Abstract
We have analyzed the mutagenic specificity of an abasic site in DNA using the yeast oligonucleotide transformation assay. Oligonucleotides containing an abasic site or its analog were introduced into B7528 or its derivatives, and nucleotide incorporation opposite abasic sites was analyzed. Cytosine was most frequently incorporated opposite a natural abasic site (O) (‘C-rule’), followed by thymine. Deletion of REV1 decreased the transformation efficiency and the incorporation of cytosine nearly to a background level. In contrast, deletion of RAD30 did not affect them. We compared the mutagenic specificity with that of a tetrahydrofuran abasic site (F), an abasic analog used widely. Its mutation spectrum was clearly different from that of O. Adenine, not cytosine, was most favorably incorporated. However, deletion of REV1 decreased the transformation efficiency with F-containing oligonucleotide as in the case of O. These results suggest that the bypass mechanism of F is different from that of O, although the bypasses in both cases are dependent on REV1. We also found that the mutagenic specificity of F can be affected by not only the adjacent bases, but also a base located two positions away from F.
INTRODUCTION
Abasic sites are common DNA lesions formed by hydrolysis of N-glycosidic bonds of nucleotides in DNA, which releases the DNA base but leaves the phosphodiester backbone intact. Hydrolysis of the glycosidic bond can occur spontaneously, or enzymatically as an intermediate step in base excision repair (1,2). Abasic sites are known to act as strong blocks to the progression of DNA polymerases in vitro, but they can also be highly mutagenic in vivo (1,3). For example, abasic sites occur during base excision repair of dU in DNA. To remove dU from DNA, uracil is released by the action of uracil-DNA glycosylase, the product of the UNG1 gene (4) in yeast. The resulting abasic site is cleaved by an AP-endonuclease, and then removal of the abnormal termini and repair DNA synthesis follow. In yeast, the major AP-endonuclease is the APN1 gene product (5), but the more recently discovered APN2 AP-endonuclease can also play a role in some cases (6,7). Although abasic sites that escape the action of the AP endonucleases are expected to block DNA polymerases strongly, translesion synthesis (TLS) also occurs. The non-instructive nature of the abasic lesion can make the TLS highly mutagenic. For TLS of an abasic site, Rev1p, the product of the REV1 gene, can insert dCMP opposite the abasic site (8,9). This terminal dCMP can then be extended by DNA polymerase ζ, composed of the REV3 and REV7 gene products (8,9). However, other pathways including other TLS polymerases, such as DNA polymerase η, the RAD30 gene product, may also contribute to TLS (10). In addition, a role for Rev1p other than its dCMP transferase activity has been suggested during TLS of abasic sites (7,11) and 6-4 photoproducts (11,12).
The mutagenic properties of abasic sites have been investigated under a variety of experimental conditions. In Escherichia coli, predominant incorporation of A opposite an abasic site has been known as ‘A-rule’ (13–21). Recent studies in eukaryotes suggest that the A-rule may not be a common mechanism for preferential incorporations opposite abasic sites. An essentially random incorporation of nucleotides opposite natural abasic sites was observed in COS7 cells (22–24). In another study in COS cells, predominant incorporation of A was observed opposite a tetrahydrofuran-type abasic site (F) (25). In human lymphoblastoid cells, G was incorporated preferentially opposite natural abasic sites (26). In NIH3T3 cell, T, and not A, was mainly incorporated opposite an F with point mutations in the adjacent positions (27).
The previous reports on the mutagenic specificity of abasic sites in budding yeast, Saccharomyces cerevisiae, are also inconsistent. C was predominantly incorporated opposite an abasic site using a shuttle vector system (8). In another study with the SUP4 gene on a plasmid, the frequency of spontaneous A:T → C:G events increased in the apn1 mutant, deficient in the AP-endonuclease, suggesting the preferential insertion of G opposite spontaneously occurring abasic sites (28). Recently, the preferential incorporation of A was shown from the mutation spectrum in the deletion strain of APN1 and APN2 after methyl methansulfonate treatment (7). In two cases it has shown that Rev1p is necessary for the bypass through abasic sites (7,8,11). However the contribution of dCMP transferase activity is not clear.
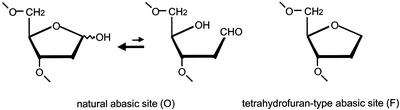
In this study, we investigated mutagenic specificities of uracil (U), a natural abasic site (O) and an F (Fig. 1) using oligonucleotide transformation method. A cyc1-31 mutation in a strain, B7528, can be reverted to wild-type phenotype Cyc+ by direct transformation with short oligonucleotides (29). We have applied this method to analyze mutagenic specificity of nucleotide analogs and a 6-4 photoproduct (12,30), using oligonucleotides with these lesions. Mutational specificity of the lesion can be easily determined by DNA sequencing analysis of the transformants under various genetic backgrounds. To improve the efficiency of transformation by F, a disruptant of APN1 gene coding the major AP-endonuclease which can process F was prepared. Deletion strains of REV1 and RAD30 of B7528 were used for the investigation of the contribution of DNA polymerases performing a TLS.
Figure 1.
Structures of abasic sites used in this study.
MATERIALS AND METHODS
Media
Escherichia coli strains were grown in LB medium that was supplemented with 100 µg/ml ampicillin when required (31). Yeast strains were grown in YPD medium (1% yeast extract, 2% peptone and 2% glucose), YPG medium (1% yeast extract, 2% peptone and 3% glycerol) or minimal selective medium (32). For solid media, 1.5% agar was added.
Strains
The E.coli strain DH5α cells were used to amplify and manipulate all plasmids described in this paper. The S.cerevisiae strains used in this study are listed in Table 1. All yeast strains were derived from B7528, which was provided by Dr Fred Sherman of Rochester University. Sequence information was provided by the Stanford Yeast Genomic Database (http://genome-www.stanford.edu/Saccharomyces/). Transformations for gene disruption of yeast were performed by the lithium acetate method (33). Transformants were streaked and purified on appropriate minimal selective medium or YPD medium containing 200 mg/l G418.
Table 1. Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| B7528 | MATa cyc1-31 cyc7-67 lys5-10 ura3-52 | Moerschell et al. (29) |
| COY7 | MATa cyc1-31 cyc7-67 lys5-10 ura3-52 apn1::hisG | This study |
| COY14 | MATa cyc1-31 cyc7-67 lys5-10 ura3-52 rad30::kanMX | This study |
| COY35 | MATa cyc1-31 cyc7-67 lys5-10 ura3-52 rev1::hisG-URA3-hisG | Otsuka et al. (12) |
| COY43 | MATa cyc1-31 cyc7-67 lys5-10 ura3-52 apn1::hisG rev1::hisG-URA3-hisG | This study |
| COY41 | MATa cyc1-31 cyc7-67 lys5-10 ura3-52 apn2::kanMX | This study |
A disruptant of APN1 was constructed by introducing hisG-URA3-hisG (34) inserted APN1 gene provided by Dr B. Demple of Harvard School of Public Health (5). URA3 marker was subsequently eliminated on 5-fluoro-orotic acid medium. Rev1 disruptants, rev1 and apn1 rev1, were constructed using hisG-URA3-hisG cassette as described (12). The rad30 disruption construct was generated using PCR-based gene disruption method with the kanMX module (35). PCR was performed using two primers, rad30DISLm (5′- TAGCGCAGGCCTGCTCATTTTTGAACGGCTTTGAT AAAACAAGACAAAGCCGTACGCTGCAGGTCGAC-3′) and rad30DSRm (5′-AGGACGTTTTAGTTGCTGAAG CCATATAATTGTCTATTTGGAATAGGATCGATGAAT TCGAGCTCG-3′). The resulting PCR product was introduced into B7528. Disruption was verified by PCR using primer sets VERRAD30L (5′-TAGTCTTCTAGCGCAGGC-3′) and VERRAD30R (5′-ATCGCCTTCAAACTTCAGAG-3′), and VERrad30RK2 (5′-AAACGATCTAATTGATTAAGTCC-3′) and KAN1 (5′-CCTCGACATCTGCCC-3′). The plasmid for apn2 disruption, pYORCYBLO19w (36), was kindly provided by Dr F. Malagon of Universidad de Sevilla. This plasmid has apn2::kanMX disruption. The apn2::kanMX fragment was released by digestion with HpaI and XhoI and used to transform B7528 and apn1. Disruption was confirmed by PCR analysis using primer sets, APN2L (5′-ATGGAGA AAAAGATGACAGGA-3′) and APN2R (5′-GAGGTTA CTGACGATGACC-3′), and KAN1 and APN2R.
A deletion strain of UNG1 was constructed from pMK201 that consists of a 9.1 kb insert containing the UNG1 gene as described (4) except that hisG-URA3-hisG was used as a selection marker (34). The plasmid pMK201 was provided by Dr Kim Gerik of Washington University School of Medicine.
Oligonucleotides
The oligonucleotides used directly for transformation in this study are listed in Table 2. Wild-type oligonucleotide (oligo-W) and 26mer dUMP-containing oligonucleotides (oligo-U) were purchased from Genset (Kyoto, Japan). To produce natural abasic site-containing oligonucleotide (oligo-O), oligo-U was treated with uracil-DNA glycosylase (Gibco BRL, Tokyo, Japan). Oligo-U (500 pmol) was incubated for 1 h at 37°C with 1 U of uracil-DNA glycosylase in 20 mM Tris–HCl (pH 8.0), 1 mM EDTA, 0.01 M DTT, 0.1 mg/l BSA. F-containing oligonucleotide (oligo-F) was synthesized using dSpacer amidite monomer purchased from Glen Research Corp. (Sterling, VA, USA).
Table 2. DNA sequences of the CYC1 target site and a list of 26mer oligonucleotides used for yeast transformation in the present study.
| CYC1 | (Met)Thr-Glu-Phe-Lys-Ala-Gly-Ser-Ala-Lys-Lys-Gly-----------ATAATGACTGAATTCAAGGCCGGTTCTGCTAAGAAAGGT------ |
| B7528 (cyc1-31) | (Met)Thr-Glu-Stop---ATAATGACTGAATA-AAGGCCGGTTCTGCTAAGAAAGGT------ |
| Oligo-W | 5′-ATAA1TGACTGAATTC12AAGGCCGGTTC23-3′ |
| Oligo-U | 5′-ATAATGACTGAATTUAAGGCCGGTTC-3′ |
| Oligo-O | 5′-ATAATGACTGAATTOAAGGCCGGTTC-3′ |
| Oligo-F | 5′-ATAATGACTGAATTFAAGGCCGGTTC-3′ |
| Oligo-AF | 5′-ATAATGACTGAACAFAAGGCCGGTTC-3′ |
| Oligo-CF | 5′-ATAATGACTGAACCFAAGGCCGGTTC-3′ |
| Oligo-GF | 5′-ATAATGACTGAACGFAAGGCCGGTTC-3′ |
| Oligo-TF | 5′-ATAATGACTGAACTFAAGGCCGGTTC-3′ |
The first and second lines show DNA sequences of wild-type CYC1 and cyc1-31 mutant, respectively. Nucleotides in the oligonucleotides are numbered by numbering the A residue in the ATG initiation codon as 1. The position of abasic analog is indicated by a bold letter. U, deoxyuridine; O, natural abasic site; F, tetrahydrofuran.
Oligonucleotide transformation and sequencing analysis of the transformants
The transformation was performed by electroporation method (12). In a typical experiment, 300 pmol of an oligonucleotide was added to 40 µl of competent cells. Colonies of transformants (Cyc+ strain) grew out of the uniform lawn of the untransformed cells (Cyc– strain) on YPD plates after 5 days incubation. Transformants that can grow on non-fermentable carbon sources were purified on YPG plates. DNA sequencing analysis of the transformants was made as described previously (12).
RESULTS
Efficiency of transformation with oligonucleotide containing abasic sites
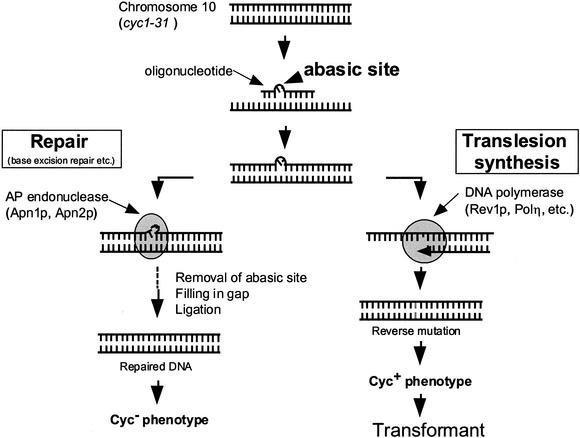
The strain B7528 and its derivatives used in this study have Cyc– phenotype because of the cyc1-31 mutation, a point mutation composed of one base change and one deletion as shown in Table 2. Oligonucleotides containing the wild-type sequence of CYC1 (oligo-W) can restore the wild-type reading frame to give Cyc+ transformants if the oligonucleotide is integrated into the correct chromosomal position. We have synthesized oligonucleotides containing an abasic site or dU at position 12. These modified oligonucleotides can still restore the wild-type reading frame if a nucleotide is incorporated opposite the modified site by TLS. Incorporation of any nucleotide at this position can restore Cyc+ phenotype (12,29,30,37). Assuming that oligonucleotides are incorporated at the same efficiency, the ratio of transformation efficiency with oligonucleotide containing a lesion to that with control oligonucleotide (oligo-W) indicates the frequency of read-through past the lesion that escapes repair (Fig. 2). The nucleotide incorporated opposite the lesion can be determined by DNA sequencing analysis.
Figure 2.
A putative mechanism of transformation with an oligonucleotide containing an abasic site. The S.cerevisiae CYC1 system was used for analyzing mutagenic specificity of abasic sites. The cyc1-31 mutant does not produce active iso-1-cytochrome c because of the frameshift/nonsense mutation on CYC1 gene. The mutant gene can be altered by transformation with synthetic oligonucleotides containing an abasic analog if a nucleotide is incorporated opposite the lesion by TLS.
B7528 and its derivatives were transformed with oligo-W, oligo-U, oligo-O or oligo-F. Transforming efficiencies are shown in Table 3. The efficiencies of oligonucleotide containing U and O were 6 and 7% of oligo-W in B7528, respectively. The transforming efficiencies of U were similar to those of O. This seems to result from the efficient removal of uracil by cellular uracil-DNA glycosylase as discussed below. In the case of oligo-F, the transforming efficiency in B7528 was 0.33% of oligo-W. The low transforming activities of modified oligonucleotides can be affected by the efficient removal of a lesion by a repair system, which causes the loss of the wild-type reading frame. To improve transformation efficiencies of the modified oligonucleotides, we disrupted either the APN1 or APN2 gene, which encode the major and minor AP-endonucleases. Indeed, the transforming efficiencies of oligo-F in apn1 were clearly increased, but not in the apn2 disruptant. This suggests that Apn1p, but not Apn2p, would cleave the 5′ end of F effectively in vivo.
Table 3. Transforming activities of oligonucleotides containing abasic analogs in B7528 and its derivatives.
| Relevant genotype | Oligonucleotide | Transformation efficiency Transformants/µg | % of control |
|---|---|---|---|
| Wild-type (B7528) | Oligo-W | 1599 ± 1172 | 100 |
| Oligo-U | 88 ± 48 | 6.0 ± 1.8 | |
| Oligo-O | 117 ± 68 | 7.2 ± 1.1 | |
| Oligo-F | 3.9 ± 2.1 | 0.33 ± 0.20 | |
| No oligo | 0.3 ± 0.8 | 0.04 ± 0.09 | |
| apn1 | Oligo-W | 3164 ± 1292 | 100 |
| Oligo-U | 313 ± 133 | 8.1 ± 2.2 | |
| Oligo-O | 339 ± 139 | 8.6 ± 2.4 | |
| Oligo-F | 217 ± 190 | 6.2 ± 4.9 | |
| No oligo | 1.3 ± 1.6 | 0.2 ± 0.3 | |
| rad30 | Oligo-W | 1510 ± 183 | 100 |
| Oligo-U | 110 ± 17 | 7.3 ± 1.0 | |
| Oligo-Oa | 105 | 7.4 | |
| Oligo-F | 4.3 ± 1.8 | 0.28 ± 0.10 | |
| No oligo | 0 | 0 | |
| rev1 | Oligo-W | 581 ± 375 | 100 |
| Oligo-U | 1.7 ± 1.7 | 0.28 ± 0.31 | |
| Oligo-O | 3.4 ± 2.9 | 0.88 ± 1.2 | |
| Oligo-F | 0.20 ± 0.30 | 0.02 ± 0.02 | |
| No oligo | 0 | 0 | |
| apn1 rev1 | Oligo-W | 2579 ± 569 | 100 |
| Oligo-U | 4.0 ± 1.4 | 0.14 ± 0.05 | |
| Oligo-O | 4.5 ± 3.1 | 0.18 ± 0.13 | |
| Oligo-F | 2.6 ± 2.5 | 0.13 ± 0.14 | |
| No oligo | 0 | 0 | |
| apn2 | Oligo-W | 1348 ± 872 | 100 |
| Oligo-U | 91 ± 58 | 8.3 ± 3.1 | |
| Oligo-Oa | 113 | 17.4 | |
| Oligo-F | 3.7 ± 2.9 | 0.30 ± 0.23 | |
| No oligo | 0.1 ± 0.2 | 0.06 ± 0.13 |
Efficiencies shown are the average of three to six independent experiments unless otherwise indicated.
aThe average of two independent experiments.
To investigate contributions of TLS polymerases, rev1 and rad30 disruptants were used. The deletion of REV1 in B7528 reduces the transforming efficiency with oligonucleotides containing abasic analogs. This effect of rev1 deletion is also seen in the comparison of apn1 and apn1 rev1 strains. These results indicate that REV1 contributes to the bypass of O and F abasic sites. In contrast, the efficiencies of oligo-U and oligo-O in rad30 deletion strain were almost the same as that in B7528.
Nucleotide incorporation frequencies opposite O
The DNA sequence of the region where the oligonucleotides were introduced was determined. Nucleotides incorporated opposite the position occupied by U and O in the transforming oligonucleotides are shown in Table 4. C was most frequently incorporated in B7528 and an apn1 mutant, and then A and T were incorporated at a much lower level. The same mutagenic specificity was observed in a rad30 strain. On the other hand, C was only poorly incorporated in rev1 and apn1 rev1 mutants. These results suggest that the deoxycytidyl transferase activity of Rev1p inserts C opposite O. G was most frequently incorporated for U and O in rev1 defective mutants. The yeast Polη, a RAD30 product, efficiently inserts G opposite a natural abasic site in vitro (38). Incorporation of G may indicate the contribution of Polη to bypass O in the absence of Rev1p. The transforming efficiencies and the mutation spectra of U were similar to those of O. This shows that U on the introduced oligonucleotide is almost completely removed in the yeast cell to create O. We confirmed that this is due to the action of uracil-DNA glycosylase coded by the UNG1 gene. Actually, the transformation efficiencies of a UNG1-deleted derivative of B7528 with oligo-U were as high as 80% of those with oligo-W oligonucleotide. A was incorporated opposite the position occupied by U in all nine transformants obtained from the ung1 disruptant.
Table 4. Efficiencies of nucleotide incorporations opposite the lesions.
| Oligonucleotide | Strain | Base-specific read-through % (N)a | ||||
|---|---|---|---|---|---|---|
| A | C | G | T | Other mutations | ||
| Oligo-U | B7528 | 1.2 (4) | 3.0 (10) | 0.6 (2) | 1.2 (4) | 0.0 (0) |
| apn1 | 1.7 (6) | 4.9 (17) | 0.0 (0) | 1.5 (5) | 0.0 (0) | |
| rad30 | 0.0 (0) | 6.5 (16) | 0.4 (1) | 0.4 (1) | 0.0 (0) | |
| rev1 | 0.0 (1) | 0.0 (0) | 0.3 (19) | 0.0 (0) | 0.0 (0) | |
| apn1 rev1 | 0.1 (7) | 0.0 (0) | 0.1 (8) | 0.0 (0) | 0.0 (0) | |
| apn2 | 0.0 (0) | 6.1 (17) | 0.7 (2) | 0.7 (2) | 0.7 (2) | |
| Oligo-O | B7528 | 0.3 (1) | 4.5 (13) | 0.3 (1) | 2.1 (6) | 0.0 (0) |
| apn1 | 0.4 (1) | 5.4 (15) | 0.4 (1) | 2.2 (6) | 0.4 (1) | |
| rad30 | 0.4 (1) | 5.0 (13) | 0.0 (0) | 1.9 (5) | 0.0 (0) | |
| rev1 | 0.1 (3) | 0.4 (12) | 0.4 (13) | 0.0 (1) | 0.0 (0) | |
| apn1 rev1 | 0.0 (1) | 0.0 (0) | 0.2 (14) | 0.0 (0) | 0.0 (0) | |
| apn2 | 1.2 (1) | 6.3 (5) | 2.4 (2) | 6.3 (5) | 1.2 (1) | |
| Oligo-F | B7528 | 0.1 (11) | 0.1 (6) | 0.0 (0) | 0.1 (6) | 0.1 (6) |
| apn1 | 2.9 (12) | 1.2 (5) | 1.9 (8) | 0.0 (0) | 0.2 (1) | |
| rad30 | 0.1 (7) | 0.1 (4) | 0.0 (0) | 0.0 (1) | 0.1 (6) | |
| rev1 | 0.0 (1) | 0.0 (0) | 0.0 (1) | 0.0 (0) | 0.0 (2) | |
| apn1 rev1 | 0.1 (18) | 0.0 (2) | 0.0 (1) | 0.0 (0) | 0.0 (0) | |
| apn2 | 0.1 (8) | 0.0 (3) | 0.0 (2) | 0.0 (1) | 0.1 (6) | |
aBase-specific read-through (%), E, indicates how frequently each base was incorporated opposite lesions. N is the number of transformants in which each base was incorporated opposite the lesion. E = N / NT × ET where NT is the number of total transformants examined for each lesion in each strain, and ET is total read-through (%) for each lesion in each strain shown in Table 2. For example, B7526 strain cells were transformed with Oligo-U, and DNA sequence opposite the position occupied by U in the oligonucleotide was determined for 20 transformants (NT). Among them, four had A opposite the position of U. This number was normalized by total transformants examined (NT) to give 0.2. The base-specific read-through efficiency (E), 1.2%, was obtained by multiplying the value by total read-through efficiency for B7528 transformed with Oligo-U (ET), 6% (Table 2). All other values were obtained in a similar way using N and transforming efficiencies shown in Table 2.
Nucleotide incorporation frequencies opposite F
Efficient read-through opposite F only takes place in an apn1 strain (Table 3 and 4). A was preferentially incorporated opposite F in oligo-F. G was the next, then C. The spectrum was clearly different from that of O in each strain. Deficiency of Rev1p suppressed the incorporation of all of A, G and C in an apn1 rev1 strain. This suggests a function of Rev1p other than deoxycytidyl transferase. However, the transferase may still be active, because the ratio of incorporation of C to A was lower in apn1 rev1 than in apn1. It is also notable that the preferential incorporation of A was clearly observed in APN1 proficient strains, B7528, rad30 and apn2, although the efficiencies were very low.
Effect of adjacent bases on mutagenicity of F
Bypass efficiency of F by human Polκ and human Polι is influenced by the sequence context 5′ to the abasic site in vitro (39–41). We used four kinds of oligonucleotides (oligo-AF, oligo-CF, oligo-GF, oligo-TF) in order to examine whether the sequence context influences bypass of an abasic site in a yeast cell. The transformation efficiency of each oligonucleotide in apn1 strain was 150–330 transformants/µg of oligonucleotide, similar to that of oligo-F. A was most favorably incorporated opposite F in oligo-AF, oligo-CF and oligo-GF, as in oligo-F (Table 5), but the efficiencies were different. C was most frequently incorporated when oligo-TF was introduced. The mutagenic specificity was influenced by a nucleotide 5′ of F. This is consistent with the previous report (40). The mutagenic specificity of F in oligo-TF is different from that in Oligo-F. Nucleotides 5′ and 3′ of F are the same in the two oligonucleotides, and only the nucleotides 2 nt away from the lesion differ, namely C in oligo-TF and T in oligo-F.
Table 5. Nucleotide incorporations opposite an F adjacent to a different nucleotide in apn1 mutant strain.
| Oligonucleotide | No. of transformants with the nucleotide opposite F (%) | ||||
|---|---|---|---|---|---|
| A | C | G | T | Other mutations | |
| Oligo-AF | 15 (68) | 6 (27) | 0 (0) | 0 (0) | 1 (4.5) |
| Oligo-CF | 13 (59) | 8 (36) | 1 (4.5) | 0 (0) | 0 (0) |
| Oligo-GF | 11 (52) | 8 (38) | 1 (4.8) | 0 (0) | 1 (4.8) |
| Oligo-TF | 8 (36) | 11 (50) | 2 (9.1) | 1 (4.5) | 0 (0) |
| Oligo-F | 12 (46) | 5 (19) | 8 (31) | 0 (0) | 1 (3.8) |
DISCUSSION
In the present study, we determined the in vivo mutagenic specificity of three types of DNA lesions in yeast: U, removal of which forms O inside cells, O and F. Surprisingly the mutagenic specificities of F were different from U or O in yeast cells, while U and O are almost the same in their mutagenic properties. C was most frequently incorporated opposite O or U. On the other hand, A was favorably incorporated opposite F, although read-through of both abasic sites required Rev1p function.
F has often been used as a structural analog of the cyclic hemiacetal form of O for the mutagenic studies of abasic sites (7,19,21,25,27). We compared F with O by side-by-side experiments using the in vivo assay system, and the results showed clear differences in the mutagenic specificity of two abasic sites. Shibutani et al. (21) reported the template activity of O, F and a deoxyribitol (a reduced form of O) in primer extension with Klenow fragment of the E.coli DNA polymerase I or calf thymus DNA polymerase α. The frequency of nucleotide insertion opposite all the three types of abasic site was in the order of A > G > C > T. The frequency of TLS past abasic sites was highest in F, O and then deoxyribitol. The results indicate that O and F are similar in miscoding specificity, but different in the ability to block DNA synthesis. If this is also the case in the yeast replication machinery, the difference in the mutational specificity may indicate differences in the DNA polymerases involved. Because F blocks replication less than O, it might be possible that the replicative polymerase can more frequently continue polymerization with the aid of Rev1p as described below. A recent report by Avkin et al. (42) is interesting from this view. They found that a replicative polymerase is involved in translesion of F in human cells and inserts A opposite F.
Another interesting difference between F, and O or U is the effect of APN1 endonuclease on the transforming activity. The transforming efficiencies of oligo-O and -U in the apn1 strain increased by only 1.2 and 1.4-fold compared with those in the wild-type, respectively, while those of oligo-F increased by 19-fold. This might be related to the fact that a natural abasic site can be repaired by not only AP endonucleases, but also AP lyases, which cannot act on F. However, the true mechanism is unknown.
We tested whether TLS polymerases, Polη and Rev1p, were required for replication past O and F. The read-through of O and F definitely depended on Rev1p, even their mutagenic properties were different from each other. Rev1p possesses a deoxycytidyl transferase activity to insert C opposite O (9) and F (7). The spectrum of nucleotide incorporation opposite F showed predominant incorporation of A in all tested strains. These results suggest that Rev1p function, other than its deoxycytidyl transferase activity, may be required for TLS past F. In other words, Rev1p has a second function as proposed before (7,11). The Rev1p second function was also observed in the mutagenesis by 6-4 photoproducts (11,12). In addition to the incorporation of C and A, we also found a significant incorporation of T opposite O. This incorporation was not observed in rev1 deletion mutants. This incorporation may again relate to Rev1p function. A further experiment using rev1 mutants lacking deoxycytidyl transferase activity is required for the clear demonstration of this role.
Deletion of RAD30 did not influence the efficiency of transformation and the mutagenic spectrum in any of U, O or F. Contribution of Polη on the bypass of the abasic sites may be low in yeast, although the polymerase may play some role which may become apparent in the bypass in the absence of Rev1p. This result agrees with the in vitro study on Polη by Haracska et al (10). These results indicate that a replicative polymerase interacting with Rev1p may insert A opposite O and F.
The experiment comparing oligonucleotides that differ in a base 5′ of F gave an unexpected result (Table 5). Surprisingly, oligo-TF and oligo-F showed clearly different mutagenic specificity. This is, to our knowledge, the first report that a base not adjacent to the lesion can affect its mutagenic property.
There are discrepancies in the reported mutagenic specificity of abasic sites in yeast. Favorable incorporation of C opposite O is found using a shuttle vector containing O (8), whereas two other reports indicated different specificity for abasic sites produced in genomic DNA intracellularly. An incorporation of G (28) and A (7) opposite abasic sites was reported. It is supposed that the inconsistency of mutagenic specificity of abasic sites might depend on whether the lesion is on a plasmid or in the genome (7). However, our results indicate that C is inserted most frequently opposite O (62% of the transformants with oligo-O), even when the translesion took place in the genome. Another possibility is that the mutagenic specificity of an abasic site already present in the DNA before transformation is different from that produced inside cells due to the interaction with the glycosylases. Our present results appear to be against this as far as uracil-DNA glycosylase is involved, because O and U showed similar mutagenic specificity.
We can find some differences between O and U, although they show similar mutagenic potential. The incorporation of A opposite the lesion in oligo-U in B7528 and apn1 was higher than in oligo-O. This might be due to the U escaped from the action of uracil-DNA glycosylase, but it is difficult to explain the absence of the incorporation of A opposite the lesion in oligo-U in other strains. A low-level incorporation of C in rev1 strains was observed for oligo-O, but not for oligo-U. Other glycosylases or base excision repair systems might interact with TLS systems more strongly than uracil-DNA glycosylase. Another difference between U and O was found in the rev1 strain. O still produced many C insertions, whereas U did not (Table 4). Table 4 also shows the clear difference between F and O (U) in the apn1 rev1 strain. However, further study is required to confirm these differences under very low transforming activities.
In conclusion, our data indicate that a very small structural difference in the DNA lesions or their environment can influence the mutation spectrum greatly. The specificity can be changed by the presence (O) or absence (F) of an OH residue at position –1 of deoxyribose in abasic sites, or a base 2 nt away from the lesion.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Roel M. Schaaper of the National Institute of Environmental Health for his advice on this manuscript. We also thank Professor Hiroshi Ide of Hiroshima University for his kind advice on the preparation of tetrahydrofuran-oligonucleotides. This work is partially supported by a Grant-in-aid for Encouragement of Young Scientists (A) (no. 12771401) and a Grant-in-aid for Scientific Research (C) (no. 14572093) from the Ministry of Education, Culture, Sport, Science and Technology, Japan.
REFERENCES
- 1.Loeb L.A. and Preston,B.D. (1986) Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet., 20, 201–230. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A. and Sancar,G.B. (1988) DNA repair enzymes. Annu. Rev. Biochem., 57, 29–67. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg E.C. and Gerlach,V.L. (1999) Novel DNA polymerases offer clues to the molecular basis of mutagenesis. Cell, 98, 413–416. [DOI] [PubMed] [Google Scholar]
- 4.Percival K.J., Klein,M.B. and Burgers,P.M. (1989) Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J. Biol. Chem., 264, 2593–2598. [PubMed] [Google Scholar]
- 5.Popoff S.C., Spira,A.I., Johnson,A.W. and Demple,B. (1990) Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc. Natl Acad. Sci. USA, 87, 4193–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson R.E., Torres-Ramos,C.A., Izumi,T., Mitra,S., Prakash,S. and Prakash,L. (1998) Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev., 12, 3137–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haracska L., Unk,I., Johnson,R.E., Johansson,E., Burgers,P.M., Prakash,S. and Prakash,L. (2001) Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev., 15, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibbs P.E. and Lawrence,C.W. (1995) Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J. Mol. Biol., 251, 229–236. [DOI] [PubMed] [Google Scholar]
- 9.Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- 10.Haracska L., Washington,M.T., Prakash,S. and Prakash,L. (2001) Inefficient bypass of an abasic site by DNA polymerase η. J. Biol. Chem., 276, 6861–6866. [DOI] [PubMed] [Google Scholar]
- 11.Nelson J.R., Gibbs,P.E., Nowicka,A.M., Hinkle,D.C. and Lawrence,C.W. (2000) Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol., 37, 549–554. [DOI] [PubMed] [Google Scholar]
- 12.Otsuka C., Kobayashi,K., Kawaguchi,N., Kunitomi,K., Moriyama,K., Hata,Y., Iwai,S., Loakes,D., Noskov,N.V., Pavlov,Y. and Negishi,K. (2002) Use of yeast transformation by oligonucleotides to study DNA lesion bypass in vivo. Mutat. Res., 504, 53–60. [DOI] [PubMed] [Google Scholar]
- 13.Schaaper R.M., Kunkel,T.A. and Loeb,L.A. (1983) Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc. Natl Acad. Sci. USA, 80, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boiteux S. and Laval,J. (1982) Coding properties of poly(deoxycytidylic acid) templates containing uracil or apyrimidinic sites: in vitro modulation of mutagenesis by deoxyribonucleic acid repair enzymes. Biochemistry, 21, 6746–6751. [DOI] [PubMed] [Google Scholar]
- 15.Sagher D. and Strauss,B. (1983) Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry, 22, 4518–4526. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel T.A. (1984) Mutational specificity of depurination. Proc. Natl Acad. Sci. USA, 5, 1494–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence C.W., Borden,A., Banerjee,S.K. and LeClerc,J.E. (1990) Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res., 18, 2153–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randall S.K., Eritja,R., Kaplan,B.E., Petruska,J. and Goodman,M.F. (1987) Nucleotide insertion kinetics opposite abasic lesions in DNA. J. Biol. Chem., 262, 6864–6870. [PubMed] [Google Scholar]
- 19.Takeshita M., Chang,C.N., Johnson,F., Will,S. and Grollman,A.P. (1987) Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem., 262, 10171–10179. [PubMed] [Google Scholar]
- 20.Strauss B.S. (1991) The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays, 13, 79–84. [DOI] [PubMed] [Google Scholar]
- 21.Shibutani S., Takeshita,M. and Grollman,A.P. (1997) Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the “A rule”. J. Biol. Chem., 272, 13916–13922. [DOI] [PubMed] [Google Scholar]
- 22.Gentil A., Renault,G., Madzak,C., Margot,A., Cabral-Neto,J.B., Vasseur,J.J., Rayner,B., Imbach,J.L. and Sarasin,A. (1990) Mutagenic properties of a unique abasic site in mammalian cells. Biochem. Biophys. Res. Commun., 173, 704–710. [DOI] [PubMed] [Google Scholar]
- 23.Gentil A., Cabral-Neto,J.B., Mariage-Samson,R., Margot,A., Imbach,J.L., Rayner,B. and Sarasin,A. (1992) Mutagenicity of a unique apurinic/apyrimidinic site in mammalian cells. J. Mol. Biol., 227, 981–984. [DOI] [PubMed] [Google Scholar]
- 24.Cabral Neto J.B., Cabral,R.E., Margot,A., Le Page,F., Sarasin,A. and Gentil,A. (1994) Coding properties of a unique apurinic/apyrimidinic site replicated in mammalian cells. J. Mol. Biol., 240, 416–420. [DOI] [PubMed] [Google Scholar]
- 25.Takeshita M. and Eisenberg,W. (1994) Mechanism of mutation on DNA templates containing synthetic abasic sites: study with a double strand vector. Nucleic Acids Res., 22, 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klinedinst D.K. and Drinkwater,N.R. (1992) Mutagenesis by apurinic sites in normal and ataxia telangiectasia human lymphoblastoid cells. Mol. Carcinog., 6, 32–42. [DOI] [PubMed] [Google Scholar]
- 27.Kamiya H., Suzuki,M., Komatsu,Y., Miura,H., Kikuchi,K., Sakaguchi,T., Murata,N., Masutani,C., Hanaoka,F. and Ohtsuka,E. (1992) An abasic site analogue activates a c-Ha-ras gene by a point mutation at modified and adjacent positions. Nucleic Acids Res., 20, 4409–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunz B.A., Henson,E.S., Roche,H., Ramotar,D., Nunoshiba,T. and Demple,B. (1994) Specificity of the mutator caused by deletion of the yeast structural gene (APN1) for the major apurinic endonuclease. Proc. Natl Acad. Sci. USA, 91, 8165–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moerschell R.P., Tsunasawa,S. and Sherman,F. (1988) Transformation of yeast with synthetic oligonucleotides. Proc. Natl Acad. Sci. USA, 85, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noskov V., Negishi,K., Ono,A., Matsuda,A., Ono,B. and Hayatsu,H. (1994) Mutagenicity of 5-bromouracil and N6-hydroxyadenine studied by yeast oligonucleotide transformation assay. Mutat. Res.,308, 43–51. [DOI] [PubMed] [Google Scholar]
- 31.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Adams A., Gottschling,D.E., Kaise,C.A. and Strearns,T. (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 33.Gietz R.D. and Woods,R.A. (1994) High efficiency transformation with lithium acetate. In Johnston,J.R. (ed.), Molecular Genetics of Yeast – A Practical Approach. IRL Press, Oxford, UK, pp. 121–134.
- 34.Alani E., Cao,L. and Kleckner,N. (1987) A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics, 116, 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wach A., Brachat,A., Poehlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 36.Malagon F. and Aguilera,A. (1999) Construction and genetic analysis of S. cerevisiae deletants of six novel ORFs from chromosome II. Yeast, 15, 955–961. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto T., Moerschell,R.P., Wakem,L.P., Ferguson,D. and Sherman,F. (1992) Parameters affecting the frequencies of transformation and co-transformation with synthetic oligonucleotides in yeast. Yeast, 8, 935–948. [DOI] [PubMed] [Google Scholar]
- 38.Yuan F., Zhang,Y., Rajpal,D.K., Wu,X., Guo,D., Wang,M., Taylor,J.S. and Wang,Z. (2000) Specificity of DNA lesion bypass by the yeast DNA polymerase η. J. Biol. Chem., 275, 8233–8239. [DOI] [PubMed] [Google Scholar]
- 39.Ohashi E., Ogi,T., Kusumoto,R., Iwai,S., Masutani,C., Hanaoka,F. and Ohmori,H. (2000) Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev., 14, 1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Yuan,F., Wu,X., Wang,M., Rechkoblit,O., Taylor,J.S., Geacintov,N.E. and Wang,Z. (2000) Error-free and error-prone lesion bypass by human DNA polymerase κ in vitro. Nucleic Acids Res., 28, 4138–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Yuan,F., Wu,X., Taylor,J.S. and Wang,Z. (2001) Response of human DNA polymerase ι to DNA lesions. Nucleic Acids Res., 29, 928–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avkin S., Adar,S., Blander,G. and Livneh,Z. (2002) Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc. Natl Acad. Sci. USA, 99, 3764–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]