Abstract
The complete genomic sequence of an intracellular bacterial pathogen, Mycoplasma penetrans HF-2 strain, was determined. The HF-2 genome consists of a 1 358 633 bp single circular chromosome containing 1038 predicted coding sequences (CDSs), one set of rRNA genes and 30 tRNA genes. Among the 1038 CDSs, 264 predicted proteins are common to the Mycoplasmataceae sequenced thus far and 463 are M.penetrans specific. The genome contains the two-component system but lacks the essential cellular gene, uridine kinase. The relatively large genome of M.penetrans HF-2 among mycoplasma species may be accounted for by both its rich core proteome and the presence of a number of paralog families corresponding to 25.4% of all CDSs. The largest paralog family is the p35 family, which encodes surface lipoproteins including the major antigen, P35. A total of 44 genes for p35 and p35 homologs were identified and 30 of them form one large cluster in the chromosome. The genetic tree of p35 paralogs suggests the occurrence of dynamic chromosomal rearrangement in paralog formation during evolution. Thus, M.penetrans HF-2 may have acquired diverse repertoires of antigenic variation-related genes to allow its persistent infection in humans.
INTRODUCTION
The family Mycoplasmataceae, which belongs to the order Mycoplasmatales under the class Mollicutes, contains the genera Mycoplasma and Ureaplasma. It is thought that the Mollicutes, which possess a notably small genome, may have evolved from a common ancestral Gram-positive bacterium with low G + C content like the Lactobacillus group, including Bacillus subtilis, by losing a considerable region of the genome (1–3).
All species of Mycoplasmataceae are obligate parasites and host specificity is quite strict for hosts including humans, rodents, artiodactyla, perissodactyla and birds. Mycoplasmal infections are most frequently associated with disease in the urogenital or respiratory tracts and, in most cases, mycoplasmas infect the host persistently. Like other parasites, many mycoplasma species display antigenic diversity, which has been noted in a variety of protein profiles (2) or colony immunoblotting (4). Antigenic variation is considered to be a strategy for persistence in the face of immune responses by the host.
Mycoplasma penetrans, a species of Mycoplasmataceae, infects humans in the urogenital and respiratory tracts. A typical feature of M.penetrans is penetration into human cells. Internalization of the organisms into the urothelium was detected in autopsy samples from an acquired immunodeficiency syndrome (AIDS) patient (5). Intracellular replication and persistence for at least 6 months has been observed in cultured cells (6). Long-term persistence of M.penetrans in patients has also been documented on the basis of its isolation from urine specimens collected at different times over the period of 1 year from the same children with human immunodeficiency virus (HIV) infection (7). In human disease, M.penetrans is associated mainly with HIV-1 infection, particularly in adults among the homosexual population in Europe and the USA, and among both homosexual and heterosexual males in South America. Anti-M.penetrans antibodies are found even in asymptomatic HIV carriers and in patients in the process of developing AIDS (5,8–14). Both the rapid decline in CD4-positive lymphocyte counts in M.penetrans-seropositive HIV-infected individuals (15) and the mitogenic effects of this organism on lymphocytes (16) imply a contribution of persistent M.penetrans infection to the deterioration of the immune system in HIV infection. On the other hand, M.penetrans infection has also been suggested to be a primary cause of human disease in non-HIV-related urethritis and respiratory disease (8,17). The M.penetrans HF-2 strain was isolated from a previously healthy HIV-negative patient suffering from severe respiratory distress caused by M.penetrans infection-associated systemic disease (17).
The whole genome sequences of Mycoplasma pneumoniae, Mycoplasma genitalium, Mycoplasma pulmonis and Ureaplasma urealyticum have been reported previously and their sizes range from 0.57 to 0.95 Mb (18–21). In contrast, the size of the M.penetrans genome was estimated to be ∼1.3 Mb (unpublished data), suggesting that this organism may possess additional genetic information involved in its unique infection process. Therefore, we determined the complete nucleotide sequence of the M.penetrans HF-2 strain. Analysis of the genomic sequence revealed that M.penetrans possesses a large number of paralogous gene repertoires and chromosomal structures allowing for antigenic variation, and hence persistent infection, in human hosts. Here, we present the complete genomic sequence of M.penetrans strain HF-2, together with computational analysis of the genome.
MATERIALS AND METHODS
Mycoplasma strain
The M.penetrans HF-2 strain was kindly provided by Dr Antonio Yáñez, Centro de Investigación Biomédica de Oriente-IMSS, Puebla City, Mexico. The HF-2 strain was isolated from the tracheal aspirate of a patient with primary M.penetrans infection. The sixth passage of organisms after primary isolation from the patient was used as the source of DNA for sequencing.
DNA manipulation
Genomic DNA was extracted from the M.penetrans HF-2 strain cultivated in PPLO broth (Becton Dickinson Microbiology Systems) supplemented with 10% (v/v) heat-inactivated horse serum (GIBCO/BRL), 0.5% glucose and 100 U/ml penicillin G (Meiji Seika Kaisha, Japan). After lysis of harvested mycoplasma cells, by incubating at 50°C for 2 h in a lysis buffer (50 mM Tris, 20 mM EDTA, pH 9.0, containing 0.1% SDS and 100 µg/ml of proteinase K), the lysate was extracted with phenol–chloroform and the DNA was precipitated with ethanol. The DNA was treated with RNaseA (Boehringer Mannheim Biochemicals) and purified by phenol–chloroform extraction and ethanol precipitation.
DNA sequencing
For the construction of shotgun DNA libraries, genomic DNA was sheared mechanically using Hydro-Shear (Gene Machines). The sheared DNA was blunt-ended, ligated into the SmaI site of the pUC18 plasmid and introduced into the Escherichia coli DH5α strain. Short (1–2 kb) and long (4–5 kb) insert libraries were constructed. Template DNA was prepared by PCR amplification of the insert DNA from colonies (22) and sequenced using the DYEnamic ET Dye Terminator Cycle Sequencing Kit and MegaBACE1000 (Amersham Biosciences).
Data assembly and finishing
We obtained 26 449 sequence reads from 1–2 kb insert templates and 9156 double-ended sequence reads from 4–5 kb insert templates. The data were assembled and edited using the Phred/Phrap/Consed package of base-calling, sequence-assembly and automated finishing software (University of Washington) (23). The assembly yielded data with an estimated redundancy of 11.73 and 35 gaps. The 35 gaps were resolved by sequencing PCR products amplified with appropriate primers. The DNA sequences covering gaps were assembled together with all other data to obtain a circular consensus sequence. For handling the sequence data, several shell and perl scripts were developed. The assembled data were validated by the pattern of pulse-field gel electrophoresis using BamHI and BglI.
Identification of CDSs, annotation and analysis
Putative coding sequences (CDSs) were identified using the GLIMMER program (24), which was trained with CDSs of M.genitalium, M.pneumoniae and U.urealyticum in which UGA was allowed to be used as tryptophan. By using the program FramePlot (25), which was optimized for the mycoplasma genome, some predicted CDSs were excluded or modified because they either overlapped with other CDSs or had some additional sequence upstream of the predicted start codon. Homology for the amino acid sequences deduced from each CDS was checked using the program BLASTP (26) and the nr database from the National Center for Biotechnology Information (NCBI). The programs HmmerPfam (GCG Wisconsin package, Accelrys Inc.) with the Pfam motif database (27), TopPredII (28) and MacStripe (29) were used for the prediction of motifs, transmembrane domains and the membrane topology of proteins. All CDSs were classified into modified COGs (clusters of orthologous groups of proteins, NCBI) categories. For analyzing metabolic pathways, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used. tRNA genes were detected by the tRNAscan-SE program (30).
Database submission
The sequence data have been submitted to the EMBL/GenBank/DDBJ database under accession numbers AP004170– AP004174.
More detailed information on the M.penetrans genome is available at our web site (http://www.nih.go.jp/Mypet/).
RESULTS AND DISCUSSION
General features of the M.penetrans genome
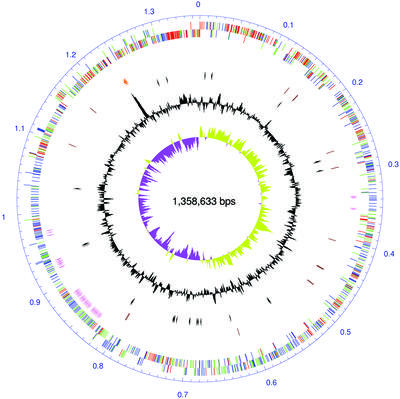
The size of the M.penetrans genome, 1 358 633 bp, is larger than those of the four Mycoplasmataceae species, M.genitalium, M.pneumoniae, M.pulmonis and U.urealyticum, sequenced so far (18–20,31). The average G + C content is 25.7%, which is close to the 25.5% of the U.urealyticum genome, which has the lowest G + C content of all bacterial species published thus far. Mycoplasma penetrans has 1038 predicted CDSs, one set of 5S-23S-16S rRNA genes (MYPE20000–MYPE20020) and 30 tRNA genes (Fig. 1).
Figure 1.
Circular representation of the M.penetrans chromosome. The dnaA gene is at position zero. Starting from the outside, the first circle indicates the starting location of the predicted coding sequences (CDSs) on the plus strand and the second circle indicates those on the minus strand. The CDSs are colored based on the results of reciprocal best-hits in a pairwise BLAST search as follows: red, common gene in Mycoplasmataceae; blue, M.penetrans-specific gene; green, other genes. The third (pink) circle indicates the location and the direction of transcription of the P35 gene family. The rRNA genes and tRNA genes are shown in orange and brown, respectively, in the fourth circle. The fifth (black) circle indicates the location and the direction of transcription of transposases. The sixth circle (black line) is a plot of the G + C content for each 2 kb interval with the magnitude of the mean value. In the seventh circle, a plot of the G + C skew for each 5 kb interval is shown in khaki (plus) and purple (minus), respectively.
In M.penetrans, dnaA (MYPE10), dnaN (MYPE20), gyrB (MYPE30) and gyrA (MYPE40) are closely linked. The dnaA box, the specific recognition site for binding dnaA that is often present near the replication origin of most bacterial genomes, was not detected near the dnaA gene, as demonstrated in M.genitalium, M.pneumoniae and U.urealyticum. On the contrary, a poly(C) sequence was found from position 696 255 to 696 269 in the genome. Both the G + C skew analysis, which is a method for predicting replication origins, and the transcriptional direction in the genome, revealed that two inversion points are clearly located near the dnaA and the poly(C) sequences. These data suggest that the replication origin is located at the inverted point near dnaA, and that the poly(C) sequence near the other inversion point could be the replication terminator (Fig. 1).
Comparison of the theoretical proteome in M.penetrans and other species
For ortholog identification, reciprocal best-hit pairs of genes were identified by pairwise BLASTP comparisons of the predicted M.penetrans proteins with those in M.genitalium, M.pneumoniae, M.pulmonis, U.urealyticum and B.subtilis. The number of reciprocal best-hit genes was 586, 433, 400, 390 and 383 for B.subtilis, M.pneumoniae, M.pulmonis, U.urealyticum and M.genitalium, respectively. Of the predicted proteins, 264 were found to be common to Mycoplasmataceae including house-keeping products such as DnaA, DNA polymerase III, DNA topoisomerase, ribosomal proteins and ATP synthase. Conversely, there were 463 M.penetrans-specific proteins compared with the four Mycoplasmataceae species, and 311 when compared with B.subtilis plus the four Mycoplasmataceae species. In addition, the number of core proteome, which is defined as functionally distinct protein families in the theoretical proteome encoded in the genome (32,33), was 847 in M.penetrans, significantly greater than the other four Mycoplasmataceae species, where 469, 590, 721 and 580 proteins were represented in M.genitalium, M.pneumoniae, M.pulmonis and U.urealyticum, respectively. From these data, the core proteome in M.penetrans is clearly richer than that of the other four Mycoplasmataceae species. The M.penetrans-specific proteins in Mycoplasmataceae included the putative two-component response regulators (MYPE3960), possible sensory transduction histidine kinase (MYPE2360), deoxyuridine 5′-triphosphate nucleotidohydrolase (dUTPase, MYPE1000), guanosine 5′-monophosphate (GMP) synthase (MYPE1220), d-lactate dehydrogenase (D-LDH, MYPE 2650), aldehyde dehydrogenase (MYPE3920, MYPE4110), anaerobic ribonucleoside-triphosphate reductase (MYPE4960), adenylosuccinate synthetase (MYPE 5830), pyruvate decarboxylase (MYPE6890), enzymes for the orotate-related pathway (MYPE7840–7900), aspartate transcarbamoylase (MYPE 7890) and flavodoxin (MYPE9120). Since no two-component signal response regulator has been found in the other four Mycoplasmataceae species sequenced (18,19,34), it should be noted that M.penetrans is the first mycoplasma predicted to possess the two-component signal system that is commonly present in other bacteria.
Predicted products of M.penetrans CDSs were categorized according to function and compared with the other four Mycoplasmataceae species (Table 1 and Supplementary Material). The results revealed that 108 genes (10.4% of the CDSs) involved in the translation process, such as ribosomal proteins, and 23 genes (2.2% of the CDSs) involved in transcription are present in M.penetrans. The relatively high number of translation- or transcription-related genes in M.penetrans is similar to the other Mycoplasmataceae (99–102 genes for translation and 14–18 genes for transcription). Of the M.penetrans CDSs, 428 were not categorized in the COG database.
Table 1. Summary of general features of the genome and functional categories of predicted CDSs of Mollicutes and B.subtilis.
| |
M.penetrans |
M.genitalium |
M.pneumoniae |
M.pulmonis |
U.urealyticum |
B.subtilis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome size (bp) | 1 358 633 | 580 074 | 816 394 | 963 879 | 751 719 | 4 214 814 | ||||||
| G + C content (mol%) | 25.7 | 32.0 | 40.0 | 26.6 | 25.5 | 43.5 | ||||||
| Total number of predicted CDSs | 1038 | 484 | 689 | 782 | 614 | 4112 | ||||||
| Functional category in COGs | % | % | % | % | % | % | ||||||
| J Translation, ribosomal structure and biogenesis | 108 | 10.4 | 99 | 20.5 | 100 | 14.5 | 102 | 13.0 | 102 | 16.6 | 151 | 3.7 |
| K Transcription | 23 | 2.2 | 14 | 2.9 | 14 | 2.0 | 18 | 2.3 | 18 | 2.9 | 276 | 6.7 |
| L DNA replication, recombination and repair | 78 | 7.5 | 41 | 8.5 | 57 | 8.3 | 72 | 9.2 | 56 | 9.1 | 134 | 3.3 |
| D Cell division and chromosome partitioning | 11 | 1.1 | 5 | 1.0 | 5 | 0.7 | 4 | 0.5 | 4 | 0.7 | 32 | 0.8 |
| O Posttranslational modification, protein turnover, chaperones | 26 | 2.5 | 20 | 4.1 | 20 | 2.9 | 18 | 2.3 | 19 | 3.1 | 87 | 2.1 |
| M Cell envelope biogenesis, outer membrane | 13 | 1.3 | 10 | 2.1 | 10 | 1.5 | 5 | 0.6 | 4 | 0.7 | 161 | 3.9 |
| N Cell motility and secretion | 12 | 1.2 | 11 | 2.3 | 12 | 1.7 | 13 | 1.7 | 15 | 2.4 | 97 | 2.4 |
| P Inorganic ion transport and metabolism | 22 | 2.1 | 17 | 3.5 | 17 | 2.5 | 15 | 1.9 | 28 | 4.6 | 148 | 3.6 |
| T Signal transduction mechanisms | 4 | 0.4 | 4 | 0.8 | 4 | 0.6 | 3 | 0.4 | 4 | 0.7 | 123 | 3.0 |
| C Energy production and conversion | 31 | 3.0 | 20 | 4.1 | 20 | 2.9 | 29 | 3.7 | 17 | 2.8 | 163 | 4.0 |
| G Carbohydrate transport and metabolism | 50 | 4.8 | 24 | 5.0 | 34 | 4.9 | 58 | 7.4 | 14 | 2.3 | 274 | 6.7 |
| E Amino acid transport and metabolism | 26 | 2.5 | 15 | 3.1 | 24 | 3.5 | 23 | 2.9 | 20 | 3.3 | 294 | 7.1 |
| F Nucleotide transport and metabolism | 39 | 3.8 | 21 | 4.3 | 21 | 3.0 | 24 | 3.1 | 21 | 3.4 | 81 | 2.0 |
| H Coenzyme metabolism | 12 | 1.2 | 13 | 2.7 | 14 | 2.0 | 12 | 1.5 | 9 | 1.5 | 109 | 2.7 |
| I Lipid metabolism | 15 | 1.4 | 8 | 1.7 | 9 | 1.3 | 8 | 1.0 | 8 | 1.3 | 85 | 2.1 |
| Q Secondary metabolites biosynthesis, transport and catabolism | 20 | 1.9 | 4 | 0.8 | 5 | 0.7 | 10 | 1.3 | 7 | 1.1 | 129 | 3.1 |
| R General function prediction only | 95 | 9.2 | 44 | 9.1 | 48 | 7.0 | 49 | 6.3 | 41 | 6.7 | 335 | 8.1 |
| S Function unknown | 25 | 2.4 | 14 | 2.9 | 15 | 2.2 | 26 | 3.3 | 22 | 3.6 | 233 | 5.7 |
| (–) not in COGs | 428 | 41.2 | 100 | 20.7 | 260 | 37.7 | 293 | 37.5 | 205 | 33.4 | 1200 | 29.2 |
To evaluate evolutionary divergence of M.penetrans from the other species of Mycoplasmataceae, the orthologous genes in M.penetrans, M.pneumoniae, M.genitalium, M.pulmonis and U.urealyticum were plotted with respect to their location. The results showed no linearity (data not shown). Similar results have been reported from comparative analysis of the U.urealyticum genome with those of M.pneumoniae and M.genitalium (19). The results suggest that rapid chromosomal rearrangement may have occurred in the Mycoplasmataceae genome during evolution.
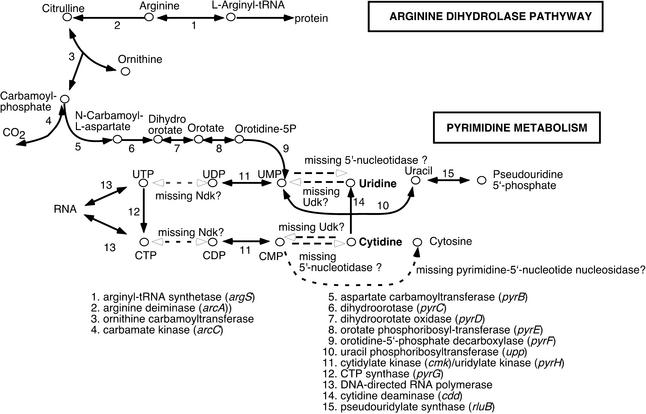
Lack of uridine kinase, an essential cellular gene involved in pyrimidine metabolism
It has been proposed that 256 genes constitute the minimal gene set necessary and sufficient for sustaining a functioning cell. This estimate was based on a comparative analysis of the genomes of M.genitalium and Haemophilus influenzae, and the study of gene knockouts of M.genitalium (21,35,36). Upon screening for gene content, we found that M.penetrans lacks uridine kinase (udk), a member of the proposed minimal gene set. Uridine kinase is a phosphotransferase that mediates phosphorylation of both uridine and cytidine to produce uridine- and cytidine-monophosphate (UMP/CMP) (Fig. 2). Mycoplasma penetrans also lacks the 5′-nucleotidase that metabolizes both UMP and CMP to uridine and cytidine, respectively. A previous study indicated that uracil phosphoribosyltransferase (Upp) is utilized in the conversion of uracil to UMP in Mycoplasma mycoides (37). The M.penetrans genome contains the upp gene (MYPE10300), so Upp-dependent uracil conversion may be the main pathway for UMP production in M.penetrans. The other pathway, via orotate-related metabolism for converting carbamoyl-phosphate to UMP, was found in M.penetrans. Mycoplasma penetrans contains a set of genes for enzymes involved in orotate-related metabolism as follows: aspartate carbamoyltransferase (pyrB, MYPE7890), dihydroorotase (pyrC, MYPE7880), dihydoorotate oxidase (pyrD, MYPE7860 and 7870), orotate phosphoribosyltransferase (pyrE, MYPE7840), orotidine-5′-phosphate decarboxylase (pyrF, MYPE7850) and the pyrimidine operon regulatory protein (pyrR, MYPE7900). Orotate-related metabolism is linked to the arginine dihydrolase pathway, which is an ATP synthesis system. Mycoplasma penetrans contains all the genes for enzymes involved in the arginine dihydrolase pathway, namely, arginine deaminase (MYPE6080), ornithine carbomoyltransferase (MYPE6090) and carbamate kinase (MYPE6100). The four Mycoplasmataceae species analyzed previously lacked either uridine kinase or 5′-nucleotidase; however, M.penetrans lacks both enzymes but has an orotate-related pathway, suggesting that UMP may be converted from both uracil and carbamoyl phosphate in this species. It should be noted that M.penetrans is the only member of Mycoplasmataceaethus far to possess orotate-related metabolism linked to the arginine dihydrolase pathway and pyrimidine metabolism (Fig. 2).
Figure 2.
Partial pathways of pyrimidine metabolism in M.penetrans. The solid lines numbered indicate the predicted metabolic pathways and enzyme names. The pathways with dotted lines indicate unidentified responsible enzymes in the M.penetrans genome.
Essential enzymes in other pathways are also missing for pyrimidine metabolism in M.penetrans. For example, the presence of pyrimidine-5′-nucleotide nucleosidase for converting cytosine to CMP remains an open question. As in other species of Mollicutes, M.penetrans also lacks nucleoside-diphosphate kinase (ndk) that converts uridine/cytidine diphosphate (UDP/CDP) to uridine/cytidine triphosphate (UTP/CTP). Non-orthologous gene displacement (38) of these essential enzymes in Mollicutes is speculated.
Paralogous gene families in M.penetrans
One of the remarkable features of the M.penetrans genome is the existence of a number of paralogous gene families. In a BLAST score-based single-linkage clustering search using BLASTCLUST, proteins with >30% amino acid identity over 70% of their length were defined as paralogs. Search results showed that 264 (25.4%) of the 1038 proteins formed 63 families, ranging from two to 44 proteins per family (Table 2). Under the same BLAST analysis conditions, the M.genitalium, M.pneumoniae, M.pulmonis and U.urealyticum genomes were found to contain 27 (5.5%), 132 (19.1%), 100 (12.7%) and 56 (8.1%) paralogs, respectively. The relatively large number of paralogs could be one of the reasons for the larger genomic size of M.penetrans compared with the other four Mycoplasmataceae species. These paralog families include P35 lipoprotein homologs, CDSs of functionally unknown proteins, the ABC transporter ATP-binding protein, oxidoreductase, predicted permease and putative lipoproteins.
Table 2. Summary of M.penetrans-specific paralog families (more than three copies) in the genome.
| Paralog families | Number of CDSs | Representative CDSs | Description | MYPE no. |
|---|---|---|---|---|
| Family 1 | 38 | MYPE 6810 | lipoprotein P35 homologs | 2620, 2630, 2690, 2700, 6490, 6500, 6510, 6520, 6530, 6540, 6550, 6560, 6570, 6580, 6590, 6630, 6640, 6650, 6660, 6670, 6680, 6690, 6710, 6720, 6730, 6740, 6750, 6780, 6790, 6800, 6810, 6820, 6830, 6840, 7330, 7370, 7380, 7400 |
| 6 | MYPE 7020 | signal-peptide-less p35 gene homologs | 7020, 7030, 7040, 7050, 7060, 7070 | |
| Family 2 | 7 | MYPE 8480 | hypothetical protein | 3410, 3420, 3430, 7350, 7360, 8470, 8480 |
| 5 | MYPE 8270 | hypothetical protein | 8270, 8280, 8290, 8300, 9860 | |
| 5 | MYPE 7490 | conserved hypothetical protein | 7480, 7490, 7500, 7510, 7520 | |
| 5 | MYPE 5680 | hypothetical protein | 4740, 4750, 4760, 5670, 5680 | |
| 2 | MYPE 110 | hypothetical protein | 110, 120 | |
| 2 | MYPE 4990 | hypothetical protein | 4990, 5000 | |
| Family 3 | 11 | MYPE 2560 | conserved hypothetical protein | 2550, 2560, 2580, 2610, 2660, 2670, 2680, 2710, 3140, 7300, 7320 |
| Family 4 | 9 | MYPE 3080 | ABC transporter ATP-binding protein | 1880, 3080, 3490, 3630, 5570, 6610, 9350, 9770, 10280 |
| Family 5 | 6 | MYPE 6970 | conserved hypothetical protein | 6950, 6960, 6970, 6980, 6990, 7010 |
| Family 6 | 6 | MYPE 2360 | predicted integral membrane protein | 2350, 2360, 2370, 2380, 2400, 2410 |
| Family 7 | 5 | MYPE 7590 | putative lipoprotein | 7570, 7590, 7600, 7610, 7620 |
| Family 8 | 5 | MYPE 2470 | ABC transporter ATP-binding protein | 2470, 4090, 4100, 7530, 7540 |
| Family 9 | 4 | MYPE 8450 | conserved hypothetical protein | 8430, 8440, 8450, 8490 |
| Family 10 | 4 | MYPE 7720 | conserved hypothetical protein | 7660, 7710, 7720, 7730 |
| Family 11 | 4 | MYPE 3580 | predicted integral membrane protein | 3580, 3590, 3600, 3610 |
| Family 12 | 4 | MYPE 4330 | hypothetical protein | 4330, 4340, 4350, 4360 |
| Family 13 | 4 | MYPE 4630 | hypothetical protein | 4630, 4650, 4660, 4680 |
| Family 14 | 4 | MYPE 4720 | oxidoreductase | 4720, 4810, 7970, 9500 |
| Family 15 | 3 | MYPE 890 | predicted permeases | 880, 890, 6130 |
| Family 16 | 3 | MYPE 7800 | conserved hypothetical protein | 7790, 7800, 7810 |
| Family 17 | 3 | MYPE 7910 | hypothetical protein | 7910, 7920, 7930 |
| Family 18 | 3 | MYPE 6060 | predicted integral membrane protein | 4070, 6060, 6070 |
| Family 19 | 3 | MYPE 5960 | conserved hypothetical protein | 5950, 5960, 5980 |
| Family 20 | 3 | MYPE 4230 | conserved hypothetical protein | 4230, 5260, 7650 |
| Family 21 | 3 | MYPE 4220 | conserved hypothetical protein | 2250, 4220, 7830 |
| Family 22 | 3 | MYPE 960 | ABC transporter ATP-binding protein | 960, 3560, 7280 |
| Family 23 | 3 | MYPE 2140 | hypothetical protein | 2140, 2150, 2160 |
| Family 24 | 3 | MYPE 6850 | hypothetical protein | 6850, 7110, 7140 |
| Family 25 | 3 | MYPE 5300 | hypothetical protein | 5300, 5390, 5430 |
| Family 26 | 3 | MYPE 950 | hypothetical protein | 950, 9070, 9080 |
The p35 gene family responsible for antigenic variation of lipoproteins
The largest paralogous gene family in the M.penetrans genome is the p35 gene family. The P35 lipoprotein is a surface-exposed molecule that is immunodominant and is used as the major antigen for serological diagnosis of M.penetrans infection (9,14,39,40). P35 has also been recognized as a phase-variable protein that has on and off phases with a frequency of variation ranging from 1.5 × 10–2 to 4 × 10–3 per cell per generation (40). In addition to the P35 protein, several different-sized lipid-associated membrane proteins are also phase variable and contribute to M.penetrans antigenic variation (39,41). In the M.penetrans genome, the gene for a unique bona fide P35 lipoprotein was detected and has been designated MYPE6810. A total of 44 CDSs, homologs to p35, including the p35 gene itself, were identified in M.penetrans. Of these 44 CDSs, 38 were predicted from their sequence similarity to the p35 lipoprotein gene to encode lipoproteins. More precisely, these 38 CDSs share a highly conserved amino acid sequence at the N-terminal containing a cysteine residue that binds to a fatty acid chain which anchors to the lipid bilayer of the cell membrane; however, the remaining six CDSs lack this sequence and are designated as signal-peptide-less p35 gene homologs. The 44 CDSs are clustered at four different loci in the genome. Among the 38 signal-peptide-containing CDSs, four are at the first locus from 335 914 to 355 279, the largest cluster consisting of 30 CDSs are at the second locus from 831 015 to 881 313, and the remaining four CDSs are at the third locus from 966 342 to 975 013. The six signal-peptide-less p35 gene homologs reside at the fourth locus, from 918 826 to 932 427 (Fig. 1). The degree of identity of each CDS to the P35 lipoprotein ranged from 34 to 70% of the amino acid sequence. The genetic tree for this p35 gene family indicated several points as follows: (i) 30 CDSs in the largest locus are closely related to each other, (ii) three CDSs (MYPE2690, 2700 and 7400) in the first and the third loci are also closely related to the 30 CDSs in the largest locus and (iii) six signal-peptide-less p35 gene homologs (MYPE7020–7070) are positioned at a great distance from p35 itself (MYPE6810), but are close to the remaining five p35 homologs (MYPE2620, 2630, 7330, 7370 and 7380) in the first and the third loci (the genetic tree is visible on our web site). In paralogous gene families other than p35 (i.e. MYPE2560 family with 11 CDSs and MYPE8480 family with 7 CDSs), there is no correlation between sequence similarity and gene cluster in the chromosome. These data imply that gene duplication may have occurred at separate positions in the chromosome and dynamic chromosomal rearrangement, including recombination, may be involved in paralog cluster formation during evolution.
One of the major antigenic epitopes in the M.penetrans P35 lipoprotein has been characterized previously and was shown to have the amino acid sequence, FTGEAYSVWSAK (42). The epitope is recognized in ∼66% of M.penetrans-seropositive individuals and two of three macaques infected with M.penetrans. The present study identified this sequence only in the p35 lipoprotein gene, suggesting that p35 paralogs identified in the genome may contain antigenic epitopes different from the P35 lipoprotein. Consequently, M.penetrans cells with a variety of antigenic epitopes may be able to escape from host antibody response, thus allowing persistent infection in humans.
In the M.penetrans genome, at least 50 lipoproteins (4.8% of all CDSs), including the P35 lipoprotein homologs were predicted. This number is comparable to the 4.3–7.0% of lipoproteins observed in the other four Mycoplasmataceae species.
Genetic factors related to chromosomal rearrangement
Since much of the data shown here suggested chromosomal rearrangement for paralog cluster formation in the M.penetrans genome, transposable elements as potential chromosome modulators were sought. Twenty-one putative transposases for insertion sequences (ISs) and five copies of IS232 ATP-binding proteins were found in the M.penetrans genome. Their types were distinguishable from each other and were categorized into four different groups as follows: orthologs of transposase for (i) IS1202 (1556 bp), (ii) IS232 (1412 bp), (iii) IS1630 (1016 bp) and (iv) other ISs including truncated forms. In all, ∼24 591 bp of sequence corresponding to 1.8% of the genome was identified as transposases for ISs. Inactivation of paralogous gene families by IS insertion was found in CDSs in paralog clusters from MYPE7110 to MYPE7140 and from MYPE9600 to MYPE9630, function unknown. Two CDSs (MYPE2900 and MYPE8180) for integrase/recombinase are present in the genome, both of which are thought to be involved in chromosomal rearrangement. As observed in many bacterial genomes, there is the possibility that horizontal transfer is involved in the emergence of paralogous gene families including the p35 family in M.penetrans. However, p35 and other paralogous gene clusters in M.penetrans are associated with neither integrase nor the tRNA gene loci that are, in some cases, hallmarks for phage-based horizontal transfer. The G + C content of these paralogous loci is similar to that of the whole chromosome. These data imply no association of mobile element such as phages with the emergence of these paralogs or, if any were involved, drastic rearrangement of the genome might have occurred after horizontal transfer.
Virulence factors
Several factors related to bacteria–host cell interactions were identified in the M.penetrans genome. Like M.pneumoniae and M.genitalium, M.penetrans is flask shaped and attaches to host epithelial cells with its attachment organelle, the tip structure. High molecular weight (HMW) proteins locate to the tip structure and associate with cytadherence in M.pneumoniae (43–46). Mycoplasma penetrans contains CDSs (MYPE470 and MYPE1570) that have similarities to either the cytadherence accessory protein HMW1 or the HMW2 ortholog in M.genitalium, but neither of them are HMW proteins. The CDS (MYPE1550), encoding a large predicted protein of 364 kDa, flanked by MYPE1570, is also an ortholog of both HMW2 of M.pneumoniae and the rhoptry protein of Plasmodium yoelli, which is involved in attachment to and invasion of red blood cells (47). Predicted proteins for both MYPE1550 and MYPE1570 contain numerous coiled-coil structures that are also found in HMW2 (48). MYPE1550 also contains Pfam domain spectrin repeats that are found in several proteins involved in cytoskeletal structure, so it might be a cytoskeletal component. FtsZ is speculated to interact with the cytoskeleton-like structure during cell division; the predicted size of ftsZ (MYPE8370) of M.penetrans is longer than that of other bacteria and both the N- and C-terminal sequences are M.penetrans specific. We also identified several candidates for adhesin including one CDS (MYPE1950), which has some similarity to M.pneumoniae’s adhesin P1 ortholog in Mycoplasma pirum (49). Mycoplasma penetrans also possesses CDSs encoding cytotoxic factors such as one similar to a predicted vacuolation-inducing cytotoxin-related molecule CagA of Helicobacter pylori (MYPE4730), hemolysin (MYPE1500), protease (MYPE700, MYPE8500) and endonuclease (MYPE1190). Induction of vacuolation by M.penetrans in HeLa cells was reported previously (50). We also observed induction of vacuolated degeneration in epithelium cells of macaque trachea a few hours after infection with the HF-2 strain (unpublished data). The function of the CagA-like protein is interesting and remains to be analyzed. The production of hydrogen peroxide has been suggested to injure host cells by inhibiting catalase activity of the host cells during mycoplasma infection (51). As for antioxidants, M.penetrans possesses orthologs for thiol peroxidase (MYPE3980) and glutathione peroxidase (MYPE5120). These enzymes may protect M.penetrans from oxidative molecules of its own making and are produced by host cells.
CONCLUSIONS
The M.penetrans HF-2 strain possesses a 1.3 Mb genome, which is the largest among the Mycoplasmataceae species thus far analyzed. We identified 1038 CDSs, among which 463 are M.penetrans specific. The relatively large M.penetrans genome may be accounted for by both its rich core proteome and the presence of paralog families, corresponding to 25.4% of all CDSs. The largest paralog family is the p35 gene family, consisting of 44 genes for surface antigenic lipoproteins, leading us to hypothesize that possession of a number of paralogs would enable this mycoplasma to express a large repertoire of different antigenic epitopes, presenting a sophisticated strategy for eluding the host immune system to maintain persistent infection. The genetic tree of the p35 gene family and dot-plot analysis of the chromosome suggested the occurrence of dynamic chromosomal rearrangement in paralogous gene cluster formation. Mycoplasma penetrans lacks the uridine kinase gene, suggesting the involvement of uracil phosphoribosyltransferase and an orotate-related pathway for UMP production in pyrimidine metabolism in this species. Several candidates for virulence factors such as a cytoskeletal component of the attachment organella, adhesin and cytotoxic factors were also identified.
The data obtained in this study should be of great use for understanding the mechanism of M.penetrans infection of humans and will also provide new insights into the regulation of virulence factors in M.penetrans as well as other Mycoplasmataceae.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to show their appreciation to Dr A. Blanchard at INRA, Centré de Recherche de Bordeaux, France, and Dr K. Hashimoto and Dr Y. Arakawa at the National Institute of Infectious Diseases for their helpful suggestions. The authors are grateful to Mr S. Minns and Dr T. D. Taylor for careful reading of the manuscript. This work was supported in part by the Research for the Future Program from the Japanese Society for the Promotion of Science (JSPS).
DDBJ/EMBL/GenBank accession nos+
REFERENCES
- 1.Maniloff J. (1992) In Maniloff,J., McElhaney,R.N., Finch,L.R. and Baseman,J.B. (eds), Mycoplasmas, Molecular Biology and Pathogenesis. American Society for Microbiology, Washington, DC, pp. 549–559.
- 2.Razin S. and Freundt,E.A. (1984) In Krieg,N.R. and Holt,J.G. (eds), Bergey’s Manual of Systematic Bacteriology. Williams & Wilkins, Baltimore, Vol. 1, pp. 740–770.
- 3.Woese C.R., Maniloff,J. and Zablen,L.B. (1980) Phylogenetic analysis of the mycoplasmas. Proc. Natl Acad. Sci. USA, 77, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosengarten R. and Wise,K.S. (1990) Phenotypic switching in mycoplasmas: phase variation of diverse surface lipoproteins. Science, 19, 315–318. [DOI] [PubMed] [Google Scholar]
- 5.Lo S.C. (1992) In Maniloff,J., McElhaney,R.N., Finch,L.R. and Baseman,J.B. (eds), Mycoplasmas, Molecular Biology and Pathogenesis. American Society for Microbiology, Washington, DC, pp. 523–545.
- 6.Dallo S.F. and Baseman,J.B. (2000) Intracellular DNA replication and long-term survival of pathogenic mycoplasmas. Microb. Pathog., 29, 301–309. [DOI] [PubMed] [Google Scholar]
- 7.Hussain A.I., Robson,W.L., Kelley,R., Reid,T. and Gangemi,J.D. (1999) Mycoplasma penetrans and other mycoplasmas in urine of human immunodeficiency virus-positive children. J. Clin. Microbiol., 37, 1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordova C.M., Takei,K., Rosenthal,C., Miranda,M.A., Vaz,A.J. and da Cunha,R.A. (1999) Evaluation of IgG, IgM and IgA antibodies to Mycoplasma penetrans detected by ELISA and immunoblot in HIV-1-infected and STD patients, in São Paulo, Brazil. Microbes Infect., 1, 1095–1101. [DOI] [PubMed] [Google Scholar]
- 9.Grau O., Slizewicz,B., Tuppin,P., Launay,V., Bourgeois,E., Sagot,N., Moynier,M., Lafeuillade,A., Bachelez,H., Clauvel,J.P., Blanchard,A., Bahraoui,E. and Montagnier,L. (1995) Association of Mycoplasma penetrans with HIV infection. J. Infect., 172, 672–681. [DOI] [PubMed] [Google Scholar]
- 10.Lo S.C., Hayes,M.M., Wang,R.Y., Pierce,P.F., Kotani,H. and Shih,J.W. (1991) Newly discovered mycoplasma isolated from patients infected with HIV. Lancet, 338, 1415–1418. [DOI] [PubMed] [Google Scholar]
- 11.Lo S.C., Hayes,M.M., Tully,J.G., Wang,R.Y., Kotani,H., Pierce,P.F., Rose,D.L. and Shih,J.W. (1992) Mycoplasma penetrans sp. nov., from the urogenital tract of patients with AIDS. Int. J. Syst. Bacteriol., 42, 357–364. [DOI] [PubMed] [Google Scholar]
- 12.Lo S.C., Hayes,M.M., Kotani,H., Pierce,P.F., Wear,D.J., Newton,P.B.,III, Tully,J.G. and Shih,J.W. (1993) Adhesion onto and invasion into mammalian cells by Mycoplasma penetrans: a newly isolated mycoplasma from patients with AIDS. Mod. Pathol., 6, 276–280. [PubMed] [Google Scholar]
- 13.Wang R.Y., Shih,J.W., Grandinetti,T., Pierce,P.F., Hayes,M.M., Wear,D.J., Alter,H.J. and Lo,S.C. (1992) High frequency of antibodies to Mycoplasma penetrans in HIV-infected patients. Lancet, 340, 1312–1316. [DOI] [PubMed] [Google Scholar]
- 14.Wang R.Y., Shih,J.W., Weiss,S.H., Grandinetti,T., Pierce,P.F., Lange,M., Alter,H.J., Wear,D.J., Davies,C.L., Mayur,R.K. and Lo,S.C. (1993) Mycoplasma penetrans infection in male homosexuals with AIDS: high seroprevalence and association with Kaposi’s sarcoma. Clin. Infect. Dis., 17, 724–729. [DOI] [PubMed] [Google Scholar]
- 15.Grau O., Tuppin,P., Slizewicz,B., Launay,V., Goujard,C., Bahraoui,E., Delfraissy,J.F. and Montagnier,L. (1998) A longitudinal study of seroreactivity against Mycoplasma penetrans in HIV-infected homosexual men: association with disease progression. AIDS Res. Hum. Retroviruses, 14, 661–667. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki Y., Blanchard,A., Watson,H.L., Garcia,S., Dulioust,A., Montagnier,L. and Gougeon,M.L. (1995) In vitro influence of Mycoplasma penetrans on activation of peripheral T lymphocytes from healthy donors or human immunodeficiency virus-infected individuals. Infect. Immun., 63, 4277–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yáñez A., Cedillo,L., Neyrolles,O., Alonso,E., Prévost,M.-C., Rojas,J., Watson,H.L., Blanchard,A. and Cassell,G.H. (1999) Mycoplasma penetrans bacteremia and primary antiphospholipid syndrome. Emerg. Infect. Dis., 5, 164–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambaud I., Heilig,R., Ferris,S., Barbe,V., Samson,D., Galisson,F., Moszer,I., Dyvig,K., Wróblewski,H., Viari,A., Rocha,E.P.C. and Blanchard,A. (2001) The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res., 29, 2145–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass J.I., Lefkowitz,E.J., Glass,J.S., Heiner,C.R., Chen,E.Y. and Cassell,G.H. (2000) The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature, 407, 757–762. [DOI] [PubMed] [Google Scholar]
- 20.Himmelreich R., Hilbert,H., Plagens,H., Pirkl,E., Li,B.C. and Herrmann,R. (1996) Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res., 24, 4420–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mushegian A.R. and Koonin,E.V. (1996) A minimal gene set for a cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA, 93, 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hattori M., Tsukahara,F., Furuhata,Y., Tanahashi,H., Hirose,M., Saito,M., Tsukuni,S. and Sakaki,Y. (1997) A novel method for making nested deletions and its application for sequencing of a 300 kb region of human APP locus. Nucleic Acids Res., 25, 1802–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon D., Desmarais,C. and Green,P. (2001) Automated finishing with autofinish. Genome Res., 11, 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delcher A.L., Harmon,D., Kasif,S., White,O. and Salzberg,S.L. (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res., 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa J. and Hotta,K. (1999) FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol. Lett., 174, 251–253. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heijne G. (1992) Membrane protein structure prediction, hydrophobicity analysis and the positive-inside rule. J. Mol. Biol., 225, 487–494. [DOI] [PubMed] [Google Scholar]
- 29.Lupas A., Van Dyke,M. and Stock,J. (1991) Predicting coiled coils from protein sequences. Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- 30.Lowe T.M. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraser C.M., Gocayne,J.D., White,O., Adams,M.D., Clayton,R.A., Fleischmann,R.D., Bult,C.J., Kerlavage,A.R., Sutton,G., Kelley,J.M., Fritchman,J.L., Weidman,J.F., Small,K.V., Sandusky,M., Fuhrmann,J., Nguyen,D., Utterback,T.R., Saudek,D.M., Phillips,C.A., Merrick,J.M., Tomb,J.-F., Dougherty,B.A., Bott,K.F., Hu,P.-C., Lucier,T.S., Peterson,S.N., Smith,H.O., Hutchison,C.A.,III and Venter,J.C. (1995) The minimal gene complement of Mycoplasma genitalium. Science, 270, 397–403. [DOI] [PubMed] [Google Scholar]
- 32.Mount D.W. (2001) Genome analysis. In Bioinformatics: Sequence and Genome Analysis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 479–532.
- 33.Rubin G.M., Yandell,M.D., Wortman,J.R., Miklos,G.L.G., Nelson,C.R., Hariharan,I.K., Fortini,M.E., Li,P.W., Apweiler,R., Fleischmann,W., Cherry,J.M., Henikoff,S., Skupski,M.P., Misra,S., Ashburner,M., Birney,E., Boguski,M.S., Brody,T., Brokstein,P., Celniker,S.E., Chervitz,S.A., Coates,D., Cravchik,A., Gabrielian,A., Galle,R.F., Gelbart,W.M., George,R.A., Goldstein,L.S.B., Gong,F., Guan,P., Harris,N.L., Hay,B.A., Hoskins,R.A., Li,J., Li,Z., Hynes,R.O., Jones,S.J.M., Kuehl,P.M., Lemaitre,B., Littleton,J.T., Morrison,D.K., Mungall,C., O’Farrell,P.H., Pickeral,O.K., Shue,C., Vosshall,L.B., Zhang,J., Zhao,Q., Zheng,X.H., Zhong,F., Zhong,W., Gibbs,R., Venter,J.C., Adams,M.D. and Lewis,S. (2000) Comparative genomics of the eukaryotes. Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Himmelreich R., Plagens,H., Hilbert,H., Reiner,B. and Herrmann,R. (1997) Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res., 25, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutchison C.A., Peterson,S.N., Gill,S.R., Cline,R.T., White,O., Fraser,C.M., Smith,H.O. and Venter,J.C. (1999) Global transposon mutagenesis and a minimal Mycoplasma genome. Science, 286, 2089–2090. [DOI] [PubMed] [Google Scholar]
- 36.Koonin E.V. (2000) How many genes can make a cell: the minimal-gene-set concept. Annu. Rev. Genomics Hum. Genet., 1, 99–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finch L.R. and Mitchell,A. (1992) In Maniloff,J., McElhaney,R.N., Finch,L.R. and Baseman,J.B. (eds), Mycoplasmas, Molecular Biology and Pathogenesis. American Society for Microbiology, Washington, DC, pp. 211–230.
- 38.Koonin E.V., Mushegian,A.R. and Bork,P. (1996) Non-orthologous displacement. Trends Genet., 12, 334–336. [PubMed] [Google Scholar]
- 39.Ferris S., Watson,H.L., Neyrolles,O., Montagnier,L. and Blanchard,A. (1995) Characterization of a major Mycoplasma penetrans lipoprotein and of its gene. FEMS Microbiol. Lett., 130, 313–319. [DOI] [PubMed] [Google Scholar]
- 40.Neyrolles O., Chambaud,I., Ferris,S., Prévost,M.C., Sasaki,T., Montagnier,L. and Blanchard,A. (1999) Phase variations of the Mycoplasma penetrans main surface lipoprotein increase antigenic diversity. Infect. Immun., 67, 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Röske K., Blanchard,A., Chambaud,I., Citti,C., Helbig,J.H., Prévost,M.C., Rosengarten,R. and Jacobs,E. (2001) Phase variation among major surface antigens of Mycoplasma penetrans. Infect. Immun., 69, 7642–7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neyrolles O., Eliane,J.P., Ferris,S., Cunha,R.A., Prévost,M.C., Bahraoui,E. and Blanchard,A. (1999) Antigenic characterization and cytolocalization of P35, the major Mycoplasma penetrans antigen. Microbiology, 145, 343–355. [DOI] [PubMed] [Google Scholar]
- 43.Baseman J.B., Cole,R.M., Krause,D.C. and Leith,D.K. (1982) Molecular basis for cytadsorption of Mycoplasma pneumoniae. J. Bacteriol., 151, 1514–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisseha M., Göhlmann,H.W.H., Herrmann,R. and Krause,D.C. (1999) Identification and complementation of frameshift mutations associated with loss of cytadherence in Mycoplasma pneumoniae. J. Bacteriol., 181, 4404–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krause D.C., Leith,D.K., Wilson,R.M. and Baseman,J.B. (1982) Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun., 35, 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krause D.C. and Balish,M.F. (2001) Structure, function, and assembly of the terminal organelle of Mycoplasma pneumoniae. FEMS Microbiol. Lett., 198, 1–7. [DOI] [PubMed] [Google Scholar]
- 47.Khan S.M., Jarra,W., Bayele,H. and Preiser,P.R. (2001) Distribution and characterisation of the 235 kDa rhoptry multigene family within the genomes of virulent and avirulent lines of Plasmodium yoellii. Mol. Biochem. Parasitol., 114, 197–208. [DOI] [PubMed] [Google Scholar]
- 48.Krause D.C., Proft,T., Hedreyda,C.T., Hilbert,H., Plagens,H. and Herrmann,R. (1997) Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytoadherence. J. Bacteriol., 179, 2668–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tham T.N., Ferris,S., Bahraoui,E., Canarelli,S., Montagnier,L. and Blanchard,A. (1994) Molecular characterization of the P1-like adhesin gene from Mycoplasma pirum. J. Bacteriol., 176, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borovsky Z., Zhang,M. and Rottem,S. (1998) Protein kinase c activation and vacuolation in HeLa cells invaded by Mycoplasma penetrans. J. Med. Microbiol., 47, 915–922. [DOI] [PubMed] [Google Scholar]
- 51.Almagor M., Yatziv,S. and Kahane,I. (1983) Inhibition of host cell catalase by Mycoplasma pneumoniae: a possible mechanism for cell injury. Infect. Immun., 41, 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.