Abstract
The cross-linking reaction described previously in the DNA and 2′-O-methyl RNA series is extended to RNA duplexes. A 17mer single-stranded RNA containing the 1,3-trans-{Pt(NH3)2[(GAG)-N7G,N7G]} intrastrand chelate, named G*AG* (* indicating a platinated base) gives, upon pairing with the complementary RNA strand, the G*AG/CUC* interstrand cross-link. The rate of the reaction in 200 mM NaClO4 is similar to that observed for DNA–RNA duplexes. It depends on the added Na+ or Mg2+ cation and on its concentration. RNA duplexes containing GA/GA or AG/AG tandem mismatches in the rearrangement triplet core were also studied. The major interstrand cross-links, G*AG/CGA* and G*AG/AGC*, are accompanied by a minor one involving the central G of the CGA or AGC complementary sequence G*AG/CG*A and G*AG/AG*C. In 200 mM NaClO4, the G*A/GA tandem mismatch does not modify the rate of the cross-linking rearrangement whereas the AG*/AG mismatch slows it down by a factor of four. Our results reflect the predominance of the local structure of the rearrangement core over the nucleophility of the cross-linking base. They also show that the reaction could be used to trap tertiary structures of naturally occurring RNAs, including those with the commonly encountered GA/GA mismatch.
INTRODUCTION
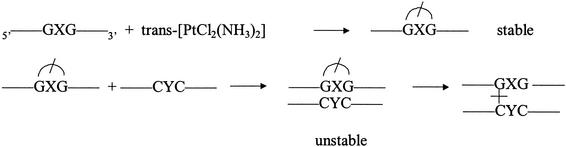
During recent years many studies have been devoted to the interactions between DNA and platinum complexes (1–5) because of the discovery of the antitumour properties of cis-diamminedichloroplatinum(II) (6,7). Furthermore, the use of platinum complexes as tools for structural studies of oligonucleotides has been investigated. Particularly, a new approach has been proposed to cross-link specifically and irreversibly two complementary oligonucleotides, based on the rearrangement of a trans-Pt(NH3)2 1,3-intrastrand chelate into an interstrand cross-link (Scheme 1) (8–11). The 1,3-trans-{Pt(NH3)2[d(GXG)-N7G,N7G]} [or G*XG* trans-Pt(NH3)2] intrastrand chelates are formed in the reaction between trans-diamminedichloroplatinum(II) and single-stranded oligonucleotides containing a triplet sequence GXG (X being any nucleotide residue but G) (1). These adducts are most often kinetically inert as long as the platinated oligonucleotides are single stranded (1,8,12,13). Hybrid isation with the complementary strands promotes the rearrangement of the intrastrand chelates into interstrand cross-links. The half-lives of the 1,3-intrastrand chelates within the duplexes depend on the nature of the nucleic acid of both the platinated strand (DNA or 2′-O-methyl RNA) and the complementary strand [DNA, RNA or RNA(-1) (10); RNA(-1) indicates an RNA strand in which the base triplet opposite the adduct is replaced by a doublet]. The influence of the X nucleotide, its complementary base Y and the nucleotides adjacent to the platinated triplet, as well as the role of mismatches and missing bases have been investigated. The fastest rate of rearrangement has been observed for the reaction between 2′-O-methyl G*UG* RNAs facing RNA(-1)s containing the AU doublet (half-reaction times of a few seconds) (10). This was applied to HBL100ras1 cells to target the coding region of Ha-ras mRNA, and led to inhibition of cell proliferation (10).
Scheme 1. Schematic representation of the interstrand cross-linking rearrangement of a 1,3-trans-{Pt(NH3)2[d(GXG)-N7G, N7G]} chelate (noted G*XG* in the text) within a duplex DNA (8). The half circle, crossed by a line, indicates the 1,3-N7G, N7G chelate of the trans-Pt(NH3)2 moiety.
Recently, platinum complexes have been used as structural tools to investigate folded structures of DNAs. Parallel-stranded DNAs with Hoogsteen base pairing were stabilised by a trans-Pt(NH3)2 cross-link (14), and quadruplex structures of telomeric sequences have been cross-linked by cis- and trans-[Pt(NH3)2(H2O)2]2+ (15). While many properties of RNAs are being discovered, including antisense-based regulation and catalysis (16–18), it seems of interest to use the rearrangement of 1,3-trans-platinum adducts to cross-link specifically and irreversibly selected RNA tertiary structures which often condition RNA activities.
In this paper, we report that the interstrand cross-linking rearrangement occurs within an RNA–RNA duplex between a 17mer containing the central G*AG*-trans-Pt(NH3)2 chelate, and three complementary oligoribonucleotides with the 5′-CUC-3′, 5′-CGA-3′ or 5′-AGC-3′ triplets respectively facing the G*AG*-platinum adduct. The influence of the tandem mismatches G*A/GA and AG*/AG in the rearrangement triplet core was studied because the former is commonly found in natural RNAs (19). Whereas the G*A/GA mismatch does not affect the rate of the cross-linking reaction, the AG*/AG mismatch slows it down by a factor of four. Several ionic strength conditions and low salt concentrations were found not to affect the half-reaction times of the rearrangements significantly.
MATERIALS AND METHODS
Chemicals
Oligoribonucleotides were purchased from Eurogentec (Belgium). Trans-diamminediaquaplatinum(II) trans-[Pt (NH3)2(H2O)2](NO3)2 was synthesised from trans-[PtCl2(NH3)2] according to the method described previously (20). T4 polynucleotide kinase, ribonuclease T1 and [γ-32P]ATP were from Amersham.
Electrophoresis
Reaction products were analysed using gel electrophoresis. Reaction mixtures were added to an equal volume of 20-80 water-formamide solution containing 100 mM EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue. The different species were separated by 20% denaturing polyacrylamide gel electrophoresis (19:1 monomer:bisacrylamide ratio) in 90 mM Tris borate buffer pH 8.3, containing 2 mM EDTA and 7 M urea. The gels were scanned and quantified using a Molecular Dynamics Phosphorimager with the ImageQuant and Kaleidagraph softwares for data processing. Then oligonucleotides were isolated from the gel, eluted into 0.15 M sodium chloride and recovered by ethanol precipitation.
RNA labelling
Oligonucleotides were 5′-end labelled with 32P by incubation with T4 polynucleotide kinase and [γ-32P]ATP at 37°C for 30 min and then purified by 20% polyacrylamide gel electrophoresis.
Platination reactions
5′-rCUUCUCUGAGUCUCUUC-3′ (250 µM) was incubated with trans-diamminediaquaplatinum(II) (250 µM) in acidified water pH 3 at 37°C for 2 h, in order to form monoplatinated adducts on N7G due to the pKa of N1A (pKa = 4.1) and N3C (pKa = 4.6) and to prevent formation of polyplatinated products. The reaction mixture was precipitated by ethanol for 2 h to remove the unreacted platinum complex. The oligonucleotides were then incubated in acidified water pH 3 at 25°C for 24 h in order to transform the monoadducts into intrastrand chelates. This multistep process was used because the chelation is slower than the first platination step (21). At the end of the reaction, platinated oligonucleotides, which migrate slower than non-platinated strands, were isolated with a 50% yield using 24% denaturing polyacrylamide gel electrophoresis.
Rearrangement reaction
5′-rCUUCUCUG*AG*UCUCUUC-3′ (where * indicates a platinated base) (10 µM) and the chosen complementary strand (50–100 µM) were incubated in 50 mM Tris–HCl buffer pH 7.5 at 25°C for 5 days, in the presence of 200 mM NaClO4, 100 mM MgCl2 or 0.5 mM MgCl2. At several time intervals, aliquots were withdrawn and analysed by polyacrylamide gel electrophoresis under denaturing conditions.
Two sets of experiments were run. In the first one, both strands were 32P-labelled. After the rearrangement reaction, removal of platinum by NaCN treatment of the interstrand cross-linked products allowed us to check that the two complementary oligonucleotides were actually bridged. In the second one, either the platinated strand or the complementary one was 32P-labelled. Then we used alkaline hydrolysis to identify the platination sites on each oligonucleotide in the cross-linked strands.
Identification of the platinum binding sites
Alkaline hydrolysis. Limited alkaline hydrolysis of oligonucleotides was carried out in 100 mM carbonate buffer pH 9.2 in the presence of 0.5 mg/ml tRNA at 100°C for 2–5 min. tRNA was added in order to obtain a partial hydrolysis of the small amount of oligonucleotide studied in the mixture.
Digestion by endonuclease T1. Oligonucleotides were digested by 0.0125 U/µl of ribonuclease T1 from Aspergillus oryzae in the presence of 0.3 mg/ml tRNA in 15 mM citrate buffer pH 5.0 for 15 min at 55°C.
NaCN treatment. Interstrand cross-linked oligonucleotides were incubated in 2 M NaCN at 37°C for 15 h in order to achieve complete removal of platinum and the resulting unbound 17mers were precipitated with ethanol (22). They were then compared with the starting single-stranded oligonucleotides on gels.
RESULTS
For the purpose of clarity, the 5′-rCUUCUCUG8A9 G10UCUCUUC-3′ strand is abbreviated to GAG, and G*AG* when the trans-Pt(NH3)2 chelate is formed. The rearrangement was studied upon annealing the platinated strand with three different complementary strands named CUC, CGA and AGC (Scheme 2). The first duplex GAG/CUC is normally paired whereas the two others, GAG/CGA and GAG/AGC, contain the two tandem mismatches GA/GA and AG/AG, respectively, within the rearrangement triplet core.
Scheme 2. (A) Ribonucleotides used in this work and their abbreviations. (B) The interstrand cross-linked products formed by the rearrangements and their abbreviations (* indicates a platinated base). They are referred to their gel migration band (Fig. 2) by the symbol of the bridged base of the complementary strand.
Formation and identification of the G*AG* platinated strand
The single-stranded 17mer GAG was incubated with trans-diamminediaquaplatinum(II) in order to form the 1,3 intrastrand chelate as described for DNA oligonucleotides containing GXG sequences (8) (see Materials and Methods). At the end of the reaction, the product was isolated, labelled with 32P, and digested by 0.025 U/µl of ribonuclease T1 in order to determine the platinated bases. We have shown that this endonuclease, which cleaves RNA oligonucleotides specifically at the 3′-side of a guanosine, is totally inhibited, when used at this concentration, by the presence of a platinum adduct on the substrate guanine. It is also inhibited to 40% on Gn–2 if a platinum adduct is present on Gn (22,23). Treatment of the 17mer platination product by ribonuclease T1 gave no digested fragments. This can account for 100% platination on G10. The absence of cleavage at G8 could be due to 100% platination, but it could also partially result from the cleavage inhibition due to a G10 adduct (23). An alkaline hydrolysis of the platinated product revealed, after migration of the products on an electrophoretic gel, a ‘gap’ between a 7mer and a platinated 10mer, which indicated that the hydrolyses after G8 or A9 did not release the expected 8mer or 9mer but two fragments still linked by the platinum complex (Supplementary Material). These results allowed us to conclude that 100% platination of G8 and G10, and to the formation of the 1,3-trans-{Pt(NH3)2[d(GXG)-N7G,N7G]} chelate, occurred.
Rearrangement of the intrastrand chelate
The platinated oligonucleotide G*AG* was incubated in 0.5 mM MgCl2 at 25°C for several days with each of the three complementary strands CUC, CGA and AGC. Gel electrophoresis revealed new products, migrating much more slowly than the starting oligonucleotides, suggesting that interstrand cross-linking had occurred (Fig. 1). Only one new product appeared for the G*AG*/CUC duplex whereas two new products appeared for the G*AG*/CGA and G*AG*/AGC mismatched sequences. In these latter cases, the slowest migrating product was formed with the highest yield. To prove the formation of interstrand cross-links, the products of the new bands were extracted from the gel and the oligonucleotides were recovered by ethanol precipitation. They were then incubated in 2 M NaCN at 37°C overnight to remove platinum complexes and loaded on denaturing electrophoresis gels to be compared with reference oligonucleotides (Fig. 2). Upon NaCN treatment, the new products regenerated GAG and the complementary strands, which indicated that platinum was bound to the paired strands. Therefore the 1,3-intrastrand chelate had rearranged into an interstrand cross-link with the complementary strand.
Figure 1.
Kinetics, in days, of formation of the interstrand cross-links within the three duplexes G*AG*/CUC, G*AG*/CGA and G*AG*/AGC in 0.5 mM MgCl2 at 25°C; both strands were 5′-end labelled. Autoradiogram of a denaturing 20% polyacrylamide gel electrophoresis. Lanes 1–5 refer to incubation times of 1–5 days respectively. Lane G*AG* indicates the platinated single strand. ‘A-, C- or G-band’ indicates, for each product, the base of the complementary strand which is cross-linked to the 5′-G* of G*AG (Scheme 2). These bases are identified in Figure 2.
Figure 2.
Determination of the nucleotides involved in the interstrand cross-links formed upon pairing G*AG* with CUC in 0.5 or 100 mM MgCl2. Denaturing gel electrophoresis of the adducts before (–) and after (+) platinum removal by treatment with 2 M NaCN at 37°C for 15 h. Initial samples GAG and CUC are shown.
The same results (formation of one or two interstrand cross-links) were obtained in 50 mM Tris–HCl pH 7.5, 100 mM MgCl2 or 200 mM NaClO4. The latter conditions were chosen for the sake of comparison with experiments reported (8). As divalent cations play an important role in RNA folding, especially Mg2+, we also tested the rearrangement in the presence of 100 mM MgCl2, which corresponds to the same ionic strength as 200 mM NaClO4.
Identification of the interstrand cross-links
To identify the platination sites involved in the interstrand cross-links, the new adducts were subjected to a mild alkaline hydrolysis to obtain one cleavage of the RNA backbone per duplex (10). The resulting fragments were analysed by gel electrophoresis under denaturing conditions. Data relative to the duplex GAG/CUC are shown in Figure 3. The fragments up to the cross-linked residue are detected, indicating that the cross-linked bases were the 5′-G* residue of the initial intrastrand chelate and the 3′-C residue facing it, i.e. G*AG/CUC*. For the duplexes containing the GA/GA and AG/AG mismatches, we observed two products on the gel (Fig. 1) corresponding to the formation of two distinct interstrand cross-links. In both cases, the major product results from the cross-link similar to that formed in G*AG*/CUC, i.e. G*AG/CGA* and G*AG/AGC* respectively. The minor product results from the cross-linking of the 5′-G* of G*AG* by the central G of the two complementary sequences, i.e. G*AG/CG*A and G*AG/AG*C respectively (Scheme 2).
Figure 3.
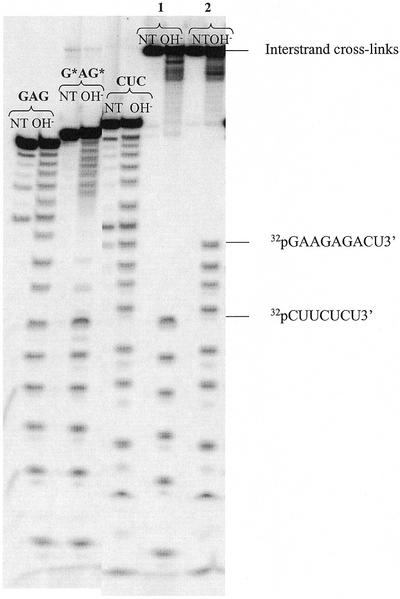
Determination of the platinum binding sites of the intrastrand cross-link G*AG* and the interstrand cross-links G*AG/CUC*, obtained upon pairing G*AG* with CUC in 0.5 mM MgCl2. Denaturing gel electrophoresis of the oligonucleotides before (lanes NT) and after (lanes OH–) limited alkaline hydrolysis in 100 mM carbonate buffer pH 9.2 in the presence of 0.5 mg/ml tRNA at 100°C for 4 min. Lanes G*AG refer to the intrastrand cross-link. Lane 1 refers to cross-linked duplexes, 32P-labelled at the 5′-end of the platinated strand. Lane 2 refers to cross-linked duplexes, 32P-labelled at the 5′-end of the complementary strand.
Half-reaction times determination
We determined the half-reaction times of the conversion of the G*AG* chelate into interstrand cross-links. G*AG*/CUC undergoes the rearrangement more quickly in 200 mM NaClO4 (37 h) than in 0.5–100 mM MgCl2 (∼100 h). In the case of the G*AG*/CGA and G*AG*/AGC duplexes, half-reaction times are between 40 and 380 h, depending on the complementary sequence, the nature and the concentration of the cation (Table 1). When G*AG* is paired with CGA, the rearrangement occurs with the same half-reaction time (∼45 h) whatever the salt and its concentration. G*AG*/AGC is the slowest system whose rearrangement has a half-reaction time >200 h, reaching 380 h, i.e. more than 15 days, in 100 mM MgCl2.
Table 1. Half-reaction times (in hours) of the rearrangement of the G*AG* 1,3-intrastrand trans-Pt(NH3)2 chelate as a function of the sequence of the complementary RNA strand and of the nature and concentration of the salt.
| 0.5 mM MgCl2 | 100 mM MgCl2 | 200 mM NaClO4 | |
|---|---|---|---|
| 5′-CUC-3′ | 86 | 105 | 37 |
| 5′-CGA-3′ | 44 | 52 | 49 |
| 5′-AGC-3′ | 216 | 377 | 202 |
DISCUSSION
A 17mer RNA containing the GAG sequence reacts with trans-diamminediaquaplatinum(II) to yield the 1,3-G*AG* chelate of the trans-Pt(NH3)22+ moiety. Such G*XG* chelates were known to rearrange into interstrand cross-links in DNA–DNA, DNA–RNA and 2′-O-methyl RNA–RNA duplexes (8–11). We have shown that this cross-linking rearrangement also occurs within RNA–RNA duplexes (Fig. 1). Three duplexes were studied, involving the same G*AG* platinated strand, paired with the CUC, CGA and AGC complementary strands, the latter two forming the tandem G*A/GA and AG*/AG mismatches respectively within the rearrangement triplet core (Scheme 2). For the G*AG*/CUC duplex, the interstrand cross-link is formed between the 5′-G* residue of the platinated strand and its complementary C as expected from previous results (8,9). For the duplexes G*AG*/CGA and G*AG*/AGC, the major cross-linking reactions by the 3′-A or 3′-C of the complementary strand, giving G*AG/CGA* and G*AG/AGC*, can be compared with those observed by N1A and N3C in the deoxy series for the two duplexes d(CTCCTG*TGTCTC)/d(GAGATA*AGGAG) (24) and d(CTCTCG*AGTCTC)/d(GAGACTC*GAGAG) (25), analysed by NMR. The major interstrand cross-links are accompanied by minor ones involving the central G of the CGA or AGC sequences facing the platinum chelate. The same type of minor interstrand cross-link has already been observed in DNA–DNA duplexes for G*XG*/CYC sequences between the 5′-G* and the Y residue when Y is a purine (8,9). The occurrence of this second type of cross-linking was not unexpected, considering the known higher nucleophilicity for platinum(II) of a N7G compared with those of the N1A or N3C (1,26). In the case of our tandem mismatched duplexes, the low proportions of the G*AG/CG*A and G*AG/AG*C cross-links probably reflect the predominance of a steric control of the interstrand attack on the platinum over the nucleophilicity of the cross-linking base.
The half-reaction times for the cross-linking rearrangements G*AG*/CUC → G*AG/CUC*, G*AG*/CGA → G*AG/CGA* and G*AG*/AGC → G*AG/AGC* in 200 mM NaClO4 at 25°C are 37, 49 and 202 h respectively (Table 1). The first two rearrangements have comparable rates and are slower than that occurring for d(G*AG*)/d(CTC) within DNA–DNA duplexes (6 h) but they occur at a similar rate to that observed for d(G*AG*)/r(CAC) in DNA–RNA duplexes (50 h) (8,10). It is noteworthy that our third reaction within G*AG*/AGC is particularly slow.
The reaction within G*AG*/CUC is slower in the presence of Mg2+ than Na+ (Table 1). This could be related to the fact that Mg2+ is known to induce the formation of secondary and tertiary structures in RNAs, which might impede the proper pairing and flexibility required by the interstrand reaction. No such cation effect is observed for the tandem mismatch duplexes G*AG*/CGA and G*AG*/AGC. The tandem GA/GA mismatch in RNAs has been shown to present a binding site for Mg2+ ions, between the guanine N7 and the phosphodiester linkage with adenine (27). Therefore, if this binding still occurs for the G*AG*/CGA duplex, it appears not to affect the helical conformation, which allows the same rearrangement rate as that observed in the presence of Na+. On the contrary, the structure induced by the AG/AG mismatch impedes the rearrangement whatever the cation and slows it down by a factor of four. Comparing the features of the different sequences studied raises the following points. (i) In the three cases, the same G*AG* intrastrand chelate of trans-Pt(NH3)2 is present. The only previously published studies concern the 1,3-trans-Pt(NH3)2 and 1,3-cis-Pt(NH3)2 chelates formed either with the deoxy trinucleotide d(GpTpG) (28) or for the second adduct with d(GTG) deoxy 11mer duplex (29). NMR concluded to the exclusion of the central T base, the maintaining of a B-type DNA helix and the adoption of a pure N conformation by the 5′-G* (29). Similar conclusions were obtained by molecular modelling of a deoxy octanucleotide with the G*TG*-trans-Pt(NH3)2 adduct (30). No such data are available on a ribo-duplex. (ii) In G*AG*/CUC and G*AG*/AGC the cross-linking base of the complementary strand is the 3′-C of the 5′-G*–3′-C pair. In G*AG*/CGA the cross-linking base is the 3′-A facing the 5′-G* within the G*A/GA mismatch. This result is in agreement with the local structure of the GA/GA tandem found in natural RNAs (19,31). NMR studies of mismatched oligonucleotides and the crystallographic structure of the hammerhead ribozyme have shown that the GA/GA mismatch within r(GGCGAGCC)2 (32), as well as within the r(-UGAGG)/r(CCGAA-) sequence of the ribozyme stem II (33,34), present the so-called ‘sheared’ conformation. It implies a non-Watson–Crick pairing N7A/NH2G and NH6A/N3G, with an anti conformation of the mismatched bases. For the AG/AG mismatch, NMR data have revealed a different non Watson–Crick pairing between NH6A/O6G and N1A/NH1G (35). The comparison of these data suggests that in G*AG*/CGA, the nucleophilic attack on platinum might be favoured by the fact that the N1A is not H-bonded. Nevertheless, G*AG*/CUC and G*AG*/CGA get cross-linked at the same rate, whereas G*AG*/AGC rearranges much more slowly. Therefore the nature of the nucleophilic base attacking platinum does not seem to be determining for the rearrangement reaction. (iii) The explanation of the different rates observed resides probably in the local structure of the rearrangement core within the duplex. Indeed, in the deoxy series, it was reported that the interstrand cross-linking rearrangement occurred efficiently, in the absence of mismatch or missing nucleotide, when the base pairing remained along the whole sequence. Such a situation is present for G*AG*/CUC but not for G*AG*/CGA and G*AG*/AGC. In G*AG*/CUC and G*AG*/CGA, the 3′-G*–5′-C pair at one end of the triplet core could favour the cross-linking reaction. This G–C pair was actually found with a typical Watson–Crick hydrogen bonding in an interstrand cross-linked dodecamer of the d(G*AG)/d(CTC*) type (25). This suggests that the 3′-G*–5′-C pair might hold the G*AG* chelate in proper ‘pairing’ with the opposite strand favouring the rearrangement reaction. On the contrary, in G*AG*/AGC, the local structure induced by the AG/AG tandem and particularly the 3′-G*-5′-A mismatch might be unfavourable to the reaching of the platinum of the G*AG* chelate by the 3′-N3C of the complementary strand.
CONCLUSION
We have shown that the rearrangement of the G*AG*-transPt(NH3)2 chelate of a 17mer RNA strand occurs upon pairing with the CUC complementary RNA strand to give the interstrand cross-link G*AG/CUC*. The reaction occurs at the same rate when the GA/GA tandem mismatch, which is common in natural RNAs, is present in the rearrangement core G*AG*/CGA. On the other end, the AG/AG mismatch in G*AG*/AGC slows down the rearrangement by a factor of four (Scheme 2). The reaction occurs at the low 0.5 mM Mg2+ concentration, which is known to be sufficient to induce the folding of some RNAs, especially those containing GA/GA tandem mismatches. This suggests that the reaction might show promise to covalently cross-link RNA tertiary structures in particular helix II of the hammerhead ribozyme (36).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Johnson-Matthey, Inc., for a generous gift of platinum complexes. This work has been supported by ARC (Association pour la Recherche sur le Cancer), contrat 9118.
REFERENCES
- 1.Lepre C.A. and Lippard,S.J. (1990) Interaction of platinum antitumor compounds with DNA. In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology. Springer-Verlag, Berlin Heidelberg, Vol. 4, pp. 9–38.
- 2.Bloemink M.J. and Reedijk,J. (1996) Cisplatin and derived anticancer drugs: mechanism and current status of DNA binding. Met. Ions Biol. Syst., 32, 641–685. [PubMed] [Google Scholar]
- 3.Gonnet F., Reeder,F., Kozelka,J. and Chottard,J.C. (1996) Kinetic analysis of the reaction between GG-containing oligonucleotides and platinum complexes. I. Reactions of single-stranded oligonucleotides with cis-[Pt(NH3)2(H2O)2]2+ and [Pt(NH3)3(H2O)]2+. Inorg. Chem., 35, 1653–1658. [DOI] [PubMed] [Google Scholar]
- 4.Jamieson E.R. and Lippard,S.J. (1999) Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev., 99, 2467–2498. [DOI] [PubMed] [Google Scholar]
- 5.Reedijk J. (1999) Why does cisplatin reach guanine-n7 with competing s-donor ligands available in the cell? Chem. Rev., 99, 2499–2510. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg B., VanCamp,L., Trosko,J.E. and Mansour,V.H. (1969) Platinum compounds: a new class of potent antitumour agents. Nature, 222, 385–386. [DOI] [PubMed] [Google Scholar]
- 7.Zamble D.B., Mu,D., Reardon,J.T., Sancar,A. and Lippard,S.J. (1996) Repair of cisplatin–DNA adducts by the mammalian excision nuclease. Biochemistry, 35, 10004–10013. [DOI] [PubMed] [Google Scholar]
- 8.Dalbiès R., Payet,D. and Leng,M. (1994) DNA double helix promotes a linkage isomerization reaction in trans-diaminedichloroplatinum(II)-modified DNA. Proc. Natl Acad. Sci. USA, 91, 8147–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boudvillain M., Dalbiès,R., Aussourd,C. and Leng,M. (1995) Intrastrand cross-links are not formed in the reaction between transplatin and native DNA: relation with the clinical inefficiency of transplatin. Nucleic Acids Res., 23, 2381–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boudvillain M., Guerin,M., Dalbies,R., Saison-Behmoaras,T. and Leng,M. (1997) Transplatin-modified oligo(2′-O-methyl ribonucleotide)s: a new tool for selective modulation of gene expression. Biochemistry, 36, 2925–2931. [DOI] [PubMed] [Google Scholar]
- 11.Colombier C., Boudvillain,M. and Leng,M. (1997) Interstrand crosslinking reaction in transplatin-modified oligo-2′-O-methyl ribonucleotide-RNA hybrids. Antisense Nucleic Acid Drug Dev., 7, 397–402. [DOI] [PubMed] [Google Scholar]
- 12.Comess K.M., Costello,C.E. and Lippard,S.J. (1990) Identification and characterization of a novel linkage isomerization in the reaction of trans-diamminedichloroplatinum(II) with 5′-d(TCTACGCGTTCT). Biochemistry, 29, 2102–2110. [DOI] [PubMed] [Google Scholar]
- 13.Dalbies R., Boudvillain,M. and Leng,M. (1995) Linkage isomerization reaction of intrastrand cross-links in trans-diamminedichloroplatinum(II)-modified single-stranded oligonucleotides. Nucleic Acids Res., 23, 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller J., Drumm,M., Boudvillain,M., Leng,M., Sletten,E. and Lippert,B. (2000) Parallel-stranded DNA with Hoogsteen base pairing stabilized by a trans-[Pt(NH3)2]2+ cross-link: characterization and conversion into a homodimer and a triplex. J. Biol. Inorg. Chem., 5, 603–611. [DOI] [PubMed] [Google Scholar]
- 15.Redon S., Bombard,S., Elizondo-Riojas,M.A. and Chottard,J.C. (2001) Platination of the (T2G4)4 telomeric sequence: a structural and cross-linking study. Biochemistry, 40, 8463–8470. [DOI] [PubMed] [Google Scholar]
- 16.Eguchi Y., Itoh,T. and Tomizawa,J. (1991) Antisense RNA. Annu. Rev. Biochem., 60, 631–652. [DOI] [PubMed] [Google Scholar]
- 17.Cech T.R. and Bass,B.L. (1986) Biological catalysis by RNA. Annu. Rev. Biochem., 55, 599–629. [DOI] [PubMed] [Google Scholar]
- 18.Gaughan D.J. and Whitehead,A.S. (1999) Function and biological applications of catalytic nucleic acids. Biochim. Biophys. Acta, 1445, 1–20. [DOI] [PubMed] [Google Scholar]
- 19.Gautheret D., Konings,D. and Gutell,R.R. (1994) A major family of motifs involving G.A mismatches in ribosomal RNA. J. Mol. Biol., 242, 1–8. [DOI] [PubMed] [Google Scholar]
- 20.Reeder F., Kozelka,J. and Chottard,J.C. (1996) Triammineplatinum(II) coordinated to a guanine does not prevent platination of an adjacent guanine in single stranded oligonucleotides. Inorg. Chem., 35, 1413–1415. [DOI] [PubMed] [Google Scholar]
- 21.Bernal-Mendez E., Boudvillain,M., Gonzalez-Vilchez,F. and Leng,M. (1997) Chemical versatility of transplatin monofunctional adducts within multiple site-specifically platinated DNA. Biochemistry, 36, 7281–7287. [DOI] [PubMed] [Google Scholar]
- 22.Bombard S., Kozelka,J., Favre,A. and Chottard,J.C. (1998) Probing the mechanism of an Mn2+-dependent ribozyme by means of platinum complexes. Eur. J. Biochem., 252, 25–35. [DOI] [PubMed] [Google Scholar]
- 23.Escaffre M., Favre,A., Chottard,J.C. and Bombard,S. (2002) Determination of platinated purines in oligoribonucleotides by limited digestion with ribonucleases T1 and U2. Anal. Biochem., in press. [DOI] [PubMed] [Google Scholar]
- 24.Andersen B., Bernal-Mendez,E., Leng,M. and Sletten,E. (2000) NMR solution structure of a DNA 12/11-mer: d(CTCCTGTGTCTC).d(GAGATA-AGGAG) containing a transplatin interstrand G-N7/A-N1 cross-link. Eur. J. Inorg. Chem., 6, 1201–1210. [Google Scholar]
- 25.Paquet F., Boudvillain,M., Lancelot,G. and Leng,M. (1999) NMR solution structure of a DNA dodecamer containing a transplatin interstrand GN7-CN3 cross-link. Nucleic Acids Res., 27, 4261–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansy S., Chu,G.Y.H., Duncan,R.E. and Tobias,R.S. (1978) Heavy metal nucleotide interaction. Competitive reaction in systems of four nucleotides with cis or trans-diamminoplatinum(II). Raman difference spectrophotometric determination of the relative nucleophilicity of guanosine, cytidine, adenosine and uridine monophosphate as well as the analogous bases in DNA. J. Am. Chem. Soc., 100, 607–616. [Google Scholar]
- 27.Tanaka Y., Morita,E.H., Hayashi,H., Kasai,Y., Tanaka,T. and Taira,K. (2000) Well-conserved tandem G-A pairs and the flanking C-G pair in hammerhead ribozymes are sufficient for capture of structurally and catalytically important metal ions. J. Am. Chem. Soc., 122, 11303–11310. [Google Scholar]
- 28.Boogaard N., Altona,C. and Reedijk,J. (1993) Conformational differences between the adducts of cis-DDP and trans-DDP with the trinucleotide d(GpTpG). A 1H and 31P NMR investigation. J. Inorg. Biochem., 49, 129–147. [Google Scholar]
- 29.Teuben J.M., Bauer,C., Wang,A.H. and Reedijk,J. (1999) Solution structure of a DNA duplex containing a cis-diammineplatinum(II) 1,3-d(GTG) intrastrand cross-link, a major adduct in cells treated with the anticancer drug carboplatin. Biochemistry, 38, 12305–12312. [DOI] [PubMed] [Google Scholar]
- 30.Prevost C., Boudvillain,M., Beudaert,P., Leng,M., Lavery,R. and Vovelle,F. (1997) Distortions of the DNA double helix induced by 1,3-trans-diamminedichloroplatinum(II)-intrastrand cross-link: an internal coordinate molecular modeling study. J. Biomol. Struct. Dyn., 14, 703–714. [DOI] [PubMed] [Google Scholar]
- 31.SantaLucia J. Jr, Kierzek,R. and Turner,D.H. (1990) Effects of GA mismatches on the structure and thermodynamics of RNA internal loops. Biochemistry, 29, 8813–8819. [DOI] [PubMed] [Google Scholar]
- 32.SantaLucia J. Jr and Turner,D.H. (1993) Structure of (rGGCGAGCC)2 in solution from NMR and restrained molecular dynamics. Biochemistry, 32, 12612–12623. [DOI] [PubMed] [Google Scholar]
- 33.Pley H.W., Flaherly,K.M. and Mc Kay,D.B. (1994) Three-dimensional structure of a hammerhead ribozyme. Nature, 372, 68–74. [DOI] [PubMed] [Google Scholar]
- 34.Scott W.G., Finch,J.T. and Klug,A. (1995) The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell, 81, 991–1002. [DOI] [PubMed] [Google Scholar]
- 35.Wu M., SantaLucia,J.,Jr and Turner,D.H. (1997) Solution structure of (rGGCAGGCC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry, 36, 4449–4460. [DOI] [PubMed] [Google Scholar]
- 36.Bassi G.S., Murchie,A.I., Walter,F., Clegg,R.M. and Lilley,D.M. (1997) Ion-induced folding of the hammerhead ribozyme: a fluorescence resonance energy transfer study EMBO J., 16, 7481–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.