Abstract
Chemically synthesized combinatorial libraries of unmodified or modified nucleic acids have not previously been used in methods to rapidly select oligonucleotides binding to target biomolecules such as proteins. Phosphorothioate oligonucleotides (S-ODNs) or phosphorodithioate oligonucleotides (S2-ODNs) with sulfurs replacing one or both of the non-bridging phosphate oxygens bind to proteins more tightly than unmodified oligonucleotides and have the potential to be used as diagnostic reagents and therapeutics. We have applied a split synthesis methodology to create one-bead one-S-ODN and one-bead one-S2-ODN libraries. Binding and selection of specific beads to the transcription factor NF-κB p50/p50 protein were demonstrated. Sequencing both the nucleic acid bases and the positions of any 3′-O-thioate/dithioate linkages was carried out by using a novel PCR-based identification tag of the selected beads. This approach allows us to rapidly and conveniently identify S-ODNs or S2-ODNs that bind to proteins.
INTRODUCTION
Synthetic phosphodiester-modified oligonucleotides such as phosphorothioate oligonucleotide (S-ODN) (1) and phosphorodithioate oligonucleotide (S2-ODN) (2,3) analogs have increased nuclease resistance and can bind to proteins with enhanced affinity (4–8). Unfortunately, ODNs possessing high fractions of phosphorothioate or phosphorodithioate linkages appear to lose some of their specificity (6–8) and are ‘stickier’ towards proteins in general than normal phosphate esters, an effect often attributed to non-specific interactions (8). This can be quite important since the recognition of nucleic acid sequences by proteins involves specific side chain and backbone interactions with both the nucleic acid bases as well as the phosphate ester backbone (9–12). We can take advantage of this ‘stickiness’ to enhance the affinity of S-ODN and S2-ODN agents for a protein target. However, we need to optimize the total number of phosphorothioate or phosphorodithioate linkages to decrease non-specific binding to the protein target and enhance only the specific favorable interactions with the target protein.
A recent advance in combinatorial chemistry has been the ability to construct and screen large random sequence nucleic acid ‘aptamer’ libraries (13–15) targeting proteins (16–18) and other molecules (19–22). However, the identification of specific S-ODN aptamers (‘thioaptamers’) that bind proteins (23,24) based upon in vitro combinatorial selection methods is limited to substrates only accepted by polymerases required for reamplification of selected libraries by PCR. One disadvantage of this polymerization of substituted nucleoside 5′-triphosphates into ODN aptamers restricts the choice of P-chirality by the enzymatic stereospecificity (1). It is known that [SP]-diastereoisomers of dNTP(αS) in Taq-catalyzed polymerization solely yield [RP]-phosphorothioate stereoisomers (1). Therefore, it is not possible to select [SP]-phosphorothioate stereoisomers along with achiral S2-ODN analogs since both [Rp]-diastereoisomers of dNTP(αS) and nucleoside dNTP(αS2) are not substrates of polymerases (25). Additionally, in these in vitro combinatorial selection methods the many iterative cycles of selection and reamplification of the bound remaining members of the library by PCR are quite time consuming.
Selection from one-bead one-compound libraries (26,27) based on split synthesis methods (28,29) offers an attractive alternative to in vitro combinatorial selection methods and has been demonstrated for organic molecule (30), peptide (26,27,31) and oligosaccharide libraries (32–34). Remark ably a one-bead one-oligonucleotide (S-ODN or S2-ODN) combinatorial library selection methodology has not been previously demonstrated for identifying a specific oligonucleotide aptamer that binds to specific proteins or other molecules. S2-ODN reagents with sulfurs replacing both of the non-bridging phosphate oxygens are isosteric and isopolar with the normal phosphorodiester. Importantly, S2-ODNs are achiral about the dithiophosphate center, which eliminates problems associated with diastereomeric mixtures generally obtained for the chemically synthesized S-ODN. Here we report the results of our initial development of the split synthesis approach to the construction of both S-ODN and S2-ODN bead-based thioaptamer libraries. Specific S-ODNs and S2-ODNs were identified by screening of the libraries against a transcription factor NF-κB p50/p50 protein. Sequencing both the nucleic acid bases and the positions of any 3′-O-thioate/dithioate linkages was carried out by using a novel PCR-based identification tag of the selected beads.
MATERIALS AND METHODS
The dA, dG, dC and dT phosphoramidites were purchased from Applied Biosystems (Palo Alto, CA) or Glen Research (Sterling, VA). The Beaucage reagent (3H-1,2-benzodithiol-3-one 1,1-dioxide) was from Glen Research. The Taq polymerase kits were from Applied Biosystems. The TOPO TA Cloning kit was from Invitrogen. The Klenow DNA polymerase I was from Promega. Polystyrene beads (60– 70 µm) with non-cleavable hexaethyleneglycol linkers with a loading of 36 µmol/g were from ChemGenes Corp. (Ashland, MA). The Alexa Fluor 488 dye was from Molecular Probes (Eugene, OR). The dA, dG, dC and dT thiophosphoramidites were synthesized as previously described (2,3,6,35,36). The ODNs and S-ODNs used in the study were synthesized on a 1 µmol scale in an Expedite 8909 System (Applied Biosystems) DNA synthesizer.
Synthesis of S-ODN and S2-ODN libraries
Standard phosphoramidite and thiophosphoramidite chemistry (2,3,36) was used for the S-ODN and S2-ODN libraries, respectively. The libraries were prepared on a 1 µmol scale on polystyrene beads. The downstream and upstream primers (5′-GGATCCGGTGGTCTG-3′ and 5′-CCTACTCGCGAATTC-3′) were synthesized in parallel on a two-column DNA synthesizer (Expedite 8909; Applied Biosystems). Following the 5′-primer, the sequences programmed on the synthesizer for the combinatorial S-ODN library were 5′-*CA*GT*TG*AG*GG*GA*CT*TT*CC*CA*GG*C-3′ on column 1 and 5′-*cC*tG*cA*cA*tC*tC*aG*gA*tG*aC*tT*t-3′ on column 2. The sequences programmed for the combinatorial S2-ODN library were 5′-ATGT*AGCC* A*GCTAGT*CTG*TCAG-3′ on column 1 and 5′-CGCC*cAGT*g*aAG GTG*gaA*CCCC-3′ on column 2. The 3′-primer sequence completed the 52mer programmed on the synthesizer. A ‘split and pool’ occurred at each position indicated by an asterisk in order to synthesize the combinatorial region for the S-ODN and S2-ODN. Lower case letters indicate a 3′-thioate linkage, upper case letters indicate a 3′-phosphate linkage, while lower case bold letters indicate a 3′-dithioate linkage. The coupling yield was typically upwards of 99% as determined by the dimethoxytrityl cation assay. Sulfurization chemistry utilized the Beaucage reagent. The fully protected S-ODN or S2-ODN combinatorial libraries with the non-cleavable linker beads were treated with concentrated ammonia at 37°C for 21 h to remove the protecting groups while allowing the ODN to remain attached to the beads. The S-ODN or S2-ODN bead-based single-stranded (ss)DNA library was washed with double distilled water. The ssDNA library (typically 1–3 mg of support beads) was converted to a double-stranded (ds)DNA by Klenow DNA polymerase I reaction in the presence of DNA polymerase buffer, dNTP mixture and reverse primer according to the manufacturer. The dsDNA library was washed twice with phosphate-buffered saline (PBS).
Labeling p50/p50 protein with Alexa Fluor 488
To 0.5 ml of p50/p50 protein [0.215 mg/ml, expressed and purified as previously described (9,10,37)] in PBS containing 30% glycerol was added 50 µl of 1 M bicarbonate. The protein was transferred to a vial of reactive Alexa Fluor 488 dye and stirred at room temperature for 1 h. Fluorescently labeled protein was purified according to procedures from Molecular Probes. The labeled protein was stored at 4°C in the dark.
Alexa Fluor 488-labeled p50/p50 binding to beads and selection of beads
A portion of the double-stranded S-ODN or S2-ODN library (∼3.0 mg of the beads) was suspended in 300 µl of blocking buffer (PBS containing 0.05% Tween-20) and incubated at room temperature for 1 h in a microcentrifuge tube. The beads were washed with 300 µl of PBS and pelleted by centrifugation. The beads were suspended in 300 µl of Alexa Fluor 488-labeled p50/p50 (0.07 µg/µl) at room temperature for 2 h and then washed with blocking buffer (2× 300 µl) and PBS (2× 300 µl). A portion of the beads was transferred to a slide and viewed under a fluorescence microscope. Individual beads with the highest fluorescence intensity were removed by a micropipette attached to a micromanipulator, sorted into PCR microcentrifuge tubes and washed with 8 M urea (pH 7.2) to remove the bound protein.
One-bead one-PCR amplification and sequencing of PCR product
A selected single bead was mixed with the following PCR components: 6 µl of 25 mM MgCl2 (8 µl for 15mer, 10mer and 8mer primers), 0.5 µl of Taq polymerase (5 U/µl), 1 µl of 8 mM dNTP, and 10 µl of PCR buffer and 1 µl of 40 mM primers. The PCR was run on a GeneAmp PCR system 2400 (Perkin Elmer). The PCR mixtures were thermal cycled using the following scheme for amplification: 94°C for 5 min (1 cycle); 94°C for 2 min, 55°C for 2 min (35°C for 10mer and 8mer primers) and 72°C for 2 min (35 cycles); 72°C for 7 min (1 cycle). The PCR products were analyzed on a 15% native polyacrylamide gel. The PCR product was cloned using the TOPO TA cloning procedure (Invitrogen) and sequenced on an ABI Prism 310 Genetic Analyzer (Applied Biosystems).
RESULTS
One-bead one-oligonucleotide libraries
A primary consideration for designing a one-bead one-ODN library using phosphoramidite chemistry is defining suitable bead linker chemistry where the ODNs can be synthesized and yet remain covalently attached to the beads after full deprotection. Additional considerations include development of the split synthesis method for construction of the ODN library, screening bead-based ODN libraries in aqueous media for one-bead binding assays and sequencing of the ODN bound on the individual bead. Although long chain alkylamine controlled pore glass (LCA-CPG) (38) (Pierce Chemical Co., Rockport, IL) has been used for many years for efficient ODN synthesis, LCA-CPG was not suitable for generation of one-bead one-ODN libraries. The size, homogeneity and swelling of CPG are not adequate for one-bead one-compound chemistry (27). Another disadvantage of the currently available CPG linker chemistry is that ODNs are cleaved from the solid support during the ammonia deprotection step (38). An advance in solid support chemistry has been the ability to synthesize ODNs on more uniform polystyrene beads. Importantly, using new chemistry developed by ChemGenes Corp. for a non-cleavable hexaethyleneglycol linker attaching the first phosphoramidite, the synthesized ODNs are still covalently attached to the beads after full base and phosphate ester deprotection. In this procedure each unique ODN chemical entity in the combinatorial library is attached to a separate support bead. Selection of a bead-based ODN combinatorial library can then be carried out by binding the bead library of ODNs to a target protein under high stringency conditions where only a few beads show binding. How do we identify the ODN sequence on the selected beads? Following the in vitro combinatorial selection method for identification of selected ODN sequences (13,14), we utilize defined 5′ and 3′ fixed ODN primer sequences flanking the combinatorial library segment of the ODN. The fixed primer regions allow PCR amplification of the sequence as well as Klenow extension of the covalently attached ssDNA to produce a combinatorial library of dsODN attached to the beads.
Primer design for one-bead one-ODN
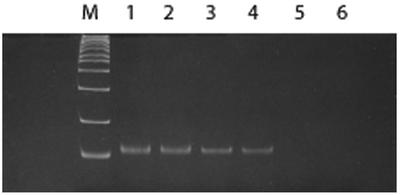
Initially we synthesized a template ODN consisting of a desired 14mer sequence region flanked by two 18mer primers on the beads (ODN 1 in Table 1). Its ability to support one-bead one-PCR amplification was tested for several individual beads. The PCR product was cloned using the TOPO TA cloning procedure and sequenced on an ABI Prism 310 Genetic Analyzer. The desired sequence was confirmed. Although 18mer or longer primers are generally used in PCR amplification (39), shorter primers are attractive since the size of the ODN is limited by the synthesis yields for long ODNs. Consequently, longer combinatorial sequence libraries would be possible with shorter primer sequences; in addition, shorter primer sections will reduce non-specific binding of target proteins to the ODN bead library. To study the primer length requirement for one-bead one-PCR amplification, a series of primers with varying lengths (8mer, 10mer and 15mer; see Table 1) were designed and synthesized to hybridize to the 52mer template ODN containing both 5′ and 3′ primer regions (ODN 2 in Table 1) on the support beads. The PCR products of these primers were monitored by 15% polyacrylamide gel electrophoresis (Fig. 1). The PCR conditions were optimized for each pair of primers of varying length. No detectable band was observed with the 8mer primers, even at the highest concentration tested (Fig. 1, lanes 5 and 6). A weak band was detected with the 10mer primers (Fig. 1, lanes 3 and 4). A strong band was observed with 15mer primers (Fig. 1, lanes 1 and 2). The fidelity in the Taq polymerase amplification yielding the ODN products was confirmed by cloning and sequencing (data not shown). These results suggest that ODNs with primer lengths of 10 nt or greater are required for efficient PCR amplification. In this study, 15mer primers were selected for the following experiments.
Table 1. ODNs on beads and primers.
| ODNs on beads used as templates | ||
| ODN 1 | 5′-ATGCCTACTCGCGAATTC-CCAGGAGATTCCAC-GGATCCGGTGGTCTGTTC-Bead | |
| ODN 2 | 5′-CCTACTCGCGAATTC-AGTTGAGGGGACTTTCCCAGGC-GGATCCGGTGGTCTG-Bead | |
| Primers | ||
| Upstream primer | Downstream primer | |
| 18mer | 5′-ATGCCTACTCGCGAATTC-3′ | 5′-GAACAGACCACCGGATCC-3′ |
| 15mer | 5′-CCTACTCGCGAATTC-3′ | 5′-CAGACCACCGGATCC-3′ |
| 10mer | 5′-CCTACTCGCG-3′ | 5′-CAGACCACCG-3′ |
| 8mer | 5′-CCTACTCG-3′ | 5′-CAGACCAC-3′ |
Figure 1.
Comparison of the PCR of different lengths of primers. The PCR products were analyzed by 15% polyacrylamide gel electrophoresis. ODN 2 was used as a template. Size of the ladders is indicated at the left. Lanes 1 and 2, 15mer primers; lanes 3 and 4, 10mer primers; lanes 5 and 6, 8mer primers.
Generation of a self-encoded S/S2-ODN library
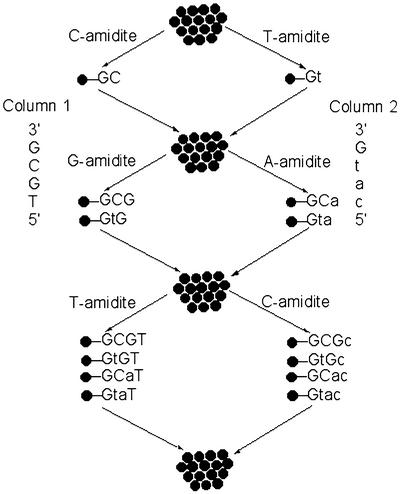
To introduce many copies of a single, chemically pure S-ODN or S2-ODN onto each bead, we utilize a ‘mix and separate’ split synthesis method (Scheme 1). A two-column DNA synthesizer was used for constructing the library. The normal phosphate backbone linkages were generated using standard phosphoramidite monomers via oxidation in column 1, while the phosphorothioate or phosphorodithioate linkages were synthesized using standard phosphoramidite or thiophosphoramidite monomers via sulfurization in column 2, respectively. Two sequences of the same length are programmed for each column and are designed such that the bases are different at every equal position not only for diversifying base compositions but also for coding a phosphate, phosphorothioate or phosphorodithioate linkage. Thus on an Expedite 8909 DNA synthesizer with dual columns, onto column 1 a phosphoramidite (for example dC) is coupled to the bead and after completion of oxidation, the resulting product is the nucleotide (dC) with a phosphotriester linkage. On column 2 a nucleoside phosphorothioate or phosphorodithioate is introduced with a different base (dT for example). The support beads from the two columns are mixed and resplit and in the second cycle, additional phosphoramidites or thiophosphoramidites are introduced, followed by oxidation and sulfurization reactions individually in columns 1 and 2. After additional coupling steps and after final split/pool synthesis is completed, the end products comprise a combinatorial library of ODNs with varying thioate/dithioate or normal phosphate ester linkages at varying positions along the ODN strand attached to the support. Each bead contains a single chemical entity with a specified backbone modification that is identified by the base. In the above example, any dC at position 1 of the sequence will be a 3′-phosphate while a dT at position 1 would indicate that it contains a 3′-thiophosphate. We applied this scheme to synthesize a library of 4096 (212) one-bead one-S-ODN. This library consists of a 22 nt combinatorial sequence (12 split/pool steps) flanked by 15 nt defined primer regions at the 5′ and 3′ ends (Table 1). The 3′ ends of the sequences were attached to the polystyrene beads. As noted above, the defined primer sequences were incorporated to allow PCR amplification and identification of the ODN sequence on the selected beads. Thus the downstream primers were first automatically synthesized in parallel on the two columns. The S-ODN sequences of the combinatorial 22mer segment on each column were programmed for each column and were generated by introducing a phosphorothioate linkage on every other base in column 2, following the ‘split and pool’ approach. The identical upstream primer sequences were then completed on both columns. As described below, a smaller S2-ODN library was created in similar fashion.
Scheme 1. General flow diagram for the solid-phase synthesis of a one-bead one-S/S2-ODN library. In the first cycle, in column 1, a phosphoramidite dC was used to form a dinucleotide phosphotriester dGC via a phosphotriester linkage; in column 2, a phosphoramidite T was used to form a dinucleotide thiophosphotriester dGt via a phosphothiotriester linkage. Upon pooling, the end products are a mixture of two kinds of bead-bound dinucleotides comprised of phosphorotriester and phosphothiotriester. After splitting and pooling through three such cycles the eight (23) possible ODN and/or S-ODN tetraoligonucleotides are represented on separate beads. A lower case letter denotes a 3′-thioate, while an upper case letter denotes a 3′-phosphate. The S2-ODN library was generated by replacing the phosphoramidite with a thiophosphoramidite globally in column 2. The sulfurization step immediately followed the thiophosphoramidite coupling step.
Selection and sequencing of the S-ODN beads
Binding of the transcription factor NF-κB p50/p50 homodimer and selection of specific beads was demonstrated by first converting the single-stranded S-ODN to dsDNA since the NF-κB transcription factor binds to DNA duplexes. The single-stranded 52mer S-ODN combinatorial library (typically 1–3 mg of beads) was converted to dsDNA by Klenow DNA polymerase I reaction in the presence of DNA polymerase buffer, dNTP mixture and reverse primer. Therefore, one strand of the duplex potentially contained thiophosphate backbone substitutions in the combinatorial library segment and the other complementary strand was composed of unmodified phosphate backbone ODN. A duplex DNA library in which both strands contain S-ODN modifications could also be generated using a Klenow reaction with no more than three dNTP(α)S (23,40). Because the S-ODN strand attached to the support was chemically synthesized utilizing phosphoramidite chemistry, each thiophosphate is a mixture of RP and SP stereoisomers. The beads were suspended in a diluted solution of NF-κB p50/p50 homodimer labeled with the Alexa Fluor 488 dye at room temperature for 2 h. After washing with PBS containing 0.1% Tween 20 and PBS to minimize non-specific binding, a portion of the beads was viewed under both light and fluorescence microscopes as shown in Figure 2. Typically, a few positive beads were intensely stained when viewed by fluorescence, while the majority of the beads remained unstained, as shown in Figure 2b. With the aid of a micropipette coupled to a micromanipulator, the intensely stained beads were retrieved as shown in Figure 2c.
Figure 2.
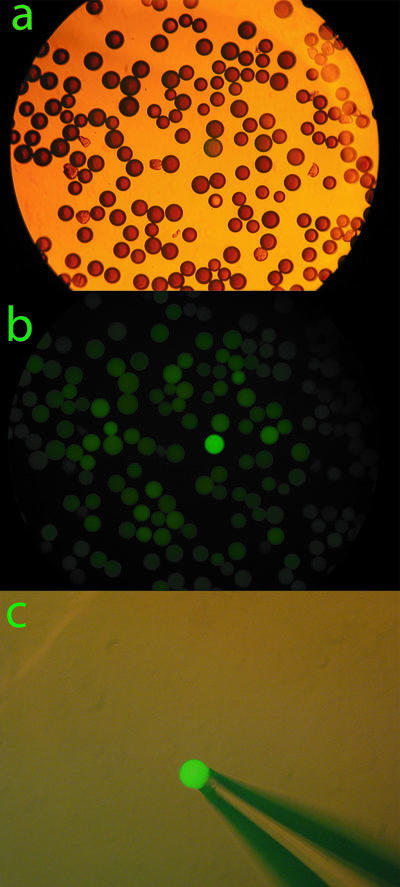
A typical representative color micrograph of a bead-based thioaptamer library screen. (a) An aliquot of S-ODN beads bound to NF-κB p50/p50 homodimer protein labeled with the Alexa Fluor 488 dye viewed under light microscopy. (b) The same beads viewed under fluorescence microscopy, in which a positive green bead stained with Alexa Fluor 488 dye can be easily identified in a background of many hundreds of non- reactive beads. (c) A single positive bead can easily be retrieved with a hand-held micropipette under a fluorescence microscope.
Five positive beads from the S-ODN library were selected. Each individual bead was washed thoroughly with urea to remove the protein and was directly used for our ‘one-bead one-PCR’ amplification using the 5′ and 3′ end primers described above. The PCR product was cloned and sequenced. Table 2 lists four of the S-ODN sequences obtained.
Table 2. S-ODN/S2-ODN sequences identified from the screen with NF-κB p50/p50 proteina.
| Automated sequence | Deduced S/S2-ODN sequence |
|---|---|
| S-ODN selection | |
| CTGTGAGTCGACTGATGACGGT | CtGTGAGtCGACTgAtGaCGGt |
| AGTTGAGTCGAAGGACCCATTT | AGTTGAGtCGAaGgACCCAtTt |
| CGTCAAGTCTCAGTTCCCATTT | CGTcAAGtCtCaGTTCCCAtTt |
| AGTCAAGTCGAAGTTCCACGGT | AGTcAAGtCGAaGTTCCaCGGt |
| S2-ODN selection | |
| ATGTAGCCAGCTAGTCTGTCAG | ATGTAGCCAGCTAGTCTGTCAGb |
| CGCCAGCCAAAGGTGCTGTCAG | CGCCAGCCAaAGGTGCTGTCAG |
| CGCCCAGTGGCTAGTGAACCCC | CGCCcAGTgGCTAGTgaACCCC |
| ATGTAGCCGAAGGTGGAACCCC | ATGTAGCCgaAGGTGgaACCCC |
| CGCCAGCCGAAGGTGGAACCCC | CGCCAGCCgaAGGTGgaACCCC |
aThe lower case letters indicate a 3′-thioate linkage. The lower case bold letters indicate a 3′-dithioate linkage.
bNo 3′-dithioate linkages are present in this strand.
Binding and selection of the combinatorial library of S2-ODN beads
As we (6) and others (7) have shown, S2-ODNs generally bind even more tightly to proteins than unsubstituted or S-ODN analogs. Thus, it is significant that this S-ODN bead-based combinatorial selection method can be applied to dithiophosphate backbone substitutions, since in vitro combinatorial selection is only possible for thiophosphate substituted ODNs with limited P-chirality. To demonstrate, a small one-bead one-S2-ODN library was synthesized consisting of a pool of 32 (25) sequences to allow further optimization of in vitro or bead-based S-ODN selected sequences. Chemical synthesis of S2-ODN avoids problems created by a mixture of diastereoisomers of chemically synthesized S-ODN. The random region (5′-CGCCcAGTgaAGGTGgaACCCC-3′) in column 2 was identified as a S-ODN sequence derived from an in vitro combinatorial selection methodology that binds the NF-κB p50/p50 protein with high affinity (<20 nM) (37) (all lower case letters indicate enzymatically synthesized chiral 3′-thioate linkages). The programmed combinatorial region sequence (5′-ATGTAGCCAGCTAGTCTGTCAG-3′) in column 1 was designed such that the bases at each 3′-dithioate position were different from the bases in column 2 at each equal position, further allowing base sequence to identify backbone substitution. Thiophosphoramidite chemistry with sulfurization was used to generate 3′-dithioate linkages. Only the previous 3′-thioate linkages were replaced with 3′-dithioate linkages. The ‘split and pool’ step followed most of the dithioate modifications. We have also identified S2-ODNs by selecting beads binding fluorescently labeled p50/p50 homodimer, followed by PCR amplification of five individually selected S2-ODN beads and cloning and sequencing of the PCR products. The sequences are also listed in Table 2.
DISCUSSION
Nucleic acid ‘aptamers’ have previously been selected by incubating the target (protein, nucleic acid or small molecule) with the combinatorial library and then separating the non-binding species from the bound. The bound fractions are then amplified using PCR and subsequently reincubated with the target in a second round of screening. These iterations are repeated (often 10–20 cycles) until the library is enhanced for sequences with high affinity for the target. However, aptamers selected from combinatorial RNA and DNA libraries have generally had normal phosphate ester backbones, and so would generally be unsuitable as drugs or diagnostics agents that are exposed to serum or cell supernatants because of their nuclease susceptibility. Rapid degradation of natural ODNs used as antisense agents or aptamers by nucleases in serum or cells necessitates chemical modification of the ODNs (41–49).
Among a large variety of modifications, S-ODN and S2-ODN render the agents more nuclease resistant. Very encouragingly, the first antisense therapeutic drug utilizes a modified S-ODN (CIBA Vision, a Novartis company). The S2-ODNs also show significant promise (6,7). However, the effect of substitution of more nuclease-resistant thiophosphates cannot be predicted, since the sulfur substitution can lead to significantly decreased (or increased) binding to a specific protein (50; X.Yang, unpublished results) as well as structural perturbations (51) and thus it is not possible to predict the effect of backbone substitution on a combinatorially selected aptamer. Thus, if at all possible, selection should be carried out simultaneously for phosphate ester backbone substitution as well as the base sequence. Recently we described an in vitro combinatorial selection of thioaptamers from random or high sequence diversity libraries based on their tight binding to the target (e.g. a protein or nucleic acid) of interest (23,37,52–55).
Oligonucleotides possessing high fractional substitutions of monothioate/dithioate internucleotide linkages appear to be ‘stickier’ towards proteins than normal phosphate esters, and therefore thioaptamers with complete thiophosphate backbone substitutions appear to lose much of their specificity (8). This increased affinity is partly due to the fact that the thioate groups only poorly coordinate hard cations such as sodium ions (56), and thus the thioaptamers serve as ‘bare’ anions and don’t require any energy to strip away the neutralizing cations to bind to proteins. This can be quite important since proteins recognize DNA at both the bases and phosphate esters. In our previous studies (6), we have demonstrated that binding of S2-ODNs to a protein target requires only a limited number of phosphorodithioate linkages in a specific ODN sequence to achieve very high affinities.
In this paper, we have developed a split and pool synthesis combinatorial chemistry method for creating combinatorial S-ODN and S2-ODN libraries (and readily extended it to unmodified ODNs, whether single strand or duplex). In this procedure each unique member of the combinatorial library is attached to a separate support bead. Targets that bind tightly to only a few of the potentially millions of different support beads can be selected by binding the targets to the beads and then identifying which beads have bound target by staining and imaging techniques. Our methodology allows us to rapidly screen and identify aptamers that bind to proteins such as NF-κB using a novel PCR-based identification tag on the selected bead.
The preliminary results obtained from two libraries in this paper indicate that the methodology can be applied to other backbone or base modifications that are compatible with templates containing these modifications. It is important that not only the S-ODNs but even the S2-ODNs are capable of acting as templates recognized by DNA polymerases for PCR amplification of selected S2-ODN beads. This demonstrates that nucleic acid analogs with phosphorodithioate linkages can be utilized as a template in the nucleotidyltransferase reaction catalyzed by DNA polymerases. Misra et al. (57) have also noted that polyamide nucleic acids lacking the phosphate backbone are recognized as templates for the polymerase reaction.
In vitro selection of combinatorial libraries of S2-ODNs is not possible because dNTP(αS2) is not a substrate for polymerases and our split synthesis, bead-based S2-ODN library selection method represents the only current combinatorial approach for identifying both optimal number and location of dithioate substitutions as well as base sequences for these S2-ODN aptamers. Additionally, even for the thioate library selection, the in vitro methods involving iterative cycles of selection, isolation and reamplification of the bound members of the library by PCR amplification are very time consuming, in contrast to a single cycle of split/pool synthesis, selection and identification. The split/pool bead-based method demonstrated in this study thus allows us to identify the positions of any 3′-monothioate/dithioate linkages.
Although the beads were screened against a target protein labeled with a fluorescent dye, the beads can also be screened directly against the unmodified transcription factor. The binding of NF-κB to a specific sequence can be detected using a primary anti-NF-κB antibody, followed by a secondary antibody conjugated to a marker molecule including fluorescein or rhodamine for fluorescence microscopy (X.Yang, unpublished results).
For verification of our selection results, the S-ODN, 5′-CtGTGAGtCGACTgAtGaCGGt-3′ (lower case letters represent location of 3′-thioates), was independently synthesized on the non-cleavable linker bead support, hybridized with its complementary ODN and then mixed again with p50/p50 protein labeled with the Alexa Fluor 488 dye. The fluorescence intensity of all of the beads viewed under the fluorescence microscope was qualitatively similar to the intensity of the selected bead containing this sequence within the combinatorial library. This result suggests that the primer regions do not contribute to the binding of p50/p50. Quantitative studies on the affinities of the selected S-ODNs and S2-ODN duplexes for the protein along with selection from a large combinatorial library (∼106–108 ODNs) to NF-κB are in progress.
In earlier studies (37) we have shown that a thioaptamer clone obtained from an in vitro combinatorial selection experiment (15 rounds of selection) bound to p50/p50 with an apparent dissociation constant <5 nM (thiophosphate modification 5′ to each dA residue, 5′-GGGGTTCCACCTTCACTGGGCG-3′·3′-CCCCAAGGTGGAAGTGACCCGC-5′). A chemically synthesized thioaptamer of the same sequence bound with a dissociation constant of <20 nM. It should be noted, however, that each chemically synthesized thioaptamer consists of a diastereomeric mixture containing 2n different stereoisomers, where n is the number of thiophosphates (27 = 128 for the p50/p50 selected thioaptamer). To test the importance of the thiophosphate substitutions in the thioaptamer toward the p50/p50 homodimer, we have previously shown that a tight binding 15th round thioaptamer clone synthesized by PCR with a nucleotide mix containing dATP instead of dATP(αS) showed no binding of the normal phosphoryl backbone aptamer to p50/p50 protein, supporting the critical role played by the thiophosphates.
Phosphoramidite chemistry has been widely used for the synthesis of S-ODNs (45) because of its automation, high coupling efficiency and ease of site-specific thioate linkage incorporation. Synthesis of S-ODNs is presently carried out by initial formation of an internucleoside phosphite linkage followed by sulfurization of the phosphite triester to a phosphorothioate. The resulting S-ODNs are a mixture of diastereoisomers and, consequently, the diastereomeric S-ODN mixtures may have variable biochemical, biophysical and biological properties. Each bead then contains a library of monothioate aptamers (a library of libraries) since each bead contains the identical sequence and position of thiophosphate substitution, but represents a mixture of diastereomers introduced through the new monothiophosphate chiral centers. We could in principle use stereocontrolled synthesis (46,58) of a stereo-defined S-ODN library to determine which is the best binder to the protein, but our ultimate goal is to select a thioaptamer (or thioaptamer library) which has high affinity for the target protein or biomolecule. Our preliminary binding data (37) suggest that diastereoisomeric mixture libraries have good selectivity and affinity, although not as high as pure stereoisomers.
Another possible solution lies in the synthesis of modifications which are achiral at phosphorus, such as the above S2-ODN thioaptamer library study. In addition, dithioates appear to have greater ‘stickiness’ to proteins than the thioates or unmodified ODN backbone (6,7,41).
The current approach demonstrated in this study requires a different nucleotide sequence to identify the backbone modification. We have also created S-ODN and S2-ODN libraries that only differ in the position of phosphate or dithioate but not in their base sequence. Eckstein has shown that the positions of thiophosphates in a mixed backbone S-ODN sequence can be determined by reaction of the S-ODN with iodoethanol followed by base catalyzed cleavage of the thiophosphate triester (59). Preliminarily, we have demonstrated the feasibility of this approach for identifying the location of thioate linkages, independent of base sequence (X.Yang, unpublished results).
The search for other split synthesis, bead-based combinatorial libraries containing base modifications and hybrid backbones with phosphate ester, thioates, dithioates or potentially neutral methylphosphonates or even peptide nucleic acid chimeras with improved properties, such as enhanced binding affinity for a specific protein, increased biological stability, and improved cellular uptake, could be achieved by the split synthesis combinatorial selection method described here.
By the split/pool method with two columns we can create 2N different members of the library for N split/pool steps. Utilizing more columns (M) would allow us to synthesize MN different beads with one unique thioaptamer sequence on each bead. The limit to the size of the combinatory library is the number of steps (N) and the number of columns (M) and of course the total number of beads, which generally is in the region of 106 or more depending upon the size of the beads and synthesizer columns. Recently we have succeeded in also synthesizing aptamer beads on 15–20 µm beads (X.Yang, unpublished results) and thus a 40-fold increase in the library size is possible. Finally, we could in principle have library sizes comparable to those created by in vitro combinatorial selection methods by utilizing mixtures of phosphoramidites/thiophosphoramidites (up to eight different species) at selected positions in a given synthesis step. This would create a library of libraries of beads, each bead containing a library of any complexity. We could thus easily create 106 beads with 108 combinatorial library members on each bead; the total diversity in principle is thus 1014, the same as in in vitro combinatorial selection libraries.
Sulfur substitution in aptamers alters the binding affinity and sequence that is obtained by in vitro combinatorial selection methods. Post-selection phosphorothioate modifications of in vitro combinatorially selected sequences can thus result in thioaptamers in which affinity cannot be reliably predicted. The simultaneous selection for both avoids this difficulty. The bead-based split synthesis, selection and PCR identification of combinatorial aptamer libraries now provides a means to combinatorially select both monothioate and dithioate variations on aptamers.
Acknowledgments
ACKNOWLEDGEMENTS
The advice and help of Dr Richard Hodge (UTMB) is much appreciated. This research was supported by DARPA (9624-107 FP), NIH (AI27744), NIEHS (ES06676), the Welch Foundation (H-1296) and the Sealy and Smith Foundation.
References
- 1.Eckstein F. (1985) Nucleoside phosphorothioates. Annu. Rev. Biochem., 54, 367–402. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen J., Brill,W.K.D. and Caruthers,M.H. (1988) Synthesis and characterization of dinucleoside phosporodithioates. Tetrahedron Lett., 29, 2911–2914. [Google Scholar]
- 3.Farschtschi N. and Gorenstein,D.G. (1988) Preparation of a deoxynucleoside thiophosphoramidite intermediate in the synthesis of nucleoside phosphorodithioates. Tetrahedron Lett., 29, 6843–6846. [Google Scholar]
- 4.Stein C.A. (1996) Exploiting the potential of antisense: beyond phosphorothioate oligonucleotides. Chem. Biol., 3, 319–323. [DOI] [PubMed] [Google Scholar]
- 5.Mou T., Gray,C.W., Terwilliger,T.C. and Gray,D.M. (2001) Ff gene 5 protein has a high binding affinity for single-stranded phosphorothioate DNA. Biochemistry, 40, 2267–2275. [DOI] [PubMed] [Google Scholar]
- 6.Yang X. Fennewald,S., Luxon,B.A., Aronson,J., Herzog,N.K. and Gorenstein,D.G. (1999) Aptamers containing thymidine 3′-O-phosphorodithioates: synthesis and binding to nuclear factor-κB. Bioorg. Med. Chem. Lett., 9, 3357–3362. [DOI] [PubMed] [Google Scholar]
- 7.Marshal W.S. and Caruthers,M.H. (1993) Phosphorodithioate DNA as a potential therapeutic drug. Science, 259, 1564–1570. [DOI] [PubMed] [Google Scholar]
- 8.Sharma H.W., Perez,J.P., Higgins-Sochaski,K., Hsiao,R. and Ratan,R.R. (1996) Transcription factor decoy approach to decipher the role of NF-κB in oncogenesis. Anticancer Res., 16, 61–69. [PubMed] [Google Scholar]
- 9.Ghosh G., Duyne,G.V., Ghosh,S. and Sigler,P.B. (1995) Structure of NF-κB p50 homodimer bound to a κB site. Nature, 373, 303–310. [DOI] [PubMed] [Google Scholar]
- 10.Muller C.W., Rey,F.A., Sodeoka,M., Verdine,G.L. and Harrison,S.C. (1995) Structure of the NF-κB p50 homodimer bound to DNA. Nature, 373, 311–317. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y.Q., Ghosh,S. and Ghosh,G. (1998) A novel DNA recognition mode by the NF-kappaB p65 homodimer. Nature Struct. Biol., 5, 67–73. [DOI] [PubMed] [Google Scholar]
- 12.Chen F.E., Huang,D.B., Chen,Y.Q. and Ghosh,G. (1998) Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature, 391, 410–413. [DOI] [PubMed] [Google Scholar]
- 13.Ellington A.D. and Szostak,J.W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 14.Tuerk C. and Gold,L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophase T4 DNA polymerase. Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 15.Bock L.C., Griffin,L.C., Latham,J.A., Vermaas,E.H. and Toole,J.J. (1992) Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature, 355, 564–566. [DOI] [PubMed] [Google Scholar]
- 16.Jellinek D., Green,L.S., Bell,C., Lynott,C.K., Gill,N., Vargeese,C., Kirschenheuter,G., McGee,D.P.C., Abesinghe,P., Pieken,W.A. et al. (1995) Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry, 34, 11363–11372. [DOI] [PubMed] [Google Scholar]
- 17.Lee S.W. and Sullenger,B.A. (1997) Isolation of a nuclease-resistant decoy RNA that can protect human acetylcholine receptors from myasthenic antibodies. Nat. Biotechnol., 15, 41–45. [DOI] [PubMed] [Google Scholar]
- 18.Lebruska L.L. and Maher,L.J.,III (1999) Selection and characterization of an RNA decoy for transcription factor NF-κB. Biochemistry, 38, 3168–3174. [DOI] [PubMed] [Google Scholar]
- 19.Koizumi M. and Breaker,R.R. (2000) Molecular recognition of cAMP by an RNA aptamer. Biochemistry, 39, 8983–8992. [DOI] [PubMed] [Google Scholar]
- 20.Gold L., Singer,B. He,Y. and Brody,E. (1997) SELEX and the evolution of genomes. Curr. Opin. Genet. Dev., 7, 848–851. [DOI] [PubMed] [Google Scholar]
- 21.Nolte A., Klußmann,S., Bald,R. Erdmann,V.A. and Furste,J.P. (1996) Mirror-design of L-oligonucleotide ligands binding to L-arginine. Nat. Biotechnol., 14, 1116–1119. [DOI] [PubMed] [Google Scholar]
- 22.Ye X., Gorin,A., Ellington,A.D. and Patel,D. (1996) Deep penetration of an α-helix into the widened RNA major groove in the HIV-1 Rev peptide-Rna aptamer complex. Nature Struct. Biol., 3, 1026–1033. [DOI] [PubMed] [Google Scholar]
- 23.King D.J., Venture,D.A., Brasier,A.R. and Gorenstein,D.G. (1998) Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry, 37, 16489–16493. [DOI] [PubMed] [Google Scholar]
- 24.Andreola M.-L., Calmels,C., Michel,J. Toulme,J.-J. and Litvak,S. (2000) Towards the selection of phosphorothioate aptamers. Eur. J. Biochem., 267, 5032–5040. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig J. and Eckstein,F. (1991) Synthesis of nucleoside 5′-O-(1,3-dithiotriphosphates) and 5′-O-(1,1-dithiotriphosphates). J. Org. Chem., 56, 1777–1783. [Google Scholar]
- 26.Lam K.S., Salmon,S.E., Hersh,E.M., Hruby,V.J., Kazmierskl,W.M. and Knapp,R. (1991) A new type of synthetic peptide library for identifying ligand-binding activity. Nature, 354, 82–84. [DOI] [PubMed] [Google Scholar]
- 27.Lam K.S., Lebl,M. and Krchnak,V. (1997) The “one-bead-one-compound” combinatorial library method. Chem. Rev., 97, 411–448. [DOI] [PubMed] [Google Scholar]
- 28.Furka A., Sebestyen,F., Asgedom,M. and Dibo,G. (1988) Highlights of modern biochemistry. In Proceedings of the 14th International Congress of Biochemistry. VSP, Utrecht, The Netherlands, Vol. 5, p. 47.
- 29.Furka A., Sebestyen,F., Asgedom,M. and Dibo,G. (1988) Proceedings of the 10th International Symposium of Medicinal Chemistry. Magyar Kemikusok Egyesulete, Budapest, Hungary.
- 30.Felder E.R. (1999) Resins, linkers and reactions for solid-phase synthesis of organic libraries. In Miertus,S. (ed.), Combinatorial Chemistry and Technology, Principles, Methods and Applications. Marcel Dekker, New York, NY, pp. 35–51.
- 31.Lam K.S. (1995) Synthetic peptide libraries. In Meyer,R.A. (ed.), Molecular Biology and Biotechnology: A Comprehensive Desk Reference. VCH, New York, NY, p. 880.
- 32.Zhu T. and Boom,G.J. (1998) A two-directional approach for the solid-phase synthesis of trisaccharide libraries. Angew. Chem. Int. Ed., 37, 1898–1900. [Google Scholar]
- 33.Liang R., Yan,L., Loebach,J., Ge,M., Uozumi,Y., Sekanina,K., Horan,N., Gildersleeve,J., Thompson,C., Smith,A. et al. (1996) Parallel synthesis and screening of a solid phase carbohydrate library. Science, 274, 1520–1522. [DOI] [PubMed] [Google Scholar]
- 34.St. Hilaire P.M. and Meldal,M. (2000) Glycopeptide and oligosaccharide libraries. Angew. Chem. Int. Ed., 39, 1162–1179. [DOI] [PubMed] [Google Scholar]
- 35.Yang X., Hodge,R.P., Luxon,B.A., Shope,R. and Gorenstein,D.G. (2002) Separation of synthetic oligonucleotide dithioates from monothiophosphate impurities by anion-exchange chromatography on a Mono Q column. Anal. Biochem., 306, 92–99. [DOI] [PubMed] [Google Scholar]
- 36.Wiesler W.T. and Caruthers,M.H. (1996) Synthesis of phosphorodithioate DNA via sulfur-linked base labile protecting groups. J. Org. Chem., 61, 4272–4281. [DOI] [PubMed] [Google Scholar]
- 37.King D., Bassett,S.E., Li,X., Fennewald,S.A. Herzog,N.K., Luxon,B.A., Shope,R. and Gorenstein,D.G. (2002) Combinatorial selection and binding of phosphorothioate aptamers targeting human NF-κB RelA(p65) and p50. Biochemistry, 41, 9696–9706. [DOI] [PubMed] [Google Scholar]
- 38.Pon R.T. (1993) Solid-phase supports for oligonucleotide synthesis. In Agarwal,S. (ed.), Methods in Molecular Biology: Protocols for Oligonucleotides and Analogs. Humana Press, Totowa, NJ, Vol. 20, pp. 465–497. [DOI] [PubMed]
- 39.Hayashi K. (1994) Manipulation of DNA by PCR. In Mullis,K.B., Ferre,F. and Gibbs,R. (eds), The Polymerase Chain Reaction. Birkhauser, Boston, MA, pp. 3–13.
- 40.Ciafre S.A., Rinaldi,M., Gasparini,P., Seripa,D. Bisceglia,L., Zelante,L. Farace,M.G. and Faziom,V.M. (1995) Stability and functional effectiveness of phosphorothioate modified duplex DNA and synthetic ‘mini-genes’. Nucleic Acids Res., 16, 4134–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhlmann E. and Peyman,A. (1990) Antisense oligonucleotides: a new therapeutic principle. Chem. Rev., 90, 543–584. [Google Scholar]
- 42.Yang X., Misiura,K., Sochacki,M. and Stec,W.J. (1997) Deoxyxylothymidine 3′-O-phosphorothioates: synthesis, stereochemistry and stereocontrolled incorporation into oligothymidylates. Bioorg. Med. Chem. Lett., 7, 2651–2656. [Google Scholar]
- 43.Yang X., Sierzchala,A., Misiura,K., Niewiarowski,W., Sochacki,M., Stec,W.J. and Wiczorek,W. (1998) The first strereocontrolled solid-phase synthesis of di-, tri- and tetra[adenosine (2′,5′)phosphorothioate]s. J. Org. Chem., 63, 7097–7100. [DOI] [PubMed] [Google Scholar]
- 44.Miller P.S., Yano,J., Yano,E., Carroll,C., Jayaraman,K. and Ts’o,P.O.P. (1979) Nonionic nucleic acid analogues. Synthesis and characterization of dideoxyribonucleoside methylphosphonates. Biochemistry, 18, 5134–5143. [DOI] [PubMed] [Google Scholar]
- 45.Zon G. and Geiser,T.G. (1991) Phosphorothioate oligonucleotides: chemistry, purification, analysis, scale-up and future directions. Anticancer Drug Des., 6, 539–568. [PubMed] [Google Scholar]
- 46.Stec W.J. and Wilk,A. (1994) Stereocontrolled synthesis of oligo(nucleoside phosphorothioate)s. Angew. Chem. Int. Ed., 33, 709–722. [Google Scholar]
- 47.Wilk A., Grajkowski,A., Phillips,L.R. and Beaucage,S.L. (2000) Deoxyribonucleoside cyclic N-acylphosphoramidites as a new class of monomers for the stereocontrolled synthesis of oligothymidylyl and oligodeoxycytidylyl phosphorothioates. J. Am. Chem. Soc., 122, 2149–2156. [Google Scholar]
- 48.Agrawal S. and Tang,J. (1992) GEM 91: an antisense oligonucleotide phosphorothioate as a therapeutic agent for AIDS. Antisense Res. Dev., 2, 261–266. [DOI] [PubMed] [Google Scholar]
- 49.Oka N. Wada,T. and Saigo,K. (2002) Diastereocontrolled synthesis of dinucleoside phosphorothioates using a novel class of activator, dialkyl(cyanomethyl)ammonium tetrafluoroborates. J. Am. Chem. Soc., 124, 4962–4963. [DOI] [PubMed] [Google Scholar]
- 50.Milligan J.F. and Uhlenbeck,O.C. (1989) Determination of RNA-protein contacts using thiophosphate substitutions. Biochemistry, 28, 2849–2290. [DOI] [PubMed] [Google Scholar]
- 51.Volk E.D., Yang.X., Fennewald,S.M., King,D.J., Bassett,S.E., Venkitachalam,S., Herzog,N., Luxon,B.A. and Gorenstein,D.G. (2002) Solution structure and design of dithiophosphate backbone aptamers targeting transcriptions factor NF-κB. Bioorg. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 52.Gorenstein D.G., King,D.J., Ventura,D.A. and Brasier,A.R. (2002) Combinatorial selection of oligonucleotide aptamers. US patent 6,423,493. [DOI] [PubMed]
- 53.Gorenstein D.G., Herzog,N.H., Aronson,J. and Luxon,B. (1999) Thio-modified aptamer synthetic methods and compositions. US patent pending.
- 54.Gorenstein D.G., Herzog,N.H., Aronson,J. and Luxon,B. (1999) Thio-modified aptamer synthetic methods and compositions. Foreign patent pending.
- 55.Gorenstein D.G., Herzog,N.H., Luxon,B. and Yang,X. (2001) Phosphoromonothioate and phosphorodithioate oligonucleotide aptamer chip for functional proteomics. US patent pending.
- 56.Volk D.E., Power,T.D., Gorenstein,D.G. and Luxon,B.A. (2002) An ab initio study of phosphorothioate and phosphorodithioate interactions with sodium cation. Tetrahedron Lett., 43, 4443–4447. [Google Scholar]
- 57.Misra H.S., Pandey,P.K., Modak,M.J., Vinayak,R. and Pandey,V.N. (1998) Polyamide nucleic acid-DNA chimera lacking the phosphate backbone are novel primers for polymerase reaction catalyzed by DNA polymerases. Biochemistry, 37, 1917–1925. [DOI] [PubMed] [Google Scholar]
- 58.Stec W.J., Karwowski,B., Boczkowska,M., Guga,P., Koziolkiewicz,M., Sochacki,M., Wieczorek,M.W. and Blaszczyk,J. (1998) Deoxyribonucleoside 3′-O-(2-thio- and 2-oxo“spiro”-4,4-pentamethylene-1,3,2-oxathiaphospholane)s: monomers for stereocontrolled synthesis of oligo(deoxyribonucleoside phosphorothioate)s and chimeric PS/PO oligonucleotides. J. Am. Chem. Soc., 120, 7156–7167. [Google Scholar]
- 59.Gish G. and Eckstein,F. (1988) DNA and RNA sequence determination based on phosphorothioate chemistry. Science, 240, 1520–1522. [DOI] [PubMed] [Google Scholar]