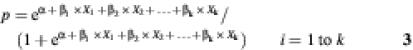Abstract
Using an empirical panel of more than 20 000 single base primer extension (SNP-IT) assays we have developed a set of statistical scores for evaluating and rank ordering various parameters of the SNP-IT reaction to facilitate high-throughput assay primer design with improved likelihood of success. Each score predicts either signal magnitude from primer extension or signal noise caused by mispriming of primers and structure of the PCR product. All scores have been shown to correlate with the success/ failure rate of the SNP-IT reaction, based on analysis of assay results. A logistic regression analysis was applied to combine all scored parameters into one measure predicting the overall success/failure rate of a given SNP marker. Three training sets for different types of SNP-IT reaction, each containing about 22 000 SNP markers, were used to assign weights to each score and optimize the prediction of the combined measure. c-Statistics of 0.69, 0.77 and 0.72 were achieved for three training sets. This new statistical prediction can be used to improve primer design for the SNP-IT reaction and evaluate the probability of genotyping success for a given SNP based on analysis of the surrounding genomic sequence.
INTRODUCTION
The most common method of SNP genotyping today is single base primer extension, which we call SNP-IT. As SNP databases grow and more groups perform chromosome- and genome-wide studies using SNPs, the relative inefficiency of SNP assay design and validation have become a major bottleneck. Here we present a strategy for improved prediction of assay success based on an empirical analysis of a large set of SNP-IT assay designs. The SNP-IT reaction has two major steps: PCR amplification of short (50–200 bp) sequences surrounding the SNP site from genomic DNA and a single base primer extension reaction using primer annealing immediately next to the SNP (1–4). The two PCR primers and one extension primer are the three primers comprising the primer set for the SNP assay. Depending on availability of adjacent sequence, several primer sets can be designed for one SNP, comprised of different PCR primer pairs in combination with the two possible extension primers. All of the SNP sites used in this study are biallelic, comprising various readout combinations such as G/T, C/A, C/T, etc. During the last 5–7 years the Orchid database has accumulated a large number of SNP markers from validation tests of the three primer sets used in the SNP-IT reaction. Two PCR primers were designed using enhanced algorithms based on standard PCR design tools such as Primer3 (5) software. The third primer used for the single base extension (SNP-IT primer) was usually designed to complement 25 bases next to the SNP site. Single base substitution by an amino group (C3-linker abasic bond) at the site of potential DNA–DNA duplex was used to destabilize SNP-IT primers capable of forming primer–primer dimers. The primer sets that failed in a functional validation of the SNP-IT reaction were stored in the database marked as ‘failed’. For each SNP, we attempted multiple rounds of primer design, with a maximum of three primer sets, until a valid marker was tested. Such a validation approach allowed an accumulation in the database of 23 525 SNP markers run on at least 44 samples, each with a known validation status of ‘passed’ or ‘failed’.
The theoretical considerations and experimental analysis of SNP-IT primer sets provided the foundation for generating a comprehensive list of possible molecular mechanisms that can lead to SNP-IT assay failure and/or predict its success. The quantitative measures of these effects (scores) were developed and used to combine all of the effects in one statistical model predicting the success/failure probability of the given primer set. The combined model uses training sets consisting of functionally validated passed and failed markers to assign a statistical weight to every score. These weights reflect the predicted contribution of each molecular mechanism or property to the overall probability of a primer set to fail or succeed in the SNP-IT assay. Examples of molecular mechanisms that were assigned scores include prediction of magnitude of noise caused by mispriming of primers in the SNP-IT assay and prediction of magnitude of a signal from primer extension. This paper describes these effects and methods to calculate the scores. We also describe the use of logistic regression to combine these individual scores into one model, calculating the complete success probability for a given primer set.
MATERIALS AND METHODS
Computation
A 933 MHz, Pentium III, 128 MB RAM PC (Professional M-933; Gateway) was used for all calculations. Software was written using the C programing language in Microsoft Visual Studio™ 6.0. Statistical Analysis System (SAS) software (6,7) was used for data analysis and statistical modeling.
Melting temperature and free energy calculations of DNA structures were calculated using the nearest neighbor method (8–16). DNA loop free energy estimates were obtained using a published model for loop stability in RNA (17–19). Corresponding table values for RNA loops initiation and bonus and first mismatch free energies were used for DNA loop calculations. However, their absolute values were adjusted to achieve the highest correlation of primary scores with the failure rate, keeping the sequence dependencies similar to RNA loops. A typical DNA structure calculation included computation of all possible consecutive match dimers between two DNA molecules and subsequent calculation of loops between found dimers. The dimer is defined as a stretch of uninterrupted matches between two DNA molecules in their alignment with each other. The same C functions were used to calculate intramolecular structures. In this case one DNA sequence was passed twice to the function. The free energy of the DNA structure was the sum of free energies of all consecutive match dimers in the structure and free energies of the loops between them.
Training sets
Three training sets were used. The first set contains 23 525 SNP markers genotyped by single-plex SNP-IT assay on at least 44 individual samples using Orchid’s SNPStream 25K instrument. The standard SNP-IT assay contains the following steps: PCR amplification of SNP marker from genomic sequence (1), targeted exonuclease digestion of the strand non-complementary to the SNP-IT primer (4), primer extension using the Klenow fragment of DNA polymerase with one biotin-labeled, one fluorescein-labeled and two unlabeled terminating dideoxynucleoside triphosphates (3).
Two other training sets were obtained using Orchid’s SNPcode platform based on the tag-array approach described earlier. The SNP-IT assay for the SNPcode platform includes 12-plex or 24-plex PCR amplification of a SNP marker, followed by multiplexed primer extension and hybridization to the solid phase hybridization SNP-IT primer for marker separation and fluorescent detection. These training sets contained 30 948 and 30 576 markers, respectively.
Logistic regression and discrimination
Logistic regression is a commonly used regression method when the dependent variable is a categorical variable (6,20–22) such as pass or fail. Logistic regression first transforms the dependent variable into a natural log of odds,
ln[p/(1 – p)]1
where p is the probability of the event (e.g. fail) (6). The effects of independent variables, called logit coefficients or weights, can then be calculated with maximum likelihood estimation from the specified model:
ln[p/(1 – p)] = α + β1 × X1 + β2 × X2 + … + βk × Xk2i = 1 to k
where α is a constant referred to as the intercept, βi values are the regression coefficients, Xi values are the independent variables, and k is the number of independent variables included in the model. In this study, p is the probability of reaction failure, βi values are the weights of primary scores to be calculated from the model using the training sets, and Xi values are the primary scores.
After weights are calculated from the model, probability of failure, p, for any new SNP-IT reaction can then be calculated as
A separate model for each training set was developed to account for different effects of primary scores in each reaction setting. All data in the training set were used to get more variations in molecular properties and smaller standard errors of estimated weights.
The c-statistic was used as a measure of discriminatory power of the logistic regression (6,23). Specifically, the c-statistic measures how many times the model assigns a higher failure probability to ‘failed’ SNP markers compared to the failure probability of ‘passed’ markers. The c-statistic ranges from around 0.5, equivalent to no discrimination power, to 1, equivalent to perfect discrimination power.
Primary score selection
All scores were designed to correlate with the failure rate of the SNP-IT reaction. The computer program contains C functions calculating an individual score for each single marker. The same C functions were used for scoring the training set markers to obtain logistic regression coefficients in the combined logistic regression model and for predicting the failure/success probability of the unknown marker. The correlation of an individual primary score with the failure rate was measured by Wald χ2 and the c-statistic of the distribution of the primary score in the training set against passes and fails of SNP markers. Only scores that showed significant correlation with failure rate and improved the prediction of the combined model were selected.
Final model construction
Selected primary scores were entered into the model based on their statistical importance. The stepwise logistic regression procedure available in SAS PROC LOGISTIC (7) was used to select several scores at a time. Stepwise logistic regression automatically selected scores that accounted for the most variation in fails and passes for a given confidence level. However, because of the large number of scores and their correlation with each other the individual effects could not be estimated reliably when all scores were entered into the model together (24). Careful attention was paid to multicolinearity each time a set of new scores was introduced to the model. When multicolinearity occurred, the scores that contributed most to the model predictability were chosen for the final model. The backward stepwise option in the logistic regression procedure with a cut-off P value <0.1 was used to find the optimal set of scores.
Several forms of scores were used in the model in addition to primary scores, depending on their relationship to probability of failure. Table 1 summarizes different scores included in the final models. A detailed description of all scores is presented in Results. The primary score itself was used if it showed a linear relationship to the log odds ratio of failure. Some scores only had a significantly elevated probability of failure in certain ranges. In that case, binary measures reflecting the cut-off in the primary score values were used. If a primary score exceeded a certain threshold, the corresponding binary score was changed from 0 to 1. In addition, the non-linear relationship between some scores and the failure rate were approximated with power functions (cube or square) or spline fit of the primary score. Such approximations were added as a new score to the model in addition to the original primary score.
Table 1. Complete list of primary scores, Wald χ2, degrees of freedom and P values from logistic models for single-plex and 12-plex SNP-IT reaction.
| Score name | Score description | Single-plex model χ2, df, P value | Multiplex model χ2, df, P value |
|---|---|---|---|
| TIN | The free energy of the most stable structure of extension primer with itself | 2.4, df = 1, P < 0.0001 | |
| Most stable 3′-end dimer | The free energy of the most stable dimer formed by annealing SNP-IT primer with itself. Only the dimers formed by last nine 3′-end bases are considered for this score. In addition a binary becoming 1 when no dimers are formed at the 3′-end is used for single-plex model | 65.2, df = 1, P < 0.0001 | |
| TDN | The free energy of the most stable structure between extension primer and PCR product | 28.1, df = 1, P < 0.0001 | 12.3, df = 1, P = 0.0005 |
| Cumulative TDN | Free energies sum of all possible dimers (no loops) between extension primer and PCR product | 36.6, df = 1, P < 0.0001 | 7.1, df = 1, P = 0.0077 |
| Number of consecutive Gs in SNP-IT primer | Binary score becomes 1 when number of consecutive Gs in SNP-IT primer is six and greater | 7.1, df = 1, P = 0.0078 | |
| Stability of last 9 bases of extension primer | The free energy of the SNP-IT primer 3′-end. In addition to this score single-plex model uses its cube approximation and multiplex model uses its square approximation: | ||
| Linear term of stability of last 9 bases of extension primer | 60.1, df = 1, P < 0.0001 | 284.1, df = 1, P < 0.0001 | |
| Square term of stability of last 9 bases of extension primer | 172.0, df = 1, P < 0.0001 | ||
| Cubic term of stability of last 9 bases of extension primer | 32.1, df = 1, P < 0.0001 | ||
| Number of C3 linkers | Number of C3 linkers in the extension primer | 149.3, df = 1, P < 0.0001 | |
| Binary score becomes one when number of C3 linkers is equal to 1 | 16.4, df = 1, P < 0.0001 | ||
| Binary score becomes one when number of C3 linkers is greater than 3 | 8.9, df = 1, P = 0.0029 | ||
| %GC of PCR primers | 29.7, d.f.=1, P < 0.0001 | 43.4, df = 1, P < 0.0001 | |
| Amplicon melting temperature | 106.0, df = 1, P < 0.0001 | ||
| Binary score becomes one when amplicon Tm is <73°C | 106.0, df = 1, P < 0.0001 | ||
| Number of ambiguous bases | Number of ambiguous bases in amplicon | 27.6, df = 3, P < 0.0001 | |
| Number of repeats | Number of repeats in amplicon | 109.1, df = 1, P < 0.0001 | |
| Binary score becomes one when number of repeats in amplicon is 8 or 9 | 22.0, 1, P < 0.0001 | ||
| Amplicon structure around SNP site | The free energy of the most stable amplicon structure (with loops) containing 5 bases of extension primer annealing site and 2 bases upstream of primer annealing site | 231.5, 1, P < 0.0001 | |
| Amplicon structure around PCR primer annealing sites | The free energy of the most stable amplicon structure (with loops) containing 5 bases of PCR primer annealing site and 2 bases upstream of primer annealing site. Only one the most stable structure between two PCR primers was considered | 80.2, df = 1, P < 0.0001 | |
| Extension mix change date and 13 interactions with combinations of extension mix and last 3′-end of SNP-IT | This binary score was introduced for the single-plex model to reflect historical assay modification. New extension nucleotides were introduced at Orchid Bioscience into SNP-IT assay | 491.6, df = 14, P < 0.0001a | |
| Extension mix + last 2 bases at 3′-end of SNP-IT primer | 24 different combinations were used for the single-plex model and 67 combinations for the multiplex model out of 96 possible. Only combinations which show significant correlations with the failure/success rate were considered. In addition, 3 interactions scores with extension mix change date score are used for the single-plex model | 187.7, df = 24, P < 0.0001a | 543.0, df = 67, P < 0.0001a |
| Extension mix + last 3 bases at 3′-end of SNP-IT primer | 42 different combinations were used for the single-plex model and 56 combinations for the multiplex model out of 384 possible. Only combinations which show significant correlations with the failure/success rate were considered. In addition, 7 interactions scores with extension mix change date score are used for the single-plex model | 208.2, df = 42, P < 0.0001a | 255.5, df = 56, P < 0.0001a |
| Extension mix + last 4 bases at 3′-end of SNP-IT primer | 34 different combinations were used for the single-plex model and 60 combinations for the multiplex model out of 1536 possible. Only combinations which show significant correlations with the failure/success rate were considered | 101.8, p < 0.0001a | 155.1, p < 0.0001a |
| Extension mix + last 5 bases at 3′-end of SNP-IT primer | 3 different combinations were used for the single-plex model and 18 combinations for the multiplex model out of 6144 possible. Only combinations which show significant correlations with the failure/success rate were considered | 9.3, P = 0.03a | 53.6, P < 0.0001a |
| Last 2 bases at the 3′-end of the PCR primer + 1 amplicon base next to PCR primer annealing site | 2 different combinations were used for the single-plex model and 7 combinations for the multiplex model out of 64 possible. Only combinations which show significant correlations with the failure/success rate were considered | 5.9, P = 0.05a | 36.0, P < 0.0001a |
| Last 3 bases at the 3′-end of the PCR primer + 1 base next to PCR primer annealing site | 22 different combinations were used for the single-plex model and 24 combinations for the multiplex model out of 256 possible. Only combinations which show significant correlations with the failure/success rate were considered | 91.1, P < 0.0001a | 70.5, P < 0.0001a |
| Last 2 bases at the 3′-end of the PCR primer + 2 amplicon bases next to PCR primer annealing site | 19 different combinations were used for the single-plex model and 18 combinations for the multiplex model out of 256 possible. Only combinations which show significant correlations with the failure/success rate were considered | 67.4, P < 0.0001a | 48.5, P = 0.0001a |
| Last 3 bases at the 3′-end of the PCR primer + 2 amplicon bases next to PCR primer annealing site | 40 different combinations were used for the single-plex model and 49 combinations for the multiplex model out of 1024 possible. Only combinations which show significant correlations with the failure/success rate were considered | 135.4, P < 0.0001a | 128.8, P < 0.0001a |
| External TIN | The free energy of the most stable structure formed between marker extension primer and other extension primers in the multiplex cluster | 15.0, df = 1, P = 0.0001 | |
| External TDN | The free energy of the most stable structure between marker extension primer and other PCR products in the multiplex cluster | 39.5, P < 0.0001 | |
| External PCR TDN | The free energy of the most stable structure formed between any of two marker PCR primers and other PCR products in the multiplex cluster | 55.4, P < 0.0001 | |
| Dispersion of amplicon melting temperature | The score is calculated as the sum of absolute differences between Tm of marker amplicon and Tm of all other amplicons in the multiplex cluster | 16.3, P < 0.0001 | |
| Dispersion of amplicon complexity | The score is calculated as the sum of absolute differences between marker amplicon complexity and complexity of all other amplicons in the multiplex cluster | 22.6, P < 0.0001 |
An empty cell for the score indicates that it was not included into the corresponding model.
aThe difference in Wald χ2 between the final model and the model without the set of binary variables, degrees of freedom and P value.
Upon selection of the primary scores, scores for multiplex PCR and primer extension (external scores) were added to the multiplex model. Keeping the primary scores in the model, the optimal set of scores including external scores was reselected using backward stepwise logistic regression.
In addition, interactions between selected scores were added into the final statistical model. An interaction term was calculated as the product of two primary scores. The interaction term reflected the fact that the effect of one score could vary depending on the value of another score. All possible interactions were tested and those improving the model predictability were selected.
RESULTS
SNP-IT primer primary scores
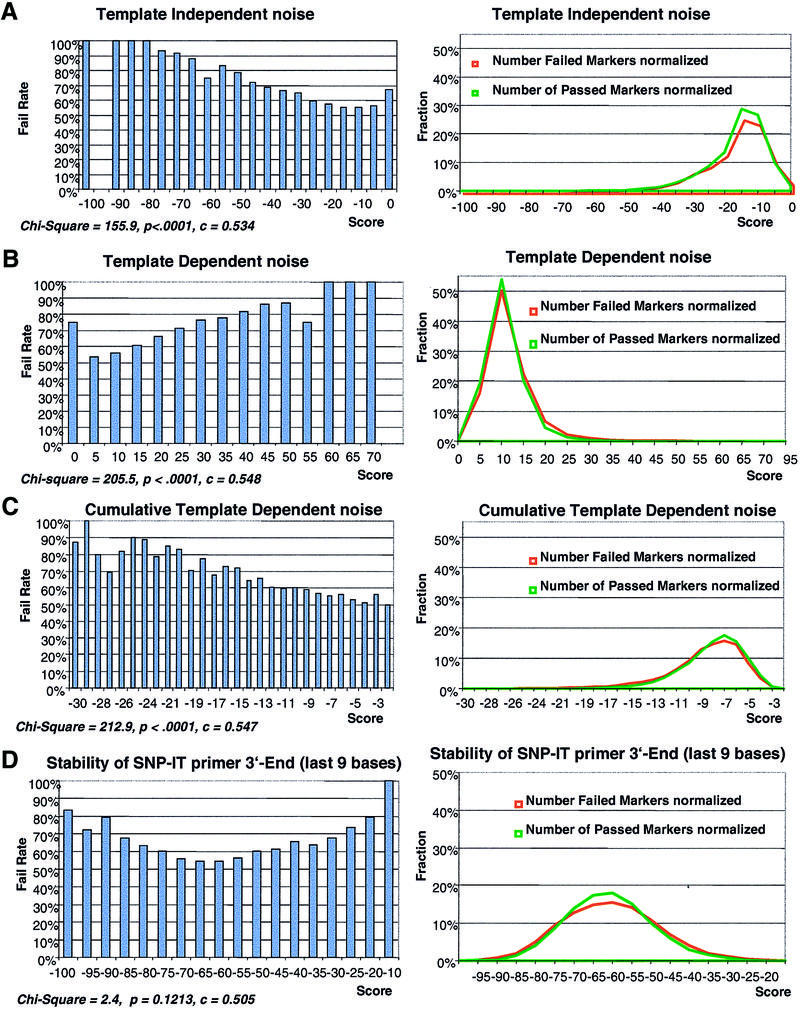
One of the major causes of failure of SNP-IT primers is what we refer to as template-independent noise (TIN). This mispriming is caused by SNP-IT primer self-extension due to secondary structures as short as two bases forming a duplex template at the 3′-end of the primer. While such primers successfully genotype in the presence of DNA template, the noise increases the potential for a false genotype call on a sample which failed to amplify in PCR. Changes in primer concentration and extension temperature modulate the formation and subsequent extension by DNA polymerase of these dimers. However, standardizing such conditions is necessary to enable consistent laboratory practices to be used on the majority of designs. To evaluate TIN probability, the free energy of the most stable dimer that can be formed by a given SNP-IT primer was calculated (Fig. 1A). Dimers close to the 3′-end obviously are more likely to enable extension by DNA polymerase than dimers at the 5′-end; therefore, we introduced a 3′-end bias and multiplied it by the dimer free energy. The optimal formula for the bias was determined to be a linear function of the distance to the 3′-end of the primer. The bias was calculated as the distance from the dimer 3′-end to the primer 5′-end normalized to maximum allowed length of the primer in the program. Thus, the closer the dimer is to the primer 3′-end the greater the weight of its free energy. The TIN score had the highest correlation with failure rate when dimers with no mismatches were used for the calculation and loop structures were not considered. In the final model, Wald χ2 for the TIN score is 92.4 with 1 degree of freedom and P < 0.0001.
Figure 1.
The correlation curve of the failure rate of the SNP-IT reaction with several primary scores (left) and distribution of failed and passed SNP markers against the same scores (right). The score values in the correlation curves (x-axis) and number of markers in the distribution curves (y-axis) are normalized for comparison. The normalization of scores is done by dividing all score values by the minimal (maximal) score. The distribution curves are normalized by plotting fraction of passed or failed markers instead of total number of markers. Distributions of passed markers are shown in green, distributions of failed markers are shown in red. (A) TIN score correlation and distribution curves. (B) Correlation and distribution curves of the ratio of TDN score to stability of the SNP-IT primer 3′-end. TDN is measured as the most stable structure formed between SNP-IT primer and PCR product. (C) Cumulative TDN: the same curves for TDN score measured as a sum of all possible dimers formed between SNP-IT primer and PCR product. (D) Correlation and distribution curves of the last nine 3′-end bases of the SNP-IT primer.
Template-dependent noise (TDN) is the second most common cause of noise in the SNP-IT system. TDN is caused by formation of dimers between the SNP-IT primer and the amplicon outside of the SNP annealing site. Two scores were developed to evaluate TDN probability of a given primer set (Fig. 1B and C). The first score calculates the free energy of the most stable structure formed between the SNP-IT primer and the PCR product outside the target site. Every structure consists of dimers formed by DNA cross-hybridization and DNA loops between adjacent dimers. The free energy of every possible structure was calculated as the sum of dimer free energies and free energies of the loops between two dimers adjacent in the structure. The second score reflects the total number of possible mispriming sites and is calculated as the sum of all possible dimers formed between the SNP-IT primer and the PCR product and is referred to as the cumulative TDN. Only dimers were considered for cumulative TDN and loop energy was not taken into account. A 3′-end bias was used as an additional weight for the free energy of primer–template structures. The model Wald χ2 values are 28.1 (P < 0.0001) and 12.3 (P = 0.0005) for the single-plex and multiplex models, respectively.
A third contributor to assay success is the 3′-end stability of the SNP-IT primer. Since thermodynamic stability of 3′ binding will vary with base composition, efficiency of extension will correlate with this stability. A very stable end of a SNP-IT primer can actually create more noise due to the increased possibility of mispriming. On the other hand, if the end is unstable it can cause a weak signal. This duality is reflected by the correlation curve of the SNP-IT primer 3′-end stability versus the failure rate, which has a parabolic shape (Fig. 1D). Because of the non-linear relationship this score and its square and cubic terms were used in the regression models. The Wald χ2 values in the final single-plex model are 60.1 (P < 0.0001) for the linear term and 32.1 (P < 0.0001) for the cubic term. The multiplex model χ2 values are 284.1 (P < 0.0001) for the linear term and 172.0 (P < 0.0001) for the squared term.
We evaluated all possible distances from the 3′-end using the correlation of 3′-end stability with the assay design failure rate to determine the window size of primer sequence that contributes most to this effect. From this analysis the 3′-most nine bases of the SNP-IT primer were found to be critical for the efficiency of the SNP-IT assay.
Last primer bases immediately 5′ upstream of the extension site were shown to be critical for the efficiency of the extension by DNA polymerase (25,26). We have calculated the failure rate for different combinations of the last 3′ bases in the SNP-IT primer and the primer extension mix, containing two labeled nucleotide terminators. A total of 103 and 201 combinations with various 3′-end lengths were found to have significant correlation with the failure rate and were included into the final models of the single-plex and 12-plex SNP-IT reactions, respectively. Only scores showing significant correlation with failure rate were kept in the model. Table 1 shows the differences in model χ2 values between the final model and the model without the selected combinations. All selected combinations are highly significant.
The number of C3 linkers inserted into the SNP-IT primer had a significant correlation with failure rate. In single-plex reactions, insertion of only one C3 linker was found to decrease the failure rate (χ2 = 16.4, P < 0.0001). However, insertion of more than two C3 linkers into a SNP-IT primer increased the failure rate, probably due to a dramatic decrease in the SNP-IT primer’s ability to anneal efficiently to the template (χ2 = 8.9, P = 0.0029). In multiplex reactions, insertion of any number of C3 linkers increased the failure rate (χ2 = 149.3, P < 0.0001). See Table 1 for details on the significance of the correlation.
PCR primers primary scores
It has been observed that certain sequences at the 3′-end of PCR primers can reduce amplification efficiency (K. Aoyagi, personal communication). The amplicon regions next to PCR primers were shown to affect PCR amplification efficiency (27). Therefore, we have introduced binary scores for the last 3′-end bases of PCR primers and their combination with amplicon bases immediately next to the PCR primer annealing site. All selected combinations show a significant contribution to the final model (see Table 1 for details).
The percent GC of the PCR primers was also selected for inclusion in the final model. χ2 values were 29.7 and 43.4 for the single-plex and multiplex models, respectively, with both P values <0.0001. Primers with increased GC content have more chances to misprime along the PCR product as well as elsewhere in the genome and contaminate the SNP-IT assay with aberrant PCR products or PCR primer dimers during the primer extension step (data not shown).
PCR product primary scores
Secondary structures of the PCR product near the SNP site can block annealing of the SNP-IT primer and inhibit the signal. To evaluate the failure probability due to template secondary structure the most stable structure containing the SNP site was calculated. Both dimer and loop energies were added to calculate the free energy of each possible template structure. The structures containing the last five 3′-end bases of the SNP-IT primer annealing site, the SNP site and one base downstream of the SNP site were found to have the strongest correlation with the failure rate. The most stable structure containing this region of PCR products was selected as the primary score for failure rate prediction. The χ2 for the final 12-plex model was 231.5 (P < 0.0001).
Similarly, the secondary structure of the region around the PCR primer annealing sites can also reduce the signal of the SNP-IT reaction due to poor amplification. By analogy with the SNP site, we chose a region containing the last 5 bases of the PCR primer annealing site and 2 bases downstream of it. The most stable structure around PCR primer annealing sites was shown to have significant correlation with the failure rate and therefore was included in the model for multiplex SNP-IT reactions. The χ2 for the final 12-plex model was 80.2 (P < 0.0001).
A variety of other scores were selected, each evaluating different sequence properties of the PCR product and showing a significant correlation with failure rate. Melting temperature of both the PCR product and PCR primers influences PCR efficiency. Presence of repeats within the template influences PCR efficiency, mispriming of the SNP-IT primer and formation of template secondary structures. Therefore, the number of repeats in the SNP-IT template was added to the model. Complexity of the template sequence reflects a similar property. Number of ambiguous bases in the template reflects the reliability of the genomic sequence information and thus the probability of an incorrect primer sequence (data not shown).
Scores for multiplexed SNP-IT reaction
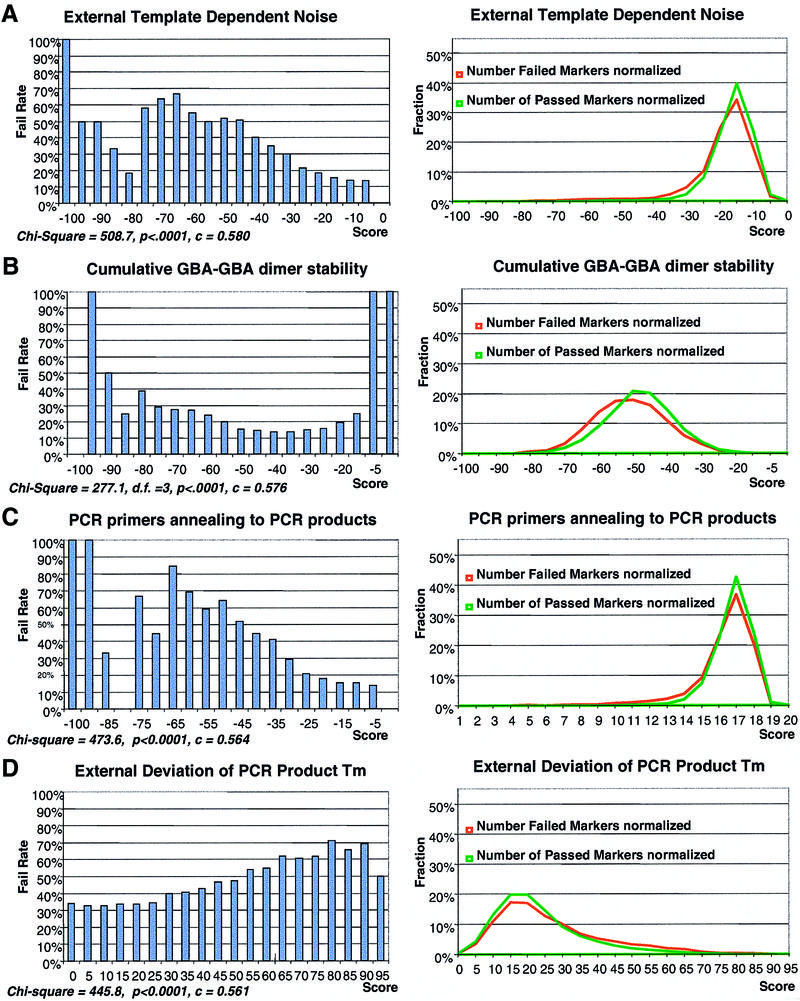
Cross-hybridization of DNA molecules from different SNP markers during multiplex PCR amplification and primer extension creates an additional factor that can contribute to the failure of individual SNP assays beyond the primary scores described in the previous section. To predict the failure probability in the multiplexed SNP-IT reaction we have introduced several ‘external’ scores in addition to ‘internal’ scores for single-plex SNP assays. By analogy with internal TDN scores, the external TDN scores measuring the mispriming of a given SNP-IT primer to other PCR products in the reaction were found to correlate with failure rate (Fig. 2A and B and Table 1).
Figure 2.
The correlation curve of the failure rate of 12-plexed SNP-IT reactions with several primary scores that measure the ‘external’ noise caused by the interference of 11 SNP markers with the one considered for calculation (left). The distribution of failed and passed SNP markers against the same scores (right). The score values in the correlation curves (x-axis) and number of markers in the distribution curves (y-axis) are normalized for comparison. The normalization of scores is done by dividing all score values by the minimal (maximal) score. The distribution curves are normalized by plotting fraction of passed or failed markers instead of total number of markers. Distributions of passed markers are shown in green, distributions of failed markers are shown in red. (A) External TDN noise measured as the free energies sum of most stable structures formed between SNP-IT primer and 11 ‘external’ PCR products. (B) External TDN noise measured as a sum of all possible dimers formed between SNP-IT primer and 11 ‘external’ PCR products. (C) Correlation and distribution curves of the deviation of the SNP-IT primer melting temperature from the other 11 SNP-IT primers. (D) Correlation and distribution curves of the deviation of the GC content of PCR primers pair from the other 11 PCR primer pairs.
Multiplex reactions are sorted into single-plex reactions prior to allele scoring in our tag-array version of the SNP-IT assay. A ‘tag’ sequence is included on the 5′-end of the primer at primer design. This tag sequence anneals to a complementary probe oligonucleotide fixed to a solid phase in the last step of the multiplex SNP-IT assay. In a multiplex reaction, SNP-IT primers for the different SNP assays can form intermolecular dimers due to intermolecular hybridizations, which prevent annealing of the given SNP-IT primer to its complementary probe. To measure this interference in the multiplex, the most stable structure of the given SNP-IT primer with all other SNP-IT primers was calculated and found to have a significant correlation with marker failure rate.
Additional ‘external’ scores include score measuring the mispriming of a given PCR primer pair along all PCR products in the multiplex reaction, the dispersion in the melting temperatures of the PCR products, reflecting both relative PCR efficiency and ‘noisiness’ of the multiplex cluster (Fig. 2D and Table 1), and dispersion of amplicon complexity. The amplicon complexity was calculated as Shanon’s entropy of its sequence (28).
Final models
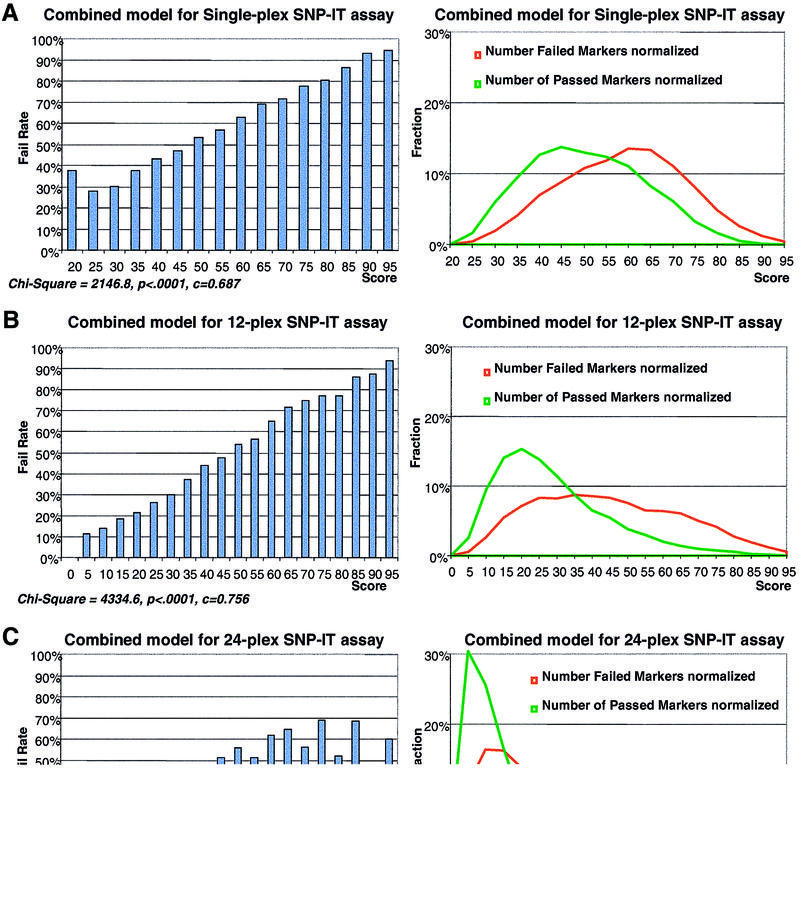
The final models for all three training sets are presented in Figure 3. All scores included in the model, their Wald χ2 values and related P values are presented in Table 1. The first model has nine primary scores, 10 interaction scores, five binary scores, one score with higher order terms, 103 binary scores corresponding to different combinations of the last two 3′-end bases of the SNP-IT primer and different extension mixes and 83 binary scores corresponding to different combinations of the last bases at the 3′-end of the PCR primers and amplicon bases immediately following PCR primer annealing site in the amplicon (Table 1). The c-statistic for the model is 0.69, which shows that the model discriminates passes and fails moderately well. The sources of noise are examined in the Discussion. Hosmer and Lemeshow’s goodness-of-fit test (6) has a χ2 of 7.9 with 8 degrees of freedom and P = 0.4471, which demonstrates that the number of passes and fails predicted from the model are not significantly different from the observed passes and fails across the whole spectrum of predicted scores.
Figure 3.
The correlation curve of the failure rate of the SNP-IT reaction with the combined score (left) and distribution of failed and passed SNP markers against the combined score (right). The score values in the correlation curves (x-axis) and number of markers in the distribution curves (y-axis) are normalized for comparison. Combined scores were calculated using logistic regression analysis of primary scores for three training sets of SNP markers. The normalization of scores is done by dividing all score values by the minimal (maximal) score. The distribution curves are normalized by plotting fraction of passed or failed markers instead of total number of markers. Distributions of passed markers are shown in green, distributions of failed markers are shown in red. (A) Model for single-plex standard SNP-IT assay (23 525 markers). (B) Model for 12-plex SNP-IT assay used on the SNPcode Orchid instrument (30 948 markers). (C) Model for 24-plex SNP-IT assay used on the SNPcode Orchid instrument (30 576 markers).
The model for the 12-plexed SNP-IT assay has 11 primary scores calculating properties of the primer set, five primary scores describing interference between different SNP markers in the multiplexed reaction and one score with higher order terms. In addition, 201 binary scores corresponding to different combinations of the last two 3′-end bases of the SNP-IT primer and different extension mixes and 98 combinations of the last bases at 3′-end of the PCR primers with amplicon bases immediately next to PCR primer annealing sites were used (Table 1). The c-statistic for the model is 0.76, which shows that the model discriminates passes and fails very well. Hosmer and Lemeshow’s goodness-of-fit test (6) has a χ2 of 14.9 with 8 degrees of freedom and P < 0.0611, which demonstrates that the number of passes and fails predicted from the model are only marginally different from observed passes and fails across all the ranges of predicted scores.
The model for 24-plexed SNP-IT assay has seven primary scores calculating properties of the primer set, five primary scores describing interference between different SNP markers in the multiplexed reaction and two scores with higher order terms. In addition, 26 of 96 possible binary scores corresponding to different combinations of the last two 3′-end bases of the SNP-IT primer and different extension mixes were used. The c-statistic for the model is 0.72, which confirms that the model discriminates passes and fails very well. Hosmer and Lemeshow’s goodness-of-fit test (6) has a χ2 of 10.2 with 8 degrees of freedom and P = 0.25, which demonstrates that the number of passes and fails predicted from the model are not different from observed passes and fails across the whole range of predicted scores.
DISCUSSION
We found that the TIN score had no significant correlation with the failure rate in the multiplex SNP-IT assay. This may be due to the fact that primer extension in the multiplexed SNP-IT assay takes place under elevated temperature cycling conditions, which prevents formation of primer–primer dimers. This is consistent with the fact that only simple consecutive match DNA dimers are responsible for TIN noise. It suggests that unstable and transient primer–primer interactions during the SNP-IT assay are sufficient to cause the noise. A high concentration of the SNP-IT primer during SNP-IT reaction causes the abundance of these short-lived structures, which are easily available for extension by DNA polymerase. In general, scores describing primer–primer and primer–template interactions have higher χ2 values and c-statistics compared with the scores describing the PCR product structure. Interactions involving DNA primers appear to be more important for noise than template interactions most likely due to the higher primer concentration.
The external scores estimating PCR product structures around the SNP site and template ends were also not significant in multiplex SNP-IT assays. These scores were calculated as the most stable structure which contains the region of interest formed due to cross-hybridization of two different PCR products. The similar score, which measures the most stable intra-molecular structure at PCR primer annealing sites formed by the PCR product itself, had a much higher statistical correlation with the failure rate. These data suggest that intramolecular looped structures are more likely to interfere with primer annealing and extension than intermolecular structures. There are probably limited intermolecular interactions between two DNA template molecules during PCR and primer extension.
There is little difference between distributions of passed and failed markers relative to each primary score (Figs 1 and 2). However, there is a significant shift between distributions of passed and failed markers relative to the combined failure probability calculated using a logistic regression model (Fig. 3). This evidence suggests that only a small portion of markers fail due to one particular reason and no single molecular mechanism or property is a principal cause of failure in the SNP-IT assay. It appears rather that the cumulative effect from all molecular characteristics of a given SNP marker results in its success or failure during the SNP-IT reaction.
A variety of other factors can contribute to the failure of a SNP marker but cannot be included in our statistical model. Orchid scientists found that the quality of synthesized oligonucleotides and other PCR reagents were critical to the success of the SNP-IT assay. Another major source of error is the absence of actual variation in the genome. Some markers could not be confirmed to contain a real SNP or had a very low population frequency of one SNP allele. Human operator and instrument calibration errors could also cause failure during the assay. Due to the large quantity of data gathered, it was not possible to record the reasons for all failures. Therefore, we could not exclude the markers, which failed for reasons other than their sequence and poor primer design from the training sets.
Due to other noise in the data, the first model for standard single-plex SNP-IT reactions has moderate predictive power. This training set was accumulated over 5–6 years in the Orchid database. During this period, several improvements were made to the biochemistry of the assay. For example, a new type of DNA polymerase was introduced and the chemistry of DNA terminators used for primer extension was modified. Changes such as these influence the role that each particular mechanism plays in the SNP-IT assay. Thus, our first model represents a historical average over several different SNP-IT assay versions. To confirm this hypothesis we introduced several binary timeline scores to reflect the date when a particular assay was performed relative to the major assay modifications. Such scores improved significantly both the χ2 and c-statistic, making the latter 0.71 (data not shown). This proved the importance of understanding the biochemical changes in the model specification and model validation.
Our statistical model for the SNP-IT genotyping assay has two applications. First, the model can be used to improve primer design by selecting three primers with the highest probability of success among all possible primer sets for a given SNP. We have used the first model for single-plex SNP-IT assays to select optimal primers in Orchid’s proprietary ‘AutoPrimer’ software for primer design, available on the World Wide Web (http://www.autoprimer.com). ‘AutoPrimer’ makes a list of different primer sets for a given SNP according to melting temperature and other standard primer design requirements. The set is then evaluated by the statistical model to select the SNP marker with the highest probability of success. We found that every SNP sequence has an optimal PCR product length selected by the statistical model. This optimal length is usually between 60 and 150 bp. Both smaller and larger PCR products have a lower probability of success. In some SNP sequences one particular DNA strand is strongly preferred for SNP-IT primer design while for other SNPs the strand does not seem to matter. The overall experimental success rate of our primer design has increased 15% from the first pass compared to primer design without the statistical model. We also found about 30 SNP markers in the database that failed in the past due to TIN or TDN noise. The primer sets designed for the same SNPs using the statistical model did not have any noise and passed the genotyping call. Current implementation of the statistical model in AutoPrimer does not allow for selection of PCR primers based on the model. Thus, the quality of primer design can be improved even more when we completely switch AutoPrimer to use the statistical model.
The second application of the statistical prediction of the failure/success rate is the selection of primer sets for genotyping in gene mapping and haplotyping projects. In cases where a large region of a chromosome is to be investigated for linkage or association many SNPs may be available for genotyping from public databases, such as dbSNP at NCBI, but not all of them are necessary to provide the marker density needed for a genotyping study. Our statistical prediction will enable efficient SNP selection for such studies, guaranteeing the overall success and reliability of the entire genotyping experiment. The same approach can be used when a particular SNP haplotype is being investigated for genetic association. Not all SNP markers in a haplotype need to be genotyped. Only those with the highest probability of success in the genotyping reaction can be selected to guarantee a successful experiment. Selection of SNP markers with a higher likelihood of success for a genome-wide scan study is another method which may benefit from failure/success probability prediction.
Acknowledgments
ACKNOWLEDGEMENT
We thank Kazuko Aoyagi for the suggestion to add scores for PCR primer ends into the model.
REFERENCES
- 1.Head S.R., Rogers,Y.H., Parikh,K., Lan,G., Anderson,S., Goelet,P. and Boyce-Jacino,M.T. (1997) Nested genetic bit analysis (N-GBA) for mutation detection in the p53 tumor suppressor gene. Nucleic Acids Res., 25, 5065–5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikiforov T.T. and Rogers,Y.H. (1995) The use of 96-well polystyrene plates for DNA hybridization-based assays: an evaluation of different approaches to oligonucleotide immobilization. Anal. Biochem., 227, 201–209. [DOI] [PubMed] [Google Scholar]
- 3.Nikiforov T.T., Rendle,R.B., Goelet,P., Rogers,Y.H., Kotewicz,M.L., Anderson,S., Trainor,G.L. and Knapp,M.R. (1994) Genetic bit analysis: a solid phase method for typing single nucleotide polymorphisms. Nucleic Acids Res., 22, 4167–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikiforov T.T., Rendle,R.B., Kotewicz,M.L. and Rogers,Y.H. (1994) The use of phosphorothioate primers and exonuclease hydrolysis for the preparation of single-stranded PCR products and their detection by solid-phase hybridization. PCR Methods Appl., 3, 285–291. [DOI] [PubMed] [Google Scholar]
- 5.Rozen S. and Skaletsky,H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol., 132, 365–386. [DOI] [PubMed] [Google Scholar]
- 6.Hosmer D.W. and Lemeshow,S. (1989) Applied Logistic Regression. John Wiley & Sons, New York, NY.
- 7.SAS Institute Inc. (1999) SAS/STAT User’s Guide, Version 8. SAS Institute Inc., Cary, NC, Vol. 1, pp. 1901–2042.
- 8.Allawi H.T. and SantaLucia,J.,Jr (1998) Thermodynamics of internal C.T mismatches in DNA. Nucleic Acids Res., 26, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allawi H.T. and SantaLucia,J.,Jr (1998) Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: sequence dependence and pH effects. Biochemistry, 37, 9435–9444. [DOI] [PubMed] [Google Scholar]
- 10.Allawi H.T. and SantaLucia,J.,Jr (1998) Nearest neighbor thermodynamic parameters for internal G.A mismatches in DNA. Biochemistry, 37, 2170–2179. [DOI] [PubMed] [Google Scholar]
- 11.Allawi H.T. and SantaLucia,J.,Jr (1998) NMR solution structure of a DNA dodecamer containing single G*T mismatches. Nucleic Acids Res., 26, 4925–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allawi H.T. and SantaLucia,J.,Jr (1997) Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 13.Bommarito S., Peyret,N. and SantaLucia,J.,Jr (2000) Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res., 28, 1929–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.SantaLucia J. Jr, Allawi,H.T. and Seneviratne,P.A. (1996) Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry, 35, 3555–3562. [DOI] [PubMed] [Google Scholar]
- 15.SantaLucia J. Jr (1996) A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peyret N., Seneviratne,P.A., Allawi,H.T. and SantaLucia,J.,Jr (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G and T.T mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 17.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 18.Dale T., Smith,R. and Serra,M.J. (2000) A test of the model to predict unusually stable RNA hairpin loop stability. RNA, 6, 608–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeder S.J. and Turner,D.H. (2000) Factors affecting the thermodynamic stability of small asymmetric internal loops in RNA. Biochemistry, 39, 9257–9274. [DOI] [PubMed] [Google Scholar]
- 20.Fienberg S.E. (1980) Fixed margins and logit models. The Analysis of Cross-classified Categorical Data. MIT Press, Cambridge, MA, Ch. 6, pp. 95–119.
- 21.Hanushek E.A. and Jackson,J.E. (1977) Models with discrete dependent variables. In Statistical Methods for Social Scientists. Academic Press, New York, NY, pp. 179–216.
- 22.Theil H. (1970) On the estimation of relationships involving qualitative variables. Am. J. Sociol., 76, 103–154. [Google Scholar]
- 23.Wilks S.S. (1963) Mathematical Statistics. Princeton University Press, Princeton, NJ.
- 24.Lewis-Beck M.S. (1989) Applied Regression, An Introduction, Quantities Applications in the Social Sciences Series, Sage University Papers Series. Sage Newbury Park, CA, pp. 58–63.
- 25.Parker L.T., Deng,Q., Zakeri,H., Carlson,C., Nickerson,D.A. and Kwok,P.Y. (1995) Peak height variations in automated sequencing of PCR products using Taq dye-terminator chemistry. Biotechniques, 19, 116–121. [PubMed] [Google Scholar]
- 26.Parker L.T., Zakeri,H., Deng,Q., Spurgeon,S., Kwok,P.Y. and Nickerson,D.A. (1996). AmpliTaq DNA polymerase, FS dye-terminator sequencing: analysis of peak height patterns. Biotechniques, 21, 694–699. [DOI] [PubMed] [Google Scholar]
- 27.McGrath A., Higgins,D.D. and McCarthy,T.V. (1998) Sequence analysis of DNA randomly amplified from the Saccharomyces cerevisiae genome. Mol. Cell Probes, 12, 397–405. [DOI] [PubMed] [Google Scholar]
- 28.Shannon C.E. (1948) A mathematical theory of communication. Bell Syst. Tech. J., 27, 379–423. [Google Scholar]