Abstract
The spondylocostal dysostoses (SCDs) are a heterogeneous group of vertebral malsegmentation disorders that arise during embryonic development by a disruption of somitogenesis. Previously, we had identified two genes that cause a subset of autosomal recessive forms of this disease: DLL3 (SCD1) and MESP2 (SCD2). These genes are important components of the Notch signaling pathway, which has multiple roles in development and disease. Here, we have used a candidate-gene approach to identify a mutation in a third Notch pathway gene, LUNATIC FRINGE (LFNG), in a family with autosomal recessive SCD. LFNG encodes a glycosyltransferase that modifies the Notch family of cell-surface receptors, a key step in the regulation of this signaling pathway. A missense mutation was identified in a highly conserved phenylalanine close to the active site of the enzyme. Functional analysis revealed that the mutant LFNG was not localized to the correct compartment of the cell, was unable to modulate Notch signaling in a cell-based assay, and was enzymatically inactive. This represents the first known mutation in the human LFNG gene and reinforces the hypothesis that proper regulation of the Notch signaling pathway is an absolute requirement for the correct patterning of the axial skeleton.
The spondylocostal dysostoses (SCDs) are a group of disorders characterized by multiple vertebral segmentation defects and rib anomalies (Turnpenny et al. 2003). SCD is most frequently seen in sporadic cases and with diverse radiological phenotypes, which are difficult both to classify and to investigate. Nevertheless, the developmental mechanisms that produce all these defects in the embryo are likely to involve abnormal formation of somites (precursors of vertebra and associated musculature). In vertebrates, all skeletal muscle of the body, the axial skeleton, the tendons, and the dorsal dermis are derived from somites. These are paired blocks of mesoderm located on either side of the neural tube that form by a regular wave of segmentation in a rostral-to-caudal direction throughout the trunk (reviewed by Pourquié [2001]). Somite formation is a reiterative process—for example, in the mouse, somites segment from the presomitic mesoderm every 2 h for 8–10.5 d post coitum (equivalent to human Carnegie stages 9–13 or days 19–29). The Notch signaling pathway is central to somite formation, and, in the mouse, mutations in several components of the pathway (Notch1, Dll3, Dll1, Lfng, Psen1, and CSL) and a downstream target gene (Hes7) result in abnormal somitogenesis (reviewed by Weinmaster and Kintner [2003]).
The Notch signaling pathway is an evolutionarily conserved signal-transduction pathway that has important roles throughout embryonic development, as well as in the onset of diseases such as cancer (Harper et al. 2003). Mechanistically, the pathway is relatively simple, with ligand-activated signaling mediated by the nuclear translocation of the intracellular portion of the receptor to form part of a transcriptional complex. Notch is a heterodimeric one-pass transmembrane receptor. The epidermal growth factor (EGF)–like repeat containing extracellular fragments mediates interactions with DSL (Delta, Serrate, and Lag-2) ligands such as Delta-like–1 (Dll1) and Jagged1. The second fragment of Notch spans the membrane and contains an intracellular domain (ICD) that mediates Notch signaling. Binding to a ligand leads to cleavage by a disintegrin and metalloproteinase and release of the extracellular domain of Notch. A second cleavage by γ-secretase releases the ICD. The ICD translocates to the nucleus, forms a complex with the CSL (CBF1/Su(H)/Lag-1) DNA-binding protein, and converts it from a repressor to an activator of transcription. Known direct targets of Notch include members of the hairy/enhancer-of-split (HES) and HES-related family of basic helix-loop-helix transcription factors (Iso et al. 2003) and the glycosyltransferase Lunatic fringe (Morales et al. 2002).
Elsewhere, we identified two genes that cause a subset of autosomal recessive forms of SCD: DLL3 (SCD1 [MIM 277300]) (Bulman et al. 2000; Sparrow et al. 2002; Turnpenny et al. 2003; Whittock et al. 2004a) and MESP2 (SCD2 [MIM 608681]) (Whittock et al. 2004b). Mutations in either of these genes account for ∼20%–25% of all cases of this disorder, with 24 distinct mutations detected in DLL3 (Bonafé et al. 2003; Turnpenny et al. 2003; Whittock et al. 2004a; authors' unpublished data) and a single mutation detected in MESP2 (Whittock et al. 2004b). In this article, we describe a patient with a skeletal phenotype more severe than that of previously reported cases of SCD1 and SCD2. We show that this patient is homozygous for a missense mutation in the LFNG gene. LFNG is a fucose-specific β 1,3 N-acetylglucosaminyltransferase that adds N-acetylglucosamine (GlcNAc) residues to O-fucose on the EGF-like repeats of Notch receptors (Bruckner et al. 2000; Moloney et al. 2000). LFNG localizes to the Golgi, where the modification of Notch receptors is believed to occur (Haines and Irvine 2003). LFNG enhances Notch1’s ability to be activated by Dll1 and reduces Notch1 signaling when Jagged1 is the activating ligand. Functional analysis revealed that the mutant LFNG was not localized to the correct compartment of the cell, was unable to modify Notch signaling in vitro, and was enzymatically inactive. Therefore, we conclude that this missense mutation is likely to be causative of SCD in this patient.
Material and Methods
Human Subjects
Appropriate informed consent was obtained from the proband and family for this study.
DNA Sequencing
Genomic DNA was sequenced as described elsewhere (Sparrow et al. 2002), by use of primers listed in table 1.
Table 1.
Primer Sequences
|
Primer Sequence(5′→3′) |
|||
| Exon(s) | Forward | Reverse | Internal Sequencing |
| 1: | |||
| Pair 1 | cgacgggcttcgggtcggtg | tcttgacagcgatgaagacg | … |
| Pair 2 | agtctgtccgagtacttcag | ctggcgtcgcccacagatgg | … |
| 2 | cacgagtggggaaaccaagg | ggcggggcatggagcaaacc | … |
| 3 | ggagggtccacaggcccaag | gggcccgcaggttgacgtag | … |
| 4 and 5 | tcatcgagtccggcaggaag | tgaaacccagagggaagtgg | caggcgctgctgcccctcac; cgagaacaaggtggtgagtg |
| 6 | tgaggagtgcagcgcctttg | tgtttagaacagtgccccac | … |
| 7 | ccagtttgggaccttattcc | catttgcagtcccacaacac | … |
| 8 | gtgttgtgggactgcaaatg | ccccttcacctgtgtgcctc | tcttaagccacagcgtccag |
RFLP Analysis
The F188L mutation produces an MseI restriction site. Control samples were tested by amplification with primers 5′-GGAGGGTCCACAGGCCCAAG-3′ (forward) and 5′-GGGCCCGCAGGTTGACGTAG-3′ (reverse) and were digested with MseI to yield fragments of 413 bp for the wild-type allele and 251 bp and 162 bp for the mutant allele.
In Vitro GlcNAc-Transferase Assays
HEK 293T cells were transfected with expression plasmids encoding full-length mouse Lfng cDNAs (wild-type and mutants) fused to human IgG1 Fc (at the carboxy terminus). Protein was affinity purified from either concentrated medium or whole-cell extract by use of Protein-G-Sepharose beads (Sigma), as described elsewhere (Moloney et al. 2000). GlcNAc-transferase activity was determined as described elsewhere (Moloney et al. 2000).
Cell Lines and Coculture Assays
C2C12 and NIH3T3 cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum. C2C12 cells, which overexpress full-length Notch1, are described elsewhere (G. Chapman, L. Liu, C. Dahlqvist, and U. Lendahl, unpublished data). Control cells, cells overexpressing mouse Dll1, and cells overexpressing mouse Jagged1 were created by stably transfecting NIH3T3 cells with pCAGIRESpuro, pCAG-mDll1-IRESpuro, and pCAG-mJagged1-IRESpuro, respectively. Cultures were grown in 1.5 μg/ml puromycin for 10 d. Individual mDll1 or mJagged1 stable clones were screened for expression by western blotting and immunocytochemistry with rabbit anti-Dll1 or anti-Jagged1 antibodies (Santa Cruz). Transfections were performed using LipofectAMINE PLUS reagent (Invitrogen) in 12-well trays with 14 ng of pCMV-Renilla, 350 ng of p6×TP1-Luc (CSL reporter [Kato et al. 1997]), and 350 ng of each expression plasmid or pCMV-CAT control plasmid. Cocultures were performed in triplicate by addition of 2×105 NIH3T3 cells stably transfected with vector (control cells), mDll1, or mJagged1. Cocultures were harvested in 200 μl of Passive Lysis Buffer (Promega) 24 h after transfection. Firefly and Renilla luciferase activities were assayed using the Dual-Luciferase reporter system (Promega) and were measured on a FLUOstar Optima Luminometer (BMG). Firefly luciferase counts were normalized against Renilla luciferase counts, to account for differences in transfection efficiency. Fold activation of ligand-expressing cells over control NIH3T3 cells was expressed relative to that of chloramphenicol transferase (CAT)–transfected cocultures. One-way analysis of variance was performed on data from four independent experiments. Significance was determined using Tukey’s post hoc test.
Western Blots and Immunocytochemistry
Western blotting was performed as described elsewhere (G. Chapman, L. Liu, C. Dahlqvist, and U. Lendahl, unpublished data). Confocal immunofluorescence of hemagglutinin (HA)–tagged Lfng and mutants was done as described elsewhere (Dahlqvist et al. 2003), by use of a TCS SP confocal microscope (Leica Microsystems). Antibodies used were obtained from Santa Cruz (Lfng), Clontech (rabbit anti-HA), Covance (mouse anti-HA), and BD transduction labs (GM130).
Results
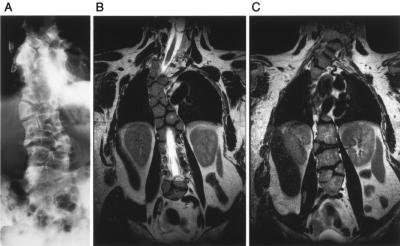
The proband was of Lebanese background and presented with extensive congenital vertebral anomalies; long, slender fingers; and camptodactyly of the left index finger. X-ray (fig. 1A) and magnetic resonance imaging (MRI) scans (fig. 1B) showed multiple vertebral ossification centers in the thoracic spine, which showed fitted angular shapes similar to those seen in the patient with SCD2 who was described elsewhere (Whittock et al. 2004b). Severe foreshortening of the spine is emphasized by the comparison of the patient’s arm span (186.5 cm) with adult height (155 cm; lower segment 92.5 cm). The patient had nonprogressive scoliosis of the cervical and thoracic spine. However, in contrast to SCD2, vertebral anomalies were also present in the cervical and lumbar spine (fig. 1A and 1C). Vertebral anomalies along the whole length of the spine are a feature of SCD1; however, in SCD1, the vertebrae have a more rounded shape (“pebble beach” sign [Turnpenny et al. 2003]) than in this case. Although vertebral shape is known to change with increasing age—with vertebrae becoming less rounded and more angular—the proband clearly had a spine more severely disorganized (throughout all the vertebral bodies) than that in the majority of SCD1 cases seen so far and much more severe than the singly reported sibling cases with an SCD2 mutation. Analysis of the entire coding region and splice sites of DLL3 revealed no deviations from the published sequence. Likewise, no mutations were found in the MESP2 coding region for this patient.
Figure 1.
Radiograph (A) and T2-weighted coronal MRI images (B and C) in the vertebral plane of the proband. A, Severe vertebral segmentation anomalies throughout the vertebral column. B, Thoracic spine, showing vertebral centers with a fitted angular shape. C, Cervical and lumbar spine, showing similar segmentation anomalies.
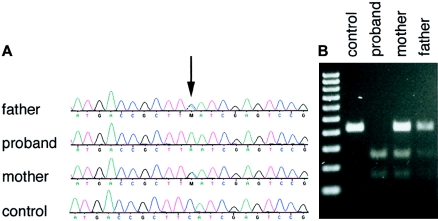
We therefore sought to identify other candidate genes for sequencing in this patient. Using the Dll3 null mouse as a model for SCD, we have shown that SCD is caused by a defect in the formation of embryonic structures called “somites,” which are the precursors of the axial skeleton, tendons, muscle, and the dorsal dermis (Dunwoodie et al. 2002). The Notch signaling pathway is central to the regulation of this process (reviewed by Weinmaster and Kintner [2003]). Indeed, both of the genes thus far implicated in causing SCD encode components of this signaling pathway: DLL3 is a ligand for the Notch family of signaling receptors, and MESP2 is a downstream target of Notch signaling. Deletion of other components of the Notch signaling pathway in the mouse also causes somite dysmorphology, and these mutants represent the majority of defects in somitogenesis reported thus far (see Sparrow et al. 2002). We therefore examined the phenotypes of such mouse lines in detail, in an effort to identify further candidate genes for sequencing in our patient. The phenotypes of Dll3 (pudgy) and Lfng null mice were virtually identical (Zhang et al. 2002), indicating that murine somitogenesis is similarly dependent on each gene. LFNG is a fucose-specific β 1,3 N-acetylglucosaminyltransferase (Bruckner et al. 2000; Moloney et al. 2000) that functions in the Golgi to posttranslationally modify the Notch receptors, altering their signaling properties (Haines and Irvine 2003). Furthermore, our previous analysis revealed that Lfng gene expression is severely disregulated in Dll3 null mice, suggesting that Lfng expression is dependent on Dll3 function (Dunwoodie et al. 2002; Kusumi et al. 2004). We sequenced the entire coding region and splice sites of the LFNG gene in the proband (fig. 2A), using primers listed in table 1. A homozygous missense mutation (c.564C→A) in exon 3 was detected that results in substitution of leucine for phenylalanine (F188L). Sequencing of DNA from the proband’s parents, who had normal spinal and hand anatomy, confirmed that they were both heterozygous for the mutant allele. A number of approaches were taken to demonstrate that this base change was not a common polymorphism unassociated with the SCD phenotype. The mutation created a novel MseI restriction enzyme site, and the resultant RFLP was used to confirm the sequencing results in the pedigree (fig. 2B). The MseI polymorphism was not found in 125 control subjects (250 chromosomes), giving ∼80% power to distinguish a normal sequence variant from a mutation (Collins and Schwartz 2002). In addition, the underlying base substitution was not present in the National Center for Biotechnology Information SNP database (dbSNP Web site).
Figure 2.
Detection of the c.564C→A mutation. A, Electropherograms documenting the affection status of the proband and parents. B, Confirmation of the presence of the c.564C→A mutation by MseI RFLP in genomic DNA isolated from an unrelated control individual, the proband, and the proband’s mother and father.
Structural Consequences of the F188L LFNG Mutation
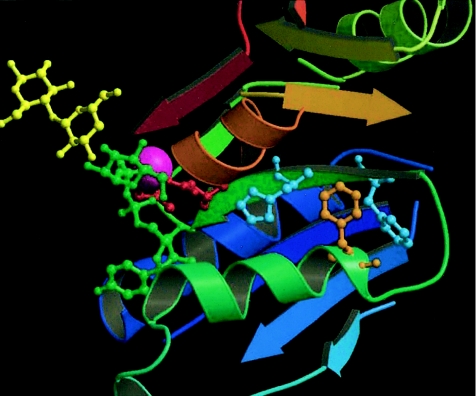
The mutated phenylalanine is absolutely conserved in all known fringe proteins, from Drosophila melanogaster to humans (Correia et al. 2003). Examination of the mutation within an LFNG model based on solved glycosyltransferase structures suggests that the conserved phenylalanine residue (F188) is not directly involved in uridine diphosphate (UDP)–GlcNAc (donor) or protein binding (fig. 3). Rather, it is likely to reside in a helix that packs against the strand containing the Mn2+ligating residues D200 and the nearby D202. F188 is predicted to form an aromatic cluster with residues F196 and H198, and thus the F188L mutation may either cause steric perturbation of the Mn2+ binding site by altered packing of the smaller mutant (leucine) residue or cause electronic disruption of the enzymatic reaction by removal of required π-π interactions associated with the aromatic ring. Further details of structural modeling techniques are given in appendix A (online only).
Figure 3.
Structural model of LFNG, showing the proximity of the mutated phenylalanine residue (orange) to the Mn2+ binding site (Mn2+ in purple). Directly interacting residues F196 and H198 are shown in cyan, and the nearby Mn2+-ligating residues D202 and D203 are in red. The position of the UDP-sugar donor group (green) and an acceptor sugar (yellow) are shown for reference. α-helices and β-sheets are colored in a gradient from dark blue (amino terminus) to red (carboxy terminus).
Functional Analysis of the F188L LFNG Mutation
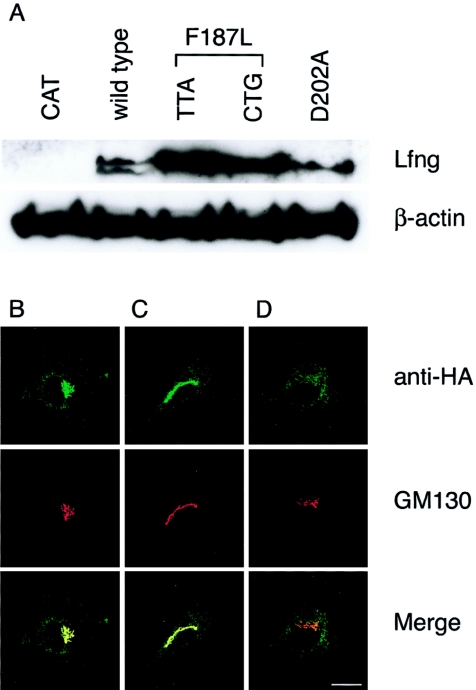
To provide evidence that this was, indeed, an SCD-causative mutation, we undertook a range of assays to assess the function of the LFNG mutant. Since no full-length human cDNA was available, we created two F187L mutations in mouse Lfng that correspond to F188L in human LFNG: a c.564C→A mutation encoding the rare leucine codon (TTA) observed in the proband and the c.562T→C + c.567C→G mutation encoding the most common human leucine codon (CTG). In addition, a previously characterized, enzymatically inactive form of Lfng (D202A) was created that disrupts the conserved DDD Mn2+ binding active site (Chen et al. 2001). We initially examined whether the mutant protein was stable and efficiently expressed, since, in addition to the potential destabilization of the protein by amino acid substitution, the presence of low-frequency codons in transcripts has been shown to cause stalling of translation machinery, transcript degradation, and lowered levels of protein expression (Lemm and Ross 2002). HA-tagged versions of Lfng were transiently transfected into C2C12 cells, and protein levels were determined by western blot (fig. 4A). Both F187L mutant Lfng proteins were expressed at higher levels than the wild-type or D202A Lfng forms, indicating that both translation efficiency and protein stability were not adversely affected by the F187L amino acid change or the presence of a low-frequency codon in the transcript. In all subsequent assays, the TTA and CTG mutants behaved identically (figs. 4A, 5A, and 5B and data not shown).
Figure 4.
A, Relative expression of transfected wild-type and mutant Lfng proteins. Shown is western blot detection of Lfng in lysates from C2C12 cells transfected with HA-tagged wild-type, TTA F187L, CTG F187L, and D202A Lfng. A total of 50 μg of each lysate was run on a 4%–12% PAGE gel and was blotted. Lfng proteins were detected using a mouse anti-HA antibody. Western blot detection of β-actin was performed to control for loading. B–D, Immunofluorescence analysis of wild-type and mutant Lfng proteins in cultured cells. F187L Lfng does not localize to the Golgi. C2C12 myoblasts were transiently transfected with constructs encoding HA-tagged wild-type Lfng (B), D202A mutant Lfng (C), and F187L Lfng (D). In each case, the top panel shows a confocal image of immunofluorescence with a rabbit anti-HA antibody, and the middle panel with the Golgi-specific antibody (GM130); the bottom panel shows a merged image. The scale bar represents 25 μm.
Figure 5.
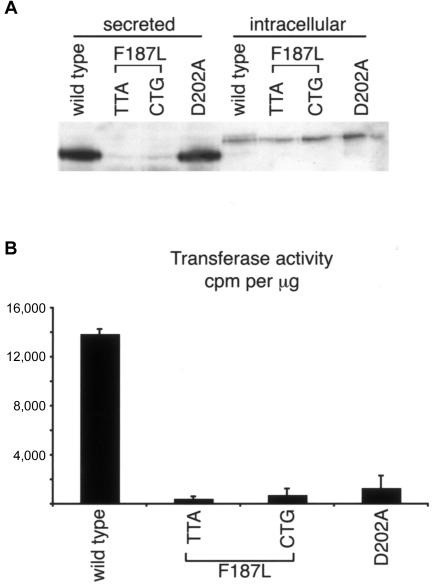
Results showing that F187 Lfng is enzymatically inactive. A, Western blot using an Lfng-specific antibody on Lfng-Fc fusion protein that was affinity purified from conditioned medium (“secreted”) or whole-cell extract (“intracellular”) derived from transiently transfected HEK 293T cells. B, GlcNAc-transferase assays performed on Lfng that was affinity purified from whole-cell extract by use of pNp-fucose (4 mM) as an acceptor and UDP-[3H]GlcNAc as a donor. Assays were performed in duplicate, and error bars represent SDs of the mean.
Lfng protein is normally present in the Golgi apparatus. To determine whether the F187L mutation affected the targeting of Lfng, intracellular protein localization was examined using immunofluorescence (fig. 4B–4D). Wild-type and D202A mutant Lfng were localized predominantly to the Golgi, as assessed by costaining with the Golgi-specific marker GM130 (fig. 4B and 4C). By contrast, the F187L mutant Lfng did not colocalize with GM130 (fig. 4D). Therefore, we conclude that the F187L mutant form of Lfng is expressed but is mislocalized within the cell.
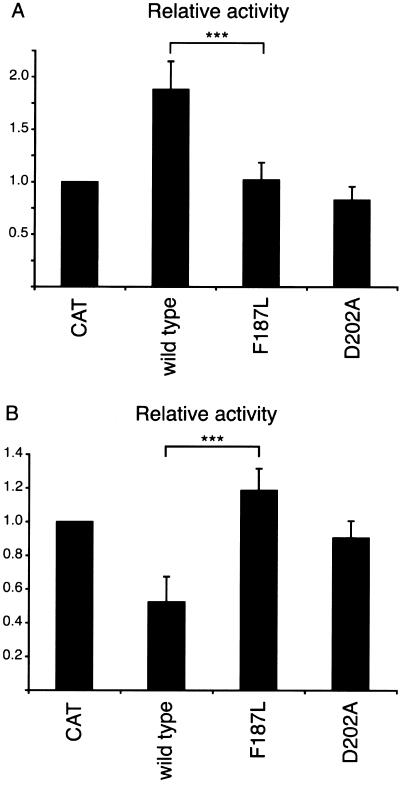
To assess the effect of the F187L mutation on Lfng function, we used a cell coculture assay described elsewhere (Hicks et al. 2000; G. Chapman, L. Liu, C. Dahlqvist, and U. Lendahl, unpublished data). In this system, C2C12 myoblasts expressing the mouse Notch1 receptor (i.e., responding cells) were cocultured with NIH3T3 cells expressing either mouse Dll1 or Jagged1 ligands. Prior to coculture, responding cells were transiently transfected with a luciferase reporter gene driven by multiple copies of the Notch signaling–responsive CSL binding site (Kato et al. 1997). After coculture for 24 h, luciferase levels were determined, to measure the extent of Notch signaling in the responding cells. Expression of wild-type Lfng in responding cells acted to potentiate Dll1 signaling (fig. 6A) and to inhibit Jagged1 signaling (fig. 6B), as reported elsewhere (Hicks et al. 2000; Shimizu et al. 2001). However, when either the F187L or the D202A mutant forms of Lfng were used in these assays, they had little or no effect on the levels of Notch signaling induced by Dll1 or Jagged1 in the responding cells. These data indicate that the F187L mutation renders Lfng functionally inactive in these Notch signaling assays.
Figure 6.
Results showing that F187L Lfng does not modulate ligand-induced Notch signaling. A, Wild-type Lfng significantly potentiates activation of Notch1 by coculture with Dll1-expressing cells, whereas F187L and D202A mutants do not alter levels of Notch1 activation. Three asterisks (***) denote P<.001 for wild-type versus F187L Lfng. B, Wild-type Lfng significantly inhibits activation of Notch1 by coculture with Jagged1-expressing cells, whereas F187L and D202A mutants do not alter levels of Notch1 activation. Three asterisks (***) denote P<.001 for wild-type versus F187L Lfng. Notch1 cells were cotransfected with plasmids encoding wild-type Lfng, mutant Lfng, or CAT (as a negative control) and a CSL reporter. Transfected cells were cocultured with either control NIH3T3, Dll1, or Jagged1 cells and were assayed for luciferase activity. Fold activation of ligand-expressing cells over control NIH3T3 cells is expressed relative to that of CAT-transfected cocultures. Notch signaling was activated 4–10-fold in these experiments. Error bars represent SDs of four independent experiments.
The incorrect subcellular localization of the F187L Lfng could be responsible for the failure of this mutant to function in Notch signaling, and thus it is possible that this form of Lfng is still capable of modifying the Notch receptors. Therefore, to assess the enzymatic activity of the wild-type and mutant Lfng proteins, we performed a GlcNAc-transferase assay in vitro. Carboxy-terminal Fc-tagged versions of wild-type or mutated forms of Lfng were expressed in 293T cells and were affinity purified from medium, as described elsewhere (Moloney et al. 2000), and protein levels were analyzed by immunoblot with an Lfng-specific antibody (fig. 5A). Although Lfng is normally considered to be localized to and function in the Golgi apparatus in vivo (Bruckner et al. 2000; Munro and Freeman 2000), when it is overexpressed in cultured cells, significant amounts of protein are secreted into the medium. This is a common feature when Golgi glycosyltransferases are overexpressed, but it is not known whether the secreted form serves any biological function, although evidence from the Drosophila homologue (Fringe) suggests that the major function of the protein occurs within the Golgi (Bruckner et al. 2000; Munro and Freeman 2000). In keeping with our subcellular localization results, wild-type and D202A mutant forms of Lfng were efficiently secreted from the cell, but only small amounts of F187L Lfng were present in conditioned medium (2%–3% of normal protein levels) (fig. 5A). This is likely to be the result of a defect in protein secretion, since the F187L mutation does not affect protein translation and stability within the cell (fig. 4A). Therefore, the protein purification was repeated using a whole-cell protein extract. As expected, all four proteins were detectable (fig. 5A). Affinity-purified protein derived from whole-cell extract was then used in an in vitro GlcNAc-transferase assay with the use of p-nitrophenyl-α-l-fucose (which structurally mimics O-linked fucose) as an acceptor and UDP-[3H]GlcNAc as a donor substrate (Moloney et al. 2000). The difference in size between protein purified from conditioned medium and protein purified from whole-cell extract may be indicative of the absence of pro-protein processing. However, there was no discernible difference in the relative in vitro enzymatic activity of wild-type protein purified from either source (data not shown). As shown in figure 5B, the F187L (TTA and CTG) and D202A mutants showed no enzymatic activity above background, whereas wild-type Lfng showed activity similar to that described elsewhere (Chen et al. 2001). This demonstrates that F187 is necessary for GlcNAc-transferase activity and that substitution of leucine at this site renders the enzyme inactive.
Discussion
Our data strongly support the hypothesis that the F188L missense mutation of LFNG is causative of SCD in the proband, because it is not localized to the correct compartment of the cell, is unable to modulate Notch signaling in the same manner as the wild-type protein, and is enzymatically inactive. We propose that this form of SCD be referred to as “SCD3” because mutation in LFNG demonstrates that this case is genetically distinct from SCD1 and SCD2. The proband has a more severely disorganized spine, compared with the majority of SCD1 cases seen so far, and certainly compared with the SCD2 cases; however, clear differentiation in phenotype between cases of SCD caused by mutation in DLL3, MESP2, and LFNG cannot be confirmed until more patients with mutations in LFNG (and MESP2) are uncovered. It is unclear how frequent such mutations are within the group of patients with costovertebral defects, since we have, to date, only sequenced the LFNG gene for 28 patients with SCD who lack mutations in both DLL3 and MESP2. Discovery that mutation in a third gene causes a discernible SCD phenotype emphasizes the importance of detailed examination of the vertebral anomalies of patients with SCD to assist diagnosis of the underlying genetic defect in this group of vertebral segmentation disorders.
In contrast to the apparent situation in humans, close analysis of the vertebral phenotypes in mice lacking Dll3 (Dunwoodie et al. 2002), Mesp2 (Saga et al. 1997), or Lfng (Zhang and Gridley 1998) shows no obvious differences in either the appearance of vertebrae or the extent of malformation along the rostral-caudal axis. Therefore, it is possible that differences are observed in the human phenotypes because the expression of these genes in humans is different from the expression in mice. In the mouse, detailed expression profiles during somitogenesis have been established for Dll3 (Dunwoodie et al. 1997), Mesp2 (Saga et al. 1997), and Lfng (Forsberg et al. 1998; Aulehla and Johnson 1999). In contrast, nothing is known about the expression of these genes during somite formation in humans. It is also possible that the differences between mouse and human phenotypes are the result of the nature of the causative mutations, rather than being intrinsic to the genes themselves. Here, we have shown that the LFNG F188L mutation is equivalent to a null allele. Likewise, the majority of DLL3 mutations are probably also null alleles (Turnpenny et al. 2003). In contrast, the only known MESP2 mutant allele produces a truncated protein that retains its DNA-binding domain and may possess some of its normal function. This may explain the lesser extent of the phenotype along the vertebral column in the patients with the MESP2 mutation than that in the patients with DLL3 and LFNG mutations.
In addition to vertebral anomalies, the proband has long, slender fingers and camptodactyly of the left index finger. We feel that this is unlikely to be a consequence of the mutation in LFNG, since mouse embryos lacking Lfng expression do not show limb defects (Zhang et al. 2002) and, within the developing vertebrate limb, Lfng expression is limited to hemangioblasts, the precursors of blood vessels. However, the exact expression pattern of LFNG in the human embryo remains to be investigated, and it is formally possible that human limb development differs from that in the mouse. It should also be noted that another member of the fringe family, Radical fringe (Rfng), is expressed in the apical ectodermal ridge in both mouse and chick. This is a key organizing center in the developing vertebrate limb that has several distinct roles in embryonic limb formation (Capdevila and Izpisua Belmonte 2001). The role of Rfng within the developing limb is controversial because studies in the chick and mouse show differences. In the chick, ectopic overexpression of Rfng in the developing limb causes a disruption of normal limb formation (Laufer et al. 1997; Rodriguez-Esteban et al. 1997). However, in mouse embryos lacking Rfng, no overt phenotype is seen in the limb or elsewhere (Moran et al. 1999; Zhang and Gridley 1999; Zhang et al. 2002). Furthermore, mouse embryos lacking both Lfng and Rfng show an identical phenotype to Lfng null embryos, with no overt limb deformity (Zhang et al. 2002). These discrepancies may be caused by differences between chick and mouse limb formation or by differences in experimental approach (i.e., overexpression vs. gene deletion). Additional patients with mutation in LFNG will show whether there is a correlation between the mutation and the presence of camptodactyly; however, an anomaly of a single finger could possibly be a secondary consequence of the cervical segmentation phenotype through nerve entrapment.
Lfng is expressed in a number of other locations in the developing mouse embryo, including the segmenting hindbrain, neural crest migrating into the branchial arches, olfactory placode, inner ear, lung, kidney, thymus, ovary, and teeth (Cohen et al. 1997; Johnston et al. 1997; Zhang et al. 2000; Koch et al. 2001; Mustonen et al. 2002; Leimeister et al. 2003; Hahn et al. 2005; van Tuyl et al. 2005). Because of a reduced viability of the Lfng null mouse line, phenotypic assessment of these organs in null mice has been limited to the inner ear (Zhang et al. 2000) and the ovary (Hahn et al. 2005). In the latter case, female null mice have been shown to be infertile because of defects in the meiotic maturation of oocytes. The availability of a null LFNG allele in humans provides a valuable opportunity to explore the consequence of a loss of LFNG beyond the axial skeleton. In particular, LFNG is expressed in restricted regions of the developing brain and nervous system, and thus clinical neurological investigation of the proband may be useful to further elucidate the importance of modulation of Notch signaling in the development of the brain.
It is not clear how the F188L mutation leads to a loss of subcellular localization of Lfng in the Golgi, especially since the D202A mutant (which also lacks enzymatic activity) is localized correctly. It is possible that this mutation has uncovered a new nonenzymatic functional domain in the Lfng protein that is required to bind another protein or chaperone to achieve proper subcellular localization. Such a dual role for a glycosyltransferase has been recently described for the Drosophila protein O-fucosyltransferase (Okajima et al. 2005). This enzyme adds an O-fucose residue to the Notch receptor to create the substrate for the fringe family of glycosyltransferases. However, this protein is likely to possess a nonenzymatic ability to facilitate folding of the Notch receptor and subsequent trafficking out of the endoplasmic reticulum to the Golgi.
Acknowledgments
We thank the family for their cooperation, U. Lendahl for cell lines and reagents, and R. Harvey for critical comments on the manuscript. This work was supported by National Health and Medical Research Council (NHMRC) (Australia) project grant 404804. S.L.D. is a Pfizer Foundation of Australia Senior Research Fellow, D.B.S. is a Westfield-Belconnen Fellow, G.C. is an NHMRC (Australia) CJ Martin Fellow (158043), M.A.W. is a Freedman Foundation Fellow, D.F. is a Sylvia and Charles Viertel Charitable Foundation Senior Research Fellow, and K.K. is a Hitchings Elion Fellow of the Burroughs Wellcome Fund.
Appendix A
Background
Fringe proteins belong to the GT31 family of inverting glycosyltransferases of the GTA structural superfamily. GTA glycosyltransferases function by transferring a sugar attached to a UDP-moiety either directly to a polypeptide chain or to the terminal sugar of a growing carbohydrate chain. In the case of Fringe, GlcNAc is transferred to a fucose moiety attached to a serine or threonine residue in the resident protein. Structures of GTA superfamily members conform to a basic Rossmann fold with β-sheet topology (β3β2β1β4β7β6β8). The first four of these β-strands, along with interspersed α-helices, form a nucleotide-binding subunit. In the case of the GTA superfamily, the nucleotide bound is UDP. The F188L mutation is found in helix α3, which lies at the crossover region of the β-sheet between β-strands 3 and 4.
Structure-Prediction Methods
Templates for LFNG were assessed by consulting SCOP (Murzin et al. 1995) and CAZy (Coutinho and Henrissat 1999; Liu and Mushegian 2003). Those selected were GTA glycosyltransferases from families GT2 (1qg8), GT27 (1xhb), and GT8 (1ll2 and 1ga8). The sequences of these enzymes were aligned using ClustalW (Thompson et al. 1994) and were checked against structure by use of DSSP (Kabsch and Sander 1983) and HERA (Hutchinson and Thornton 1990). Modifications to the alignment were made with Cameleon (Oxford Molecular). Structural templates were aligned using STAMP (Russell and Barton 1992). Human LFNG and Drosophila Fringe were aligned using ClustalW. LFNG homologs were aligned with the structure-based alignment by use of functionally important residues identified in LFNG and other glycosyltransferases. Ligplot and the literature were used to determine important protein-ligand interactions (Wallace et al. 1995). LFNG was modeled using the Homology module of the Insight II 2000 package (Accelrys). Two conformations of the UDP-sugar donor have been observed in ligand-glycosyltransferase structure complexes. In the retaining glycosyltransferase LgtC (1ga8), the UDP galactose analog is bound with the galactose in a bent position. In the other Protein Data Bank structures, the UDP-sugar ligand is bound fully extended. Persson et al. (2001) ascribed the difference in binding modes to the identity of the enzyme as a retaining or inverting glycosyltransferase. Accordingly, the UDP-sugar donor in the inverting glycosyltranferase LFNG was modeled in the extended conformation.
Web Resources
The URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SCD1 and SCD2)
References
- Aulehla A, Johnson RL (1999) Dynamic expression of lunatic fringe suggests a link between notch signaling and an autonomous cellular oscillator driving somite segmentation. Dev Biol 207:49–61 10.1006/dbio.1998.9164 [DOI] [PubMed] [Google Scholar]
- Bonafé L, Giunta C, Gassner M, Steinmann B, Superti-Furga A (2003) A cluster of autosomal recessive spondylocostal dysostosis caused by three newly identified DLL3 mutations segregating in a small village. Clin Genet 64:28–35 10.1034/j.1399-0004.2003.00085.x [DOI] [PubMed] [Google Scholar]
- Bruckner K, Perez L, Clausen H, Cohen S (2000) Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature 406:411–415 10.1038/35019075 [DOI] [PubMed] [Google Scholar]
- Bulman MP, Kusumi K, Frayling TM, McKeown C, Garrett C, Lander ES, Krumlauf R, Hattersley AT, Ellard S, Turnpenny PD (2000) Mutations in the human Delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat Genet 24:438–441 10.1038/74307 [DOI] [PubMed] [Google Scholar]
- Capdevila J, Izpisua Belmonte JC (2001) Patterning mechanisms controlling vertebrate limb development. Annu Rev Cell Dev Biol 17:87–132 10.1146/annurev.cellbio.17.1.87 [DOI] [PubMed] [Google Scholar]
- Charnock SJ, Davies GJ (1999) Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38:6380–6385 10.1021/bi990270y [DOI] [PubMed] [Google Scholar]
- Chen J, Moloney DJ, Stanley P (2001) Fringe modulation of Jagged1-induced Notch signaling requires the action of β4galactosyltransferase-1. Proc Natl Acad Sci USA 98:13716–13721 10.1073/pnas.241398098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B, Bashirullah A, Dagnino L, Campbell C, Fisher WW, Leow CC, Whiting E, Ryan D, Zinyk D, Boulianne G, Hui CC, Gallie B, Phillips RA, Lipshitz HD, Egan SE (1997) Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nat Genet 16:283–288 10.1038/ng0797-283 [DOI] [PubMed] [Google Scholar]
- Collins JS, Schwartz CE (2002) Detecting polymorphisms and mutations in candidate genes. Am J Hum Genet 71:1251–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia T, Papayannopoulos V, Panin V, Woronoff P, Jiang J, Vogt TF, Irvine KD (2003) Molecular genetic analysis of the glycosyltransferase Fringe in Drosophila. Proc Natl Acad Sci USA 100:6404–6409 10.1073/pnas.1131007100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Henrissat B (1999) The modular structure of cellulases and other carbohydrate-active enzymes: an integrated database approach. In: Ohmiya K, Hayashi K, Sakka K, Kobayashi Y, Karita S, Kimura T (eds) Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers, Tokyo, pp 15–23 [Google Scholar]
- Dahlqvist C, Blokzijl A, Chapman G, Falk A, Dannaeus K, Ibanez CF, Lendahl U (2003) Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development 130:6089–6099 10.1242/dev.00834 [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RS (2002) Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129:1795–1806 [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Henrique D, Harrison SM, Beddington RS (1997) Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development 124:3065–3076 [DOI] [PubMed] [Google Scholar]
- Forsberg H, Crozet F, Brown NA (1998) Waves of mouse Lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. Curr Biol 8:1027–1030 10.1016/S0960-9822(07)00424-1 [DOI] [PubMed] [Google Scholar]
- Fritz TA, Hurley JH, Trinh LB, Shiloach J, Tabak LA (2004) The beginnings of mucin biosynthesis: the crystal structure of UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase-T1. Proc Natl Acad Sci USA 101:15307–15312 10.1073/pnas.0405657101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons BJ, Roach PJ, Hurley TD (2002) Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin. J Mol Biol 319:463–477 10.1016/S0022-2836(02)00305-4 [DOI] [PubMed] [Google Scholar]
- Hahn KL, Johnson J, Beres BJ, Howard S, Wilson-Rawls J (2005) Lunatic fringe null female mice are infertile due to defects in meiotic maturation. Development 132:817–828 10.1242/dev.01601 [DOI] [PubMed] [Google Scholar]
- Haines N, Irvine KD (2003) Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol 4:786–797 [DOI] [PubMed] [Google Scholar]
- Harper JA, Yuan JS, Tan JB, Visan I, Guidos CJ (2003) Notch signaling in development and disease. Clin Genet 64:461–472 10.1046/j.1399-0004.2003.00194.x [DOI] [PubMed] [Google Scholar]
- Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF, Weinmaster G (2000) Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol 2:515–520 10.1038/35019553 [DOI] [PubMed] [Google Scholar]
- Hutchinson EG, Thornton JM (1990) HERA—a program to draw schematic diagrams of protein secondary structures. Proteins 8:203–212 10.1002/prot.340080303 [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y (2003) HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol 194:237–255 10.1002/jcp.10208 [DOI] [PubMed] [Google Scholar]
- Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF (1997) A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development 124:2245–2254 [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–25637 10.1002/bip.360221211 [DOI] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T (1997) Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124:4133–4141 [DOI] [PubMed] [Google Scholar]
- Koch U, Lacombe TA, Holland D, Bowman JL, Cohen BL, Egan SE, Guidos CJ (2001) Subversion of the T/B lineage decision in the thymus by Lunatic Fringe-mediated inhibition of Notch-1. Immunity 15:225–236 10.1016/S1074-7613(01)00189-3 [DOI] [PubMed] [Google Scholar]
- Kusumi K, Mimoto MS, Covello KL, Beddington RS, Krumlauf R, Dunwoodie SL (2004) Dll3 pudgy mutation differentially disrupts dynamic expression of somite genes. Genesis 39:115–121 10.1002/gene.20034 [DOI] [PubMed] [Google Scholar]
- Laufer E, Dahn R, Orozco OE, Yeo CY, Pisenti J, Henrique D, Abbott UK, Fallon JF, Tabin C (1997) Expression of Radical fringe in limb-bud ectoderm regulates apical ectodermal ridge formation. Nature 386:366–373 10.1038/386366a0 [DOI] [PubMed] [Google Scholar]
- Leimeister C, Schumacher N, Gessler M (2003) Expression of Notch pathway genes in the embryonic mouse metanephros suggests a role in proximal tubule development. Gene Expr Patterns 3:595–598 10.1016/S1567-133X(03)00114-5 [DOI] [PubMed] [Google Scholar]
- Lemm I, Ross J (2002) Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol Cell Biol 22:3959–3969 10.1128/MCB.22.12.3959-3969.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Mushegian A (2003) Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci 12:1418–1431 10.1110/ps.0302103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, Vogt TF (2000) Fringe is a glycosyltransferase that modifies Notch. Nature 406:369–375 10.1038/35019000 [DOI] [PubMed] [Google Scholar]
- Morales AV, Yasuda Y, Ish-Horowicz D (2002) Periodic Lunatic fringe expression is controlled during segmentation by a cyclic transcriptional enhancer responsive to Notch signaling. Dev Cell 3:63–74 10.1016/S1534-5807(02)00211-3 [DOI] [PubMed] [Google Scholar]
- Moran JL, Levorse JM, Vogt TF (1999) Limbs move beyond the Radical fringe. Nature 399:742–743 10.1038/21560 [DOI] [PubMed] [Google Scholar]
- Munro S, Freeman M (2000) The notch signalling regulator fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD. Curr Biol 10:813–820 10.1016/S0960-9822(00)00578-9 [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247:536–540 10.1006/jmbi.1995.0159 [DOI] [PubMed] [Google Scholar]
- Mustonen T, Tummers M, Mikami T, Itoh N, Zhang N, Gridley T, Thesleff I (2002) Lunatic fringe, FGF, and BMP regulate the Notch pathway during epithelial morphogenesis of teeth. Dev Biol 248:281–293 10.1006/dbio.2002.0734 [DOI] [PubMed] [Google Scholar]
- Okajima T, Xu A, Lei L, Irvine KD (2005) Chaperone activity of protein O-fucosyltransferase 1 promotes Notch receptor folding. Science 307:1599–1603 10.1126/science.1108995 [DOI] [PubMed] [Google Scholar]
- Persson K, Ly HD, Dieckelmann M, Wakarchuk WW, Withers SG, Strynadka NC (2001) Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat Struct Biol 8:166–175 10.1038/84168 [DOI] [PubMed] [Google Scholar]
- Pourquié O (2001) Vertebrate somitogenesis. Annu Rev Cell Dev Biol 17:311–350 10.1146/annurev.cellbio.17.1.311 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Esteban C, Schwabe JW, De La Pena J, Foys B, Eshelman B, Belmonte JC (1997) Radical fringe positions the apical ectodermal ridge at the dorsoventral boundary of the vertebrate limb. Nature 386:360–366 10.1038/386360a0 [DOI] [PubMed] [Google Scholar]
- Russell RB, Barton GJ (1992) Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins 14:309–323 10.1002/prot.340140216 [DOI] [PubMed] [Google Scholar]
- Saga Y, Hata N, Koseki H, Taketo MM (1997) Mesp2: a novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev 11:1827–1839 [DOI] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Saito T, Kumano K, Takahashi T, Hirai H (2001) Manic fringe and lunatic fringe modify different sites of the Notch2 extracellular region, resulting in different signaling modulation. J Biol Chem 276:25753–25758 10.1074/jbc.M103473200 [DOI] [PubMed] [Google Scholar]
- Sparrow DB, Clements M, Withington SL, Scott AN, Novotny J, Sillence D, Kusumi K, Beddington RS, Dunwoodie SL (2002) Diverse requirements for Notch signalling in mammals. Int J Dev Biol 46:365–374 [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnpenny PD, Whittock N, Duncan J, Dunwoodie S, Kusumi K, Ellard S (2003) Novel mutations in DLL3, a somitogenesis gene encoding a ligand for the Notch signalling pathway, cause a consistent pattern of abnormal vertebral segmentation in spondylocostal dysostosis. J Med Genet 40:333–339 10.1136/jmg.40.5.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tuyl M, Groenman F, Kuliszewski M, Ridsdale R, Wang J, Tibboel D, Post M (2005) Overexpression of lunatic fringe does not affect epithelial cell differentiation in the developing mouse lung. Am J Physiol Lung Cell Mol Physiol 288:L672–L682 10.1152/ajplung.00247.2004 [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8:127–134 [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Kintner C (2003) Modulation of Notch signaling during somitogenesis. Annu Rev Cell Dev Biol 19:367–395 10.1146/annurev.cellbio.19.111301.115434 [DOI] [PubMed] [Google Scholar]
- Whittock NV, Ellard S, Duncan J, de Die-Smulders CE, Vles JS, Turnpenny PD (2004a) Pseudodominant inheritance of spondylocostal dysostosis type 1 caused by two familial delta-like 3 mutations. Clin Genet 66:67–72 10.1111/j.0009-9163.2004.00272.x [DOI] [PubMed] [Google Scholar]
- Whittock NV, Sparrow DB, Wouters MA, Sillence D, Ellard S, Dunwoodie SL, Turnpenny PD (2004b) Mutated MESP2 causes spondylocostal dysostosis in humans. Am J Hum Genet 74:1249–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Gridley T (1998) Defects in somite formation in lunatic fringe-deficient mice. Nature 394:374–377 10.1038/28625 [DOI] [PubMed] [Google Scholar]
- ——— (1999) Reply to “Limbs move beyond the Radical fringe.” Nature 399:744 [DOI] [PubMed] [Google Scholar]
- Zhang N, Martin GV, Kelley MW, Gridley T (2000) A mutation in the Lunatic fringe gene suppresses the effects of a Jagged2 mutation on inner hair cell development in the cochlea. Curr Biol 10:659–662 10.1016/S0960-9822(00)00522-4 [DOI] [PubMed] [Google Scholar]
- Zhang N, Norton CR, Gridley T (2002) Segmentation defects of Notch pathway mutants and absence of a synergistic phenotype in lunatic fringe/radical fringe double mutant mice. Genesis 33:21–28 10.1002/gene.10081 [DOI] [PubMed] [Google Scholar]