Abstract
Mass spectrometry plays a central role in the characterisation of modified nucleotides, but pseudouridine is a mass-silent post-transcriptional modification and hence not detectable by direct mass spectrometric analysis. We show by the use of matrix-assisted laser desorption/ionisation (MALDI) mass spectrometry that pseudouridines in tRNA can be specifically cyanoethylated by acrylonitrile without affecting the uridines. The tRNA was cyanoethylated and then subjected to digestion with either RNase A or RNase T1. Cyanoethylated digestion fragments were identified by mass spectrometric comparison of untreated and acrylonitrile-treated samples, where the addition of one acrylonitrile resulted in a mass increment of 53.0 Da. The exact modified nucleotide could be identified by tandem mass spectrometry on the cyanoethylated digestion fragment. The methodology was used to identify additional one 4-thiouridine and one pseudouridine in tRNATyrII from Escherichia coli. Furthermore, we observed that RNase A is highly tolerant towards nucleotide modifications, only being inhibited by 2′-O-methylation, whereas RNase T1 cleavage is affected by most nucleotide modifications.
INTRODUCTION
Many cellular RNAs become heavily modified after synthesis of the primary transcript. These modifications may be additions or deletions at the terminals, excision or addition of internal sequences, intermolecular splicing and specific modifications at the single nucleotide level. The latter type of modification is generally termed post-transcriptional modification. The post-transcriptional modifications are primarily encountered in stable RNAs such as transfer RNA (tRNA) and ribosomal RNA (rRNA). The precise molecular role of post-transcriptional modification is unclear, but there are numerous examples of modifications being crucial for function of the translational apparatus (1–3). Microorganisms also use post-transcriptional modification of their rRNAs as a means to acquire antibiotic resistance (4,5).
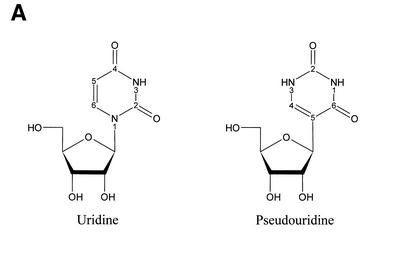
The most frequently occurring post-transcriptional modification, pseudouridine, is an isomer of its precursor uridine (Fig. 1A). In pseudouridine, the C5 of the uracil nucleobase is coupled via a C–C bond to the 1′-position of a ribose, whereas the coupling goes from N1 in uracil to the 1′-position of ribose in normal uridine. Formation of pseudouridine confers this nucleobase with noticeable new features. First, the pseudouridinylation makes N1 of uracil available as a hydrogen bond donor, which can participate in novel molecular interactions (6). Secondly, the C5–C1′ bond between uracil and ribose in pseudouridine confers this nucleotide with less rotational freedom in a tRNA context than the normal N1–C1′ bond in uridine (7), and pseudouridine consequently stabilises local RNA secondary structure (8,9). Thirdly, Ψ’s N1 is a particularly reactive atom for certain chemical modifications (see below). The function of pseudouridines on a molecular level is not known, but several studies on eukaryotes and prokaryotes show that deletion of pseudouridine-synthesising genes has a negative impact on cell growth and protein synthesis rate (10,11).
Figure 1.
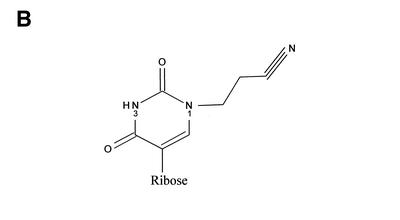
(A) Chemical structure of uridine and pseudouridine with numbering of selected atoms in the pyrimidine rings. (B) The proposed reaction product between ψ and acrylonitrile.
Mass spectrometry plays a central role in the localisation and identification of post-transcriptional modifications (12,13). However, pseudouridine is mass-neutral compared with its uridine precursor, and consequently its mass spectrometric identification is not straightforward. Currently, the most frequently used Ψ-detection method involves RNA modification with N-cyclohexyl-N′-β-(4-methylmorpholinium) ethylcarbodiimide p-tosylate (CMC), a carbodiimide reaction that modifies guanosine- and uridine-like nucleotides (14). An alkaline treatment (pH 10.4) will remove all CMC groups except those that are coupled to the N3 position in pseudouridine (15). The CMC-modified RNA is used as template for the DNA polymerase, reverse transcriptase. The reverse transcriptase terminates its polymerisation when it encounters a CMC-modified ψ, and the resulting termination product(s) are size/sequence determined by gel electrophoresis. This method is widely used with a significant success, but it has a couple of limitations. In addition to CMC-modified ψ, other naturally occurring nucleotide modifications inhibit the reverse transcriptase, thus heavily modified RNAs, such as tRNA, are not readily amenable to ψ detection by CMC-modification/reverse transcriptase termination. Dihydrouridine and 4-thiouridine are also modified by the reported CMC procedure in ways that confer reverse transcriptase termination (14). Furthermore, the 3′-end of an RNA molecule cannot be investigated because a primer annealing site is required.
The CMC modification of ψ may also be combined with mass spectrometric identification of the modified nucleotides as reported recently (16). RNase T1 digests of CMC-modified tRNA were analysed by matrix assisted laser desorption/ionisation (MALDI) mass spectrometry in order to locate digestion fragments harbouring a CMC group. We have chosen a single-reaction chemistry to modify the ψs based on selective cyanoethylation of its uniquely accessible N1 with acrylonitrile (Fig. 1B). Specific cyanoethylation of ψ-monophosphate was reported about 30 years ago (17), and this chemistry has subsequently been applied to modification of ψs in tRNA (18,19). Normal uridine is only ∼5% cyanoethylated under conditions where ψ is completely modified (19). Weakly alkaline conditions lead to the formation of the reactive nucleobase anion with either N1 or N3 being deprotonated (20,21), followed by a nucleophilic Michael-type addition to acrylonitrile. The ratio of N1 to N3 deprotonation is approximately 4:1 (20), which partly explains the selectivity for N1 cyanoethylation. However, the ratio of N1 to N3 cyanoethylation is much higher than 4:1, suggesting that N1 is inherently more reactive than N3 perhaps due to steric hindrance from the two carbonyl oxygens flanking the N3 position. We chose to pursue this chemistry because it appeared experimentally simple and could be performed under conditions that, a priori, were not expected to degrade the RNA under investigation. We report here optimised conditions for selective ψ-modification in tRNA, and the subsequent MALDI- and tandem-mass spectrometric analysis of RNase-digested tRNA.
MATERIALS AND METHODS
Cyanoethylation
tRNA, obtained from Sigma (phenylalanine specific from brewers yeast and tyrosine specific II from Escherichia coli), was subjected to an ethanol/ammonium acetate precipitation in order to remove alkali ions. Optimal cyanoethylation conditions were determined by reacting tRNA with acrylonitrile (Aldrich) as follows. (i) 2 µl of 1 µg/µl tRNA plus 30 µl of 41% ethanol/1.1 M triethylammoniumacetate (pH 8.6) and 4 µl acrylonitrile (20). (ii) 18 µg of lyophilised tRNA dissolved in 10 µl of 2.7 M N,N-dimethyl formamide/0.5 M sodium phosphate (pH 8.8)/2.3 M acrylonitrile (18). (iii) 8 µg of lyophilised tRNA dissolved in 50 µl of 6 M urea/10 mM sodium phosphate (pH 8.8) and pre-heated to 75°C for 2 min followed by addition of acrylonitrile to 2.2 M. Various incubation times between 1 and 48 h and temperatures between 20 and 70°C were tested. The optimal conditions were found to be method (i) with incubation at 70°C for 2 h. After cyanoethylation, the reaction mixture was lyophilised, redissolved in water and the tRNA recovered by precipitation with 2 M ammonium-acetate and 2.5 vol ethanol.
RNase digestion and mass spectrometry
tRNA (1–2 pmol) was digested in 1–2 µl 50 mM 3-hydroxypicolinic acid (3-HPA). RNase A (Sigma-Aldrich) digestions were performed for 2–4 h at 37°C with 0.1 µg of enzyme. RNase T1 digestions were carried out for 2–16 h at 37°C with 20 U of enzyme (United States Biochemicals); short digestion times resulted in predominantly 2′-3′ cyclic phosphates whereas longer time gave a gradual conversion to ‘linear’ phosphates (18.01 Da heavier).
Details on the MALDI mass spectrometry have been published previously (22). Briefly, an RNase digest was directly mixed with 0.7 µl of a 0.5 M 3-HPA matrix in 50% acetonitrile and ammonium-loaded ion exchange beads on the target plate; it was then allowed to air dry. The spectra were recorded in positive ion mode on either Perseptive STR or Bruker Reflex IV MALDI reflector Time-of-Flight (TOF) mass spectrometers. All MALDI-TOF spectra were smoothed using the ‘m/z’ free software (Proteometrics Inc.).
The MALDI tandem mass spectrometry data were obtained on a MicroMass MALDI Q-TOF Ultima mass spectrometer. The sample preparations were made with 0.5 µl of a 0.5 M 3-HPA matrix and 1 µl of RNase digest. All spectra were recorded in positive ion mode. MALDI ions were generated at a backing pirani pressure reading of 2.5 × 10–1 mBar N2 to provide collisional cooling, and extracted with 50 V into the instrument. The window for ion selection was between 2 and 4 m/z values in the quadrupole during MS/MS experiment. Argon was used as collision gas at an indicated manifold pressure of 3 × 10–5 mBar in the hexapole, and the collision energy (Elab) was 10 eV for MS experiments and ranged from 50 to 80 eV for MS/MS experiments. An acceleration potential of 9.1 kV was used to introduce the ions into the orthogonal reflector TOF mass analyser. All MALDI Q-TOF spectra have been smoothed using the manufacturer supplied MassLynx software.
RESULTS
Derivatisation and mass spectrometric analysis
Because ψ is a mass-neutral post-transcriptional modification, derivatisation of this nucleotide is necessary before mass spectrometric detection. Specific cyanoethylation of N1 in ψ without concomitant modification of the uridine precursor has been known for numerous years, both with pure nucleotides (17) and in a tRNA context (18,19). The chemical reaction can be performed under mildly alkaline conditions (pH 8.5–9.0) (20), which should not significantly affect the alkali-labile phosphodiester backbone of RNA.
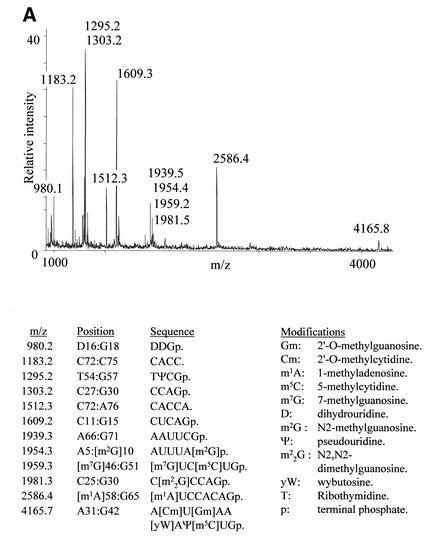
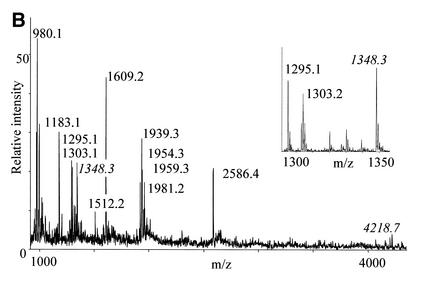
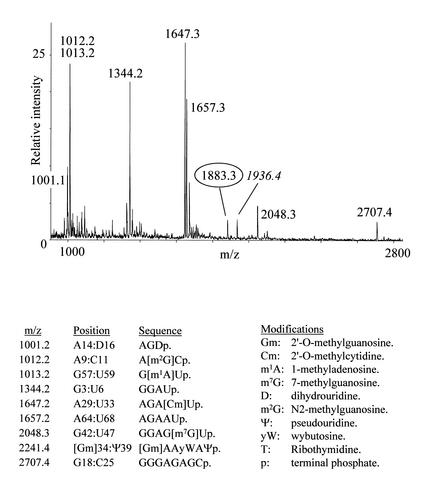
tRNA has previously been subjected to cyanoethylation, but the degree to which different ψs in the same molecule are modified is highly dependent on the reaction conditions. This is probably a result of the three-dimensional structure of the tRNA (18). Using yeast tRNAPhe as substrate, we tested ethanol/triethylamine (20), dimethylformamide/sodium phosphate (18) and urea/sodium phosphate buffers at different reaction times and temperatures to determine the best conditions for efficient but specific cyanoethylation of both ψs in the tRNA. By far the most efficient cyanoethylation was observed with the ethanol/triethylamine buffer (20), and we found a relatively short reaction time at an elevated temperature (2–4 h at 70°C) to yield the overall best results. To evaluate the cyanoethylation reactions, the tRNA was digested to completion with either RNase T1 or RNase A, and direct MALDI mass spectrometry of the digests was performed (22). Addition of a single cyanoethyl group will result in a mass increment of 53.0 Da (monoisotopic mass) compared with the unmodified digestion fragment. Figure 2 compares unmodified and cyanoethylated tRNAPhe that have both been subjected to RNase T1 digestion, and the accompanying table gives the assignment of the observed signals. Note that only tri-nucleotides or larger can be reliably mass analysed, because the signal from smaller digestion products is generally obscured by signals from matrix, buffers, etc. There are some interesting features already on the RNase T1 digestion of the chemically unmodified tRNAPhe (Fig. 2A). The fragment pattern tells us that RNase T1 cleaves efficiently after m2G, because the 1954.3 m/z (Th) species 5′-AUUUA[m2G]p-3′ is readily produced (see figures for nucleotide abbreviations). m22G is not equally well recognised by the enzyme as manifested by the simultaneous presence of the 1981.2 Th species 5′-C[m22G]CCAGp-3′ and the product of m22G recognition, 5′-CCAGp-3′, at 1303.2 Th. Complete resistance to enzyme cleavage was observed after the modified guanosine nucleotides, m7G, Gm and wybutosine. The signal at 1183.2 Th corresponds to the 3′-terminal RNase T1 digestion fragment without the distal acceptor adenosine (5′-CACC-3′); a minor signal at 1512.30 Th corresponds to the intact species 5′-CACCA-3′. We believe this degradation is present in the supplied tRNAPhe, because we have not observed a similar phenomenon with any other RNase T1-digested RNA (including the tRNATyrII described in the following section). When comparing the spectra in Figure 2A and B, it is evident that the two fragments, which each harbour one ψ (at 1295.2 and 4165.8 Th, respectively), give rise to unambiguous signals ∼53.0 Th higher upon cyanoethylation (labels in italics). The cyanoethylation is not complete as can be seen by the remaining 1295.2 Th species in the magnification in Figure 2B. However, the specificity of the cyanoethylation for ψ is noteworthy: not even the 1954.3 Th species 5′-AUUUA[m2G]p-3′, containing three uridines, exhibits a noticeable degree of reaction with the acrylonitrile. Thus, the cyanoethylation reaction is very selective for ψ under the conditions used. Preliminary experiments indicate that cyanoethylated ψ does not inhibit the polymerising activity of reverse transcriptase (data not shown), so this derivatisation method probably cannot be used to detect ψs by primer extension approaches.
Figure 2.
MALDI-TOF mass spectra of yeast tRNAPhe digested with RNase T1 before (A) and after (B) reaction with acrylonitrile. Peaks labelled with italics contain ψs and have increased by 53.0 Da upon cyanoethylation. The insert is a zoom on the region containing the TΨCGp fragment. The table lists the expected RNase T1 fragments based on sequence and modification data present in the tRNA database (27).
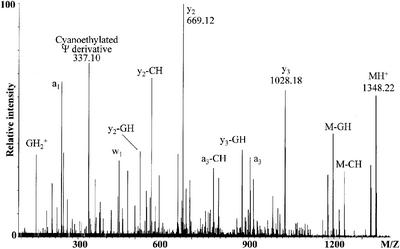
The cyanoethylation approach only locates ψs to a defined RNase digestion fragment, but does not reveal the exact modified position. We therefore performed a tandem mass spectrometry experiment (23) on the cyanoethylated 5′-TψCGp-3′ RNase T1 fragment (1348.25 Th). The mass spectrometric fragmentation was performed on a hybrid MALDI quadrupole-TOF mass spectrometer that allows selective isolation and subsequent gas-phase fragmentation by collisional activation of the ion of interest. The outcome of this experiment is shown in Figure 3, a spectrum rich in information about the structure of the analyte. Amongst others, a series of y-fragment ions [originating from cleavage of the 5′ P–O bonds of the phosphodiester groups; nomenclature according to (24)] is observed. The y-ions are prominent in the experimental set-up used (23). The difference between the parent ion (MH+) and the y3 ion corresponds to the expected 5′-T nucleotide (320.04 Th observed, 320,04 Th calculated). Between the y3 and the y2 ions is a difference of 359.06 Th, which fits precisely with the presence of a cyanoethylated ψ at nucleotide two in the selected digestion fragment. Thus, tandem mass spectrometry can be used to pinpoint the chemically modified nucleotide. The intense signal at 337.10 Th may be explained as a cyanoethylated ψ nucleotide, which has lost CH3CN, a commonly observed neutral fragment loss (25). More work is required to verify this interpretation and to determine whether the 337.10 Th fragment is generally diagnostic for cyanoethylated ψ.
Figure 3.
MALDI Q-TOF tandem mass spectrum of the 1348.22 Th singly protonated species, 5′-TØCGp-3′ + 53.03 Th, obtained through RNase T1 digestion of cyanoethylated E.coli tRNATyrII. The prominent fragment ions are assigned, and m/z-values used to locate the site of cyanoethylation are given.
In our standard protocol for RNase digestion of 5S rRNA prior to mass spectrometry, we used the addition of 3-HPA in order to partly denature the secondary structure of the substrate and thereby obtain a more complete cleavage (22). The acid was omitted in the digestions used to obtain the Figure 2 spectra because the N-glycosidic bond of post-transcriptionally modified nucleotide wybutosine is hydrolysed under mildly acidic conditions (26). Figure 4 depicts an RNase A digestion of cyanoethylated tRNAPhe in the presence of 50 mM 3-HPA. The calculated value for the wybutosine-containing fragment is 2241.4 Th, but instead a fragment at 1883.3 Th appeared, reflecting a hydrolysis of the N-glycosidic bond of wybutosine. The wybutosine-containing fragment ends in a ψ, and therefore its cyanoethylated product at 1936.4 Th is also observed. The presence of the cyanoethylated RNase A digestion fragment demonstrates that RNase A can cleave after this chemically modified pyrimidine nucleotide. We also observed RNase A cleavage after thymine, m5C, dihydrouridine and ψ, but not Cm with this tRNA.
Figure 4.
MALDI-TOF mass spectrum of cyanoethylated yeast tRNAPhe digested with RNase A in the presence of 3-HPA. The circled peak represents the 34–39 fragment that has lost the nucleobase from the wybutosine by acidic hydrolysis. The cyanoethylated version of this fragment is labelled in italics (see text for details). The table lists the expected RNase A fragments based on data present in the tRNA database (27).
Escherichia coli tRNATyrII
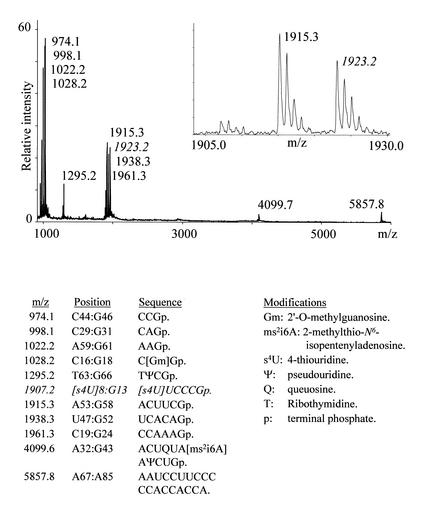
The tRNAPhe from yeast is probably the best characterised tRNA, with all post-transcriptional modifications identified. We therefore investigated the less characterised E.coli tRNATyrII to see if our method could reveal additional modifications to the ones reported in the tRNA database (27) (http://www.uni-bayreuth.de/departments/biochemie/trna/. The database is now maintained by M. Sprinzl and K. S. Vassilenko). Figure 5 presents the outcome of an RNase T1 digestion on the unmodified tRNA. The spectrum clearly has one difference compared with the expected fragment pattern: the calculated fragment at 1907.2 Th (position 8 to 13; 5′-[s4U]UCCCGp-3′, where s4U is 4-thiouridine) is essentially replaced by a signal at 1923.2 Th (Fig. 5 insert). This mass increment of ∼16.0 Da may reflect an extra oxygen atom (as a hydroxyl group) or the replacement of an oxygen atom with a sulfur atom as in s4U, but neither the exact nature nor the position of the modification could be determined from MALDI-TOF data. Digestion of the tRNATyrII with RNase A ruled out location of the 16.0 Da post-transcriptional modification on nucleotides 8 and 13, because these nucleotides occurred in RNase A digestion fragments at the predicted m/z ratios (data not shown). Thus, the detected modification must be located between positions 9 and 12 (5′-UCCC-3′ + 16.0 Da). The RNase A digestion did not reveal any further unreported modifications.
Figure 5.
MALDI-TOF mass spectrum of E.coli tRNATyrII digested with RNase T1. The fragment revealing an additional 16.0 Da post-transcriptional modification is labelled with italics in the mass spectrum and the table; the insert is a zoom of the corresponding region. The table lists the expected RNase T1 fragments based on sequence and modification data present in the tRNA database (27).
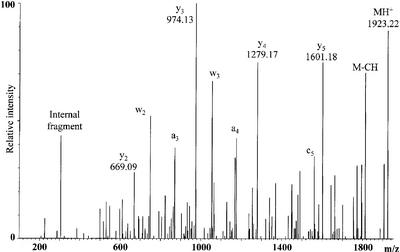
To obtain more information on the location of the +16 Da post-transcriptional modification, we performed a mass spectrometric fragmentation study on the m/z 1923.22 RNase T1 digestion product (Fig. 6), and again the y-fragment ions predominate. The y5 ion corresponds to the loss of 322.04 Th, as expected from the presence of 4-thiouridine at the 5′-end of the selected ion. The y4 ion originates from an additional loss of 322.01 Th. This unveils that the +16.0 Da post-transcriptional modification is located in the second nucleotide of the selected ion (position 9 in the tRNA), because a uridine at this position would have resulted in loss of calculated 306.03 Th going from y5 to y4. Of the known uridine modifications in E.coli (28), the modified nucleotide could be 2-thiouridine, 4-thiouridine or 5-hydroxyuridine.
Figure 6.
MALDI Q-TOF tandem mass spectrum of the 1923.22 Th singly protonated species obtained through RNase T1 digestion of E.coli tRNATyrII. The y-ion series is given together with the m/z values. Other prominent fragment ions are assigned.
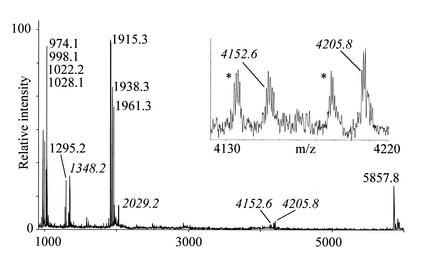
We then cyanoethylated the tRNATyrII to see if we could detect ψs additional to the ones in positions 40 and 64 (27). Digestion of the cyanoethylated tRNA with RNase T1 (Fig. 7) shows that the fragment 5′-TψCGp-3′ (1295.19 Th unmodified) exhibits the expected mass shift of 53.0 Da to 1348.2 Th, and that the other ψ-harbouring fragment, position 32 to 43, appears as a doublet at 4152.6 and 4205.8 Th (in addition to the two associated species ending in 2′-3′ cyclic phosphates ∼18.0 Da lighter) with one and two cyanoethyl groups coupled, respectively. This suggests that an extra, hitherto unreported, ψ is present in the latter fragment. Because the position 32 to 43 fragment in the RNase T1 digest turns up as a doublet upon cyanoethylation, the reaction of the two ψs with acrylonitrile has not run to completion. It is possible that either the queuosine or the 2-methylthio-N6-isopentenyladenosine in the position 32–43 RNase T1 fragment had reacted with acrylonitrile, but an RNase A digestion of the cyanoethylated tRNATyrII did not support this possibility (data not shown). Sequence elucidation by tandem mass spectrometry is not feasible with this fragment, because the MALDI Q-TOF instrument only allows specific ion selection up to 4000 Th.
Figure 7.
MALDI-TOF mass spectrum of E.coli tRNATyrII digested with RNase T1 after reaction with acrylonitrile. Peaks corresponding to cyanoethylated species are labelled in italics. The insert is a zoom on the region containing the putatively doubly pseudo-uridinylated fragment; the asterisk-labelled peaks are the accompanying 2′-3′ cyclic phosphate- harbouring species. See Figure 5 for a table of expected chemically unmodified fragments.
Figure 7 also yields additional information about the position of the 8–13 fragment (5′-[s4U]XCCCGp-3′) with the unknown +16.0 Da modification, because it becomes doubly cyanoethylated to yield a 2029.2 Th signal. Thus, the unknown nucleotide in this fragment also reacted with acrylonitrile. A priori we had expect this fragment to be singly cyanoethylated, because 4-thiouridine reacts readily with acrylonitrile forming an S-cyanoethyl product (21). The double cyanoethylation strongly indicates that the nucleotide at position 9 in tRNATyrII is also a 4-thiouridine: 2-thiouridine is reported not to react with acrylonitrile (21), and 5-hydroxyuridine does not have the chemical properties to perform a nucleophilic addition to acrylonitrile.
DISCUSSION
As an extension of previous work, where we reported a MALDI mass spectrometric method to screen for post-transcriptional modifications (22), we have implemented a specific cyanoethylation of ψ, which is then amenable to mass spectrometric detection. The reaction is generally very selective for ψ versus its uridine precursor; no appreciable cyanoethylation of uridines was observed in any of the shown spectra. The degree of ψ cyanoethylation under the applied conditions is 50% or better as estimated from the signal intensities in the mass spectra (note that signal intensities are generally analyte-dependent, and quantifications are therefore not very reliable with our experimental set-up). However, if we aimed at a quantitative conversion of ψ to its cyanoethylated form, undesired cyanoethylated fragments started to appear, probably from reaction of uridine nucleotides. Yoshida and Ukita (18) noted that the degree of cyanoethylation is dependent on the sequence context of a given ψ, which we have also observed. This is likely to be due to dissimilar accessibility of the ψs in tRNA’s three-dimensional structure. Thus, different RNA molecules may require different reaction times in order to attain the optimal degree of ψ modification. We have also performed the RNase digestions prior to the cyanoethylation in order to avoid effects of RNA higher order structure. The approach was, in principle, successful, but it introduced difficulties in the subsequent purification: whereas vacuum drying followed by ethanol precipitation sufficed for the intact cyanoethylated tRNA, purification of the cyanoethylated digestion fragments required a chromatographic step due to the small size of the digestion fragments.
The cyanoethylation of a given RNase digestion fragment does not tell us where the ψ is located if the fragment contains more than one uridine. This is often the case in RNase T1 generated fragments. The application of RNase A sometimes resolves the ambiguity, but not for sequence stretches consisting of many pyrimidine nucleotides. We therefore successfully used MALDI tandem mass spectrometry as a means to pinpoint the chemically modified nucleotide. The theoretical upper limit for this approach is 4000 Th, determined by the mass spectrometric hardware, but it is, in practice, probably lower due to reduced sequence information from larger singly charged species (23). Limbach and co-workers have used HPLC isolation of CMC-modified RNase T1 digestion fragments followed by sequential exonuclease digestion and mass spectrometry to determine the position of modified ψs (16). This method should also be applicable here. An alternative approach would be to perform tandem mass spectrometry on multiply charged ions generated by electrospray ionisation.
The RNase digestion pattern is dependent on whether post-transcriptionally modified positions are refractory to RNase cleavage. The natural cleavage site for RNase T1 is on the 3′-side of guadinylates, but in the present study we also observed cleavage after N2-methylguanine and less efficiently after N2,N2-dimethylguanine. Cleavage after prolonged incubation with RNase T1 has also been observed after N1-methylguanine (S.Douthwaite, M.F.Liu and F.Kirpekar, unpublished results). In contrast, we observed complete RNase T1 inhibition by the guadinylate derivatives 2′-O-methyl guanine, N7-methylguanine, queuosine and wybutosine. X-ray crystal data show that the both the N1, N2 and N7 positions of guanosine are involved in the substrate recognition by the enzyme (29), but there is apparently only an absolute requirement for unmodified N7 in the RNase T1-mediated cleavage. RNase A, which cleaves after pyrimidine nucleotides, is much less selective in its choice of substrate in that we have only observed inhibition by 2′-O-methylated pyrimidine nucleotides. The 2′-O-methyl inhibition is understandable in light of the RNase cleavage mechanism, where hydrolysis of the phosphodiester backbone proceeds over a 2′-3′ cyclic phosphate intermediate. RNase A cleavage was observed after 5-methylcytidine, thymidine, dihydrouridine, pseudouridine and 4-thiouridine, as well as the cyanoethylated derivatives of the latter two nucleotides.
The ‘traditional’ CMC-derivatisation affects post-transcriptionally modified nucleotides other than ψ (14), and acrylonitrile likewise reacted with 4-thiouridine [as well as inosine (21)] in the present study. This was not a problem but rather aided the identification of the additional 4-thiouridine in E.coli tRNATyrII. Analysing the unreacted tRNA already revealed the presence of an additional modification, and cyanoethylation gave a strong indication of the identity of the modification. In conclusion, mass spectrometry in combination with cyanoethylation as a means to analyse post-transcriptional modifications is experimentally simple and chemically gentle, and the methodology complements the CMC/alkali approach with either mass spectrometric (16) or reverse transcriptase/electrophoretic (14) detection.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Birte Vester for suggesting the use of acrylonitrile and Michael Lund Nielsen, MDS Proteomics, Denmark for initial help with the MALDI MS/MS analysis. This work was supported by the Danish Natural Science Research Council and the Danish Biotechnology Instrument Centre.
References
- 1.Cunningham P.R., Richard,R.B., Weitzmann,C.J., Nurse,K. and Ofengand,J. (1991) The absence of modified nucleotides affects both in vitro assembly and in vivo function of the 30S ribosomal subunit of Escherichia coli. Biochimie, 73, 789–796. [DOI] [PubMed] [Google Scholar]
- 2.Green R. and Noller,H.F. (1996) In vitro complementation analysis localizes 23S rRNA post-transcriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA, 2, 1011–1021. [PMC free article] [PubMed] [Google Scholar]
- 3.Sirum-Connolly K., Peltier,J.M., Crain,P.F., McCloskey,J.A. and Manson,T.L. (1995) Implications of a functional large ribosomal RNA with only three modified nucleotides. Biochimie, 77, 30–39. [DOI] [PubMed] [Google Scholar]
- 4.Skinner R.H. and Cundliffe,E. (1982) Dimethylation of adenine and the resistance of Streptomyces erythraeus to erythromycin. J. Gen. Microbiol., 128, 2411–2416. [Google Scholar]
- 5.Liu M., Kirpekar,F., van Wezel,G.P. and Douthwaite,S. (2000) The tylosin resistance gene tlrB of Streptomyces fradiae encodes a methyl transferase that targets G748 in 23S rRNA. Mol. Microbiol., 37, 811–820. [DOI] [PubMed] [Google Scholar]
- 6.Davis D.R. (1995) Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res., 23, 5020–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis D.R., Veltri,C.A. and Nielsen,L. (1998) An RNA model for investigation of pseudouridine stabilization of the codon–anticodon interaction in tRNA(Lys), tRNA(His) and tRNA(Tyr). J. Biomol. Struct. Dyn., 15, 1121–1132. [DOI] [PubMed] [Google Scholar]
- 8.Durant P.C. and Davis,D.R. (1999) Stabilization of the anticodon stem–loop of tRNALys,3 by an A+-C base pair and by pseudouridine. J. Mol. Biol., 285, 115–131. [DOI] [PubMed] [Google Scholar]
- 9.Meroueh M., Grohar,P.J., Qui,J., SantaLucia,J.,Jr, Scaringe,S.A. and Chow,C.S. (2000) Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA. Nucleic Acids Res., 28, 2075–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lecointe F., Simos,G., Sauer,A., Hurt,E.C., Motorin,Y. and Grosjean,H. (1999) Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of C38 and C39 in tRNA anticodon loop. J. Biol. Chem., 273, 1316–1323. [DOI] [PubMed] [Google Scholar]
- 11.Raychaudhuri S., Conrad,J. and Ofengand,J. (1998) A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA, 4, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen M.A., Kirpekar,F., Ritterbusch,W. and Vester,B. (2002) Post-transcriptional modifications in the A-loop of 23S rRNA from selected archaea and eubacteria. RNA, 8, 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowalak J.A., Bruenger,E., Crain,P.F. and McCloskey,J.A. (2000) Identification and phylogenetic comparison of post-transcriptional modifications in 16S ribosomal RNA form Haloferax volcanii. J. Biol. Chem., 275, 24484–24489. [DOI] [PubMed] [Google Scholar]
- 14.Bakin A. and Ofengand,J. (1993) Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyl transferase center: Analysis by the application of a new sequencing technique. Biochemistry, 32, 9754–9762. [DOI] [PubMed] [Google Scholar]
- 15.Ho N.W.Y. and Gilham,P.T. (1971) Reaction of pseudouridine and inosine with N-cyclohexyl-N′-beta-(4-methylmorpholinium) ethylcarbodiimide. Biochemistry, 10, 3651–3657. [PubMed] [Google Scholar]
- 16.Patteson K.G., Rodicio,L.P. and Limbach,P.A. (2001) Identification of the mass-silent post-transcriptionally modified nucleoside pseudouridine in RNA by matrix-assisted laser desorption/ionisation mass spectrometry. Nucleic Acids Res., 29, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chambers R.W., Kurkov,V. and Shapiro,R. (1963) The chemistry of pseudouridine. Synthesis of pseudouridine-5′-diphoshate. Biochemistry, 2, 1192–1203. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida M. and Ukita,T. (1968) Modification of nucleosides and nucleotides VIII. The reaction rate of pseudouridine residues with acrylonitrile and its relation to the secondary structure of transfer nucleic acid. Biochim. Biophys. Acta, 157, 466–475. [PubMed] [Google Scholar]
- 19.Yoshida M. and Ukita,T. (1968) Modification of nucleosides and nucleotides VII. Selective cyanoethylation of inosine and pseudouridine in yeast transfer nucleic acid. Biochim. Biophys. Acta, 157, 455–465. [PubMed] [Google Scholar]
- 20.Chambers R.W. (1965) The chemistry of pseudouridine IV. Cyanoethylation. Biochemistry, 4, 219–226. [Google Scholar]
- 21.Ofengand J. (1967) The function of pseudouridylic acid in transfer ribonucleic acid I. The specific cyanoethylation of pseudouridine, inosine and 4-thiouridine by acrylonitrile. J. Biol. Chem., 242, 5034–5045. [PubMed] [Google Scholar]
- 22.Kirpekar F., Douthwaite,S. and Roepstorff,P. (2000) Mapping post-transcriptional modifications in 5S ribosomal RNA by MALDI mass spectrometry. RNA, 6, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirpekar F. and Krogh,T.N. (2001) RNA fragmentation studied on a MALDI Qq-TOF mass spectrometer. Rapid Commun. Mass Spectrom., 15, 8–14. [DOI] [PubMed] [Google Scholar]
- 24.McLuckey S.A., Van Berkel,G.J. and Glish,G.L. (1992) Tandem mass spectrometry of small multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom., 3, 60–70. [DOI] [PubMed] [Google Scholar]
- 25.Hamming M.C. and Foster,N.G. (1972) Interpretation of Mass Spectra of Organic Compounds. Academic Press, New York and London.
- 26.Thiebe R. and Zachau,H.G. (1968) A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur. J. Biochem., 5, 546–555. [DOI] [PubMed] [Google Scholar]
- 27.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozenski J., Crain,P.F. and McCloskey,J.A. (1999) The RNA Modification Database: 1999 update. Nucleic Acids Res., 27, 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugio S., Amisaki,T., Ohishi,H. and Tomita,K. (1988) Refined X-ray structure of the low pH form of ribonuclease T1-2′-guanylic acid complex at 1.9 Å resolution. J. Biochem. (Tokyo), 103, 354–366. [DOI] [PubMed] [Google Scholar]