DOI] [PubMed] [Google Scholar]
- 63.Kimura K., Hirano,M., Kobayashi,R. and Hirano,T. (1998) Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science, 282, 487–490. [DOI] [PubMed] [Google Scholar]
- 64.Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. [DOI] [PubMed] [Google Scholar]
- 65.Warburton P.E. and Earnshaw,W.C. (1997) Untangling the role of DNA topoisomerase II in mitotic chromosome structure and function. Bioessays, 19, 97–99. [DOI] [PubMed] [Google Scholar]
- 66.Earnshaw W.C. and Heck,M.M.S. (1985) Localization of topoisomerase II in mitotic chromosomes. J.Cell Biol., 100, 1716–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sumner A.T. (1996) The distribution of topoisomerase II on mammalian chromosomes. Chromosome Res., 4, 5–14. [DOI] [PubMed] [Google Scholar]
- 68.Swedlow J.R., Sedat,J.W. and Agard,D.A. (1993) Multiple chromosomal populations of topoisomerase II detected in vivo by time-lapse, three dimensional wide-field microscopy. Cell, 73, 97–108. [DOI] [PubMed] [Google Scholar]
- 69.Rattner J.B., Hendzel,M.J., Furbee,C.S., Muller,M.T. and Bazett-Jones,D.P. (1996) Topoisomerase II alpha is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J. Cell Biol., 134, 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]