Abstract
Hereditary hypophosphatemic rickets with hypercalciuria (HHRH) is a rare disorder of autosomal recessive inheritance that was first described in a large consanguineous Bedouin kindred. HHRH is characterized by the presence of hypophosphatemia secondary to renal phosphate wasting, radiographic and/or histological evidence of rickets, limb deformities, muscle weakness, and bone pain. HHRH is distinct from other forms of hypophosphatemic rickets in that affected individuals present with hypercalciuria due to increased serum 1,25-dihydroxyvitamin D levels and increased intestinal calcium absorption. We performed a genomewide linkage scan combined with homozygosity mapping, using genomic DNA from a large consanguineous Bedouin kindred that included 10 patients who received the diagnosis of HHRH. The disease mapped to a 1.6-Mbp region on chromosome 9q34, which contains SLC34A3, the gene encoding the renal sodium-phosphate cotransporter NaPi-IIc. Nucleotide sequence analysis revealed a homozygous single-nucleotide deletion (c.228delC) in this candidate gene in all individuals affected by HHRH. This mutation is predicted to truncate the NaPi-IIc protein in the first membrane-spanning domain and thus likely results in a complete loss of function of this protein in individuals homozygous for c.228delC. In addition, compound heterozygous missense and deletion mutations were found in three additional unrelated HHRH kindreds, which supports the conclusion that this disease is caused by SLC34A3 mutations affecting both alleles. Individuals of the investigated kindreds who were heterozygous for a SLC34A3 mutation frequently showed hypercalciuria, often in association with mild hypophosphatemia and/or elevations in 1,25-dihydroxyvitamin D levels. We conclude that NaPi-IIc has a key role in the regulation of phosphate homeostasis.
Hereditary hypophosphatemic rickets with hypercalciuria (HHRH [MIM 241530]) is a rare disorder of autosomal recessive inheritance that was first described in a large consanguineous Bedouin kindred (Tieder et al. 1985). This disorder is characterized by the presence of hypophosphatemia secondary to renal phosphate wasting, radiographic and/or histological evidence of rickets, limb deformities, and bone pain (Tieder et al. 1985, 1987; Gazit et al. 1991). Hypophosphatemia in patients with HHRH leads to appropriate stimulation of renal 1α-hydroxylase causing increased synthesis and serum levels of the biologically active vitamin D metabolite 1,25-dihydroxyvitamin D (1,25(OH)2D). As a result, intestinal absorption of calcium is enhanced, resulting in increased urinary calcium excretion (Tieder et al. 1985). Increased serum 1,25(OH)2D levels and hypercalciuria distinguish HHRH from other hypophosphatemic disorders, such as X-linked hypophosphatemia (XLH [MIM 307800]), which is caused by mutations in PHEX (phosphate-regulating gene with homology to endopeptidases on the X chromosome [HYP Consortium 1995; Holm et al. 1997]) and autosomal dominant hypophosphatemic rickets (ADHR [MIM 193100]), which is caused by mutations in the gene encoding fibroblast growth factor 23 (FGF23) (ADHR Consortium 2000).
The kidney is a major regulator of phosphate homeostasis. Three classes of sodium-phosphate cotransporters are expressed in the kidney. The type I cotransporter, NaPi-I (SLC17A1), and the type II cotransporters NaPi-IIa (SLC34A1) and NaPi-IIc (SLC34A3) are expressed predominantly at the renal brush border membrane of the proximal tubules, where the bulk of filtered phosphate is reabsorbed (Segawa et al. 2002; Murer et al. 2000, 2004). The type III transporters, Glvr-1 (SLC20A1) and Ram-1 (SLC20A2), are two ubiquitously expressed retroviral receptors that account for <1% of the mRNAs encoding the different renal sodium-phosphate cotransporters (Tenenhouse et al. 1998) and likely serve as “housekeeping” transporters (Kavanaugh and Kabat 1996). The type II cotransporter NaPi-IIb (SLC34A2) is not expressed in the kidney but is present in several other tissues, particularly the small intestine, where it is involved in the absorption of dietary phosphate (Hilfiker et al. 1998).
Both NaPi-IIa and NaPi-IIc, as well as their respective murine orthologs Npt2a and Npt2c, are regulated in a similar fashion by parathyroid hormone (PTH), FGF23, and dietary phosphate. PTH decreases the abundance of Npt2a protein in the renal brush border membrane (Kempson et al. 1995), and preliminary data have shown that Npt2c is equivalently regulated by this hormone (Miyamoto et al. 2002, 2004). Similarly, renal expression of both cotransporters is reduced by FGF23, a recently identified important regulator of phosphate homeostasis (Bai et al. 2004; Larsson et al. 2004; Shimada et al. 2004a, 2004c). Furthermore, Hyp mice, which have elevated levels of endogenous FGF23 (Liu et al. 2005), show reduced expression of both renal sodium-dependent phosphate cotransporters (Tenenhouse et al. 2003), whereas mice lacking FGF23 show increased expression of Npt2a (Shimada et al. 2004b; Sitara et al. 2004) consistent with the renal phosphate retention seen in these animals. Lastly, phosphate deprivation increases Npt2a and Npt2c expression in the proximal tubule (Ohkido et al. 2003; Madjdpour et al. 2004). Thus, both renal sodium-dependent phosphate cotransporters are important targets for hormonal and dietary regulation of renal tubular phosphate reabsorption.
Homozygous ablation of the murine gene encoding Npt2a (Slc34a1) leads to hypophosphatemia, renal phosphate-wasting, increased serum 1,25(OH)2D levels, and hypercalciuria (Beck et al. 1998). These findings are evident at weaning, but the magnitude of these changes decreases with increasing age. Furthermore, Npt2a-ablated mice lack the typical features of rickets (Beck et al. 1998; Gupta et al. 2001; Tenenhouse 2005). It was, therefore, not too surprising that SLC34A1 mutations were excluded in genomic DNA from affected members of several kindreds with HHRH, including the original Bedouin kindred (Jones et al. 2001; van den Heuvel et al. 2001). Mutations in SLC17A1, another plausible candidate gene that is located in chromosome region 6p23-p21.3, were also excluded in patients with HHRH (Kos 1998). To identify the genetic defect responsible for HHRH, we performed a genomewide search for linkage combined with homozygosity mapping with the use of genomic DNA from a consanguineous Bedouin kindred described elsewhere (Tieder et al. 1985, 1987). A homozygous deletion in SLC34A3, c.228delC, was found in all affected individuals and is predicted to result in a complete loss of function of the encoded sodium-dependent cotransporter NaPi-IIc. Different mutations in the same gene were also found in affected members of three small and unrelated kindreds with HHRH that were described elsewhere (Jones et al. 2001; Sermet-Gaudelus et al. 2001).
Methods
Human Subjects
For the present study, we used genomic DNA samples from the previously described Bedouin kindred (fig. 1A) and kindreds A, C, and D (fig. 2) (Tieder et al. 1985, 1987; Jones et al. 2001; Sermet-Gaudelus et al. 2001). For consistency, all codes identifying individuals are the same as those used by Jones et al. (2001). The studies have been approved by institutional review committees, and informed consent has been obtained for all participants.
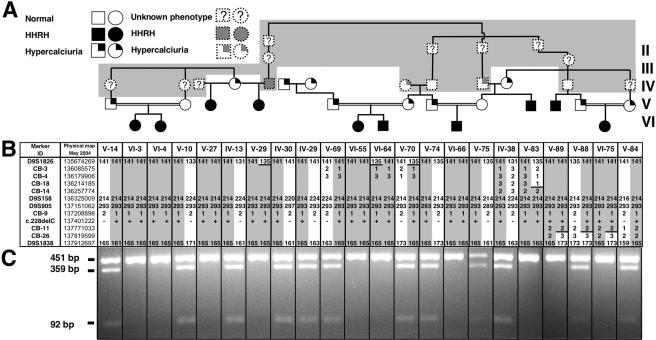
Figure 1.
Genealogical, clinical, and genetic findings in a previously published Bedouin kindred with HHRH. A, Partial pedigree of the Bedouin kindred. Blackened circles or squares indicate individuals who developed rickets during childhood, along with renal phosphate wasting, hypophosphatemia, and hypercalciuria. Unblackened symbols without a question mark (?) indicate individuals who were healthy; the question mark indicates unknown phenotype. A symbol with only the right upper quadrant blackened indicates an individual affected with only hypercalciuria. Samples for individuals with symbols with dashed outlines on the gray background were unavailable for genotyping. B, Haplotypes for chromosome region 9q34 between markers D9S1826 and D9S1838. Alleles for microsatellite markers are designated in bp or are coded; for the c.228delC mutation, “+” and “−” indicate the presence and absence of the deletion, respectively. The haplotype associated with HHRH is depicted by numbers on a gray background; regions that were excluded because of inferred ancestral recombination events are shown by numbers on a white background that are delineated with a horizontal black line. C, Endonuclease digestion to identify the c.228delC mutation. The deletion abolishes an StuI site. Therefore, PCR-amplified DNA from the mutant allele (451 bp) could not be digested with this endonuclease. In contrast, DNA amplified from the wild-type allele yielded 359- and 92-bp fragments (see also table 2).
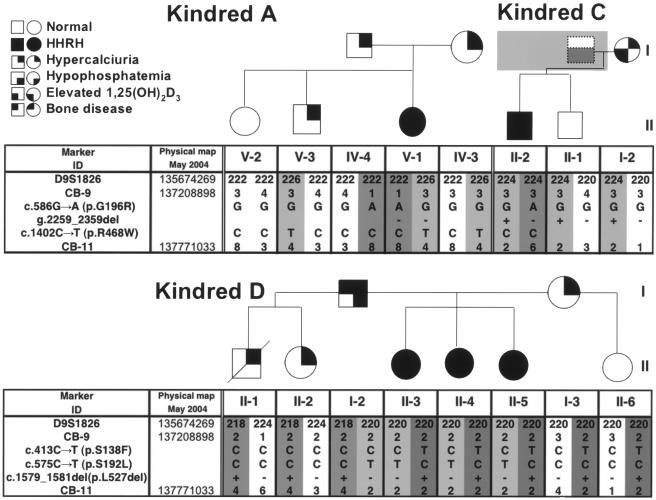
Figure 2.
Genetic findings in three previously published kindreds with HHRH (A, C, and D). In the pedigrees, the use of symbols and shading is the same as in figure 1 unless otherwise indicated. Haplotypes associated with HHRH are depicted by numbers on a dark-gray or light-gray background. Mutations were identified by nucleotide sequence analysis and were confirmed by restriction endonuclease digestion of the appropriate PCR products when possible (see table 2). Note that, in kindred D, c.1579_1581del cannot be unambiguously phased.
Search for the Genetic Locus of HHRH
A genomewide scan was performed on samples from 63 individuals from the Bedouin kindred, including 10 individuals with HHRH. A set of 389 microsatellite markers, described by Mira et al. (2003), with an average intermarker distance of 9.1 cM was used. To follow up suggestive regions of linkage from the genomewide scan, 13 additional markers from chromosomes 1 and 9 were selected from the deCODE Genetics (Kong et al. 2002) or the Marshfield (Marshfield Center for Medical Genetics Web site) genetic maps. Genotyping was performed at the McGill University and Genome Quebec Innovation Centre, as described elsewhere (Mira et al. 2003).
To further define the candidate region on chromosome 9, seven new markers were developed on the basis of the May 2004 freeze of the UCSC Genome Browser database (Karolchik et al. 2003), as described elsewhere (Bastepe et al. 1999; Bastepe and Jüppner 2000). These markers were genotyped by PCR amplification of 50 ng of DNA in 96-well plates (Corning) in a final volume of 10 μl/well by use of 32P-labeled forward oligonucleotide primers and the Qiagen PCR kit (see table 1 for primer sequences and thermal cycler conditions). After the addition of 10 μl of 2× loading dye, 5 μl of each PCR product were separated on a 5% Long Ranger (Cambrex)/7 M urea gel, as described elsewhere (Bastepe et al. 2001). After the gel was dried and exposed to Kodak MR film for 3–18 h, the films were developed and the alleles were scored.
Table 1.
New Microsatellite Markers Used for Linkage Analysis[Note]
|
Primer Sequence(5′→3′) |
||||
| Marker | Repeat | Forward | Reverse | Position onChromosome 9a |
| CB-3 | GT | CGAAACTCAACAAGCAGCAG | ACACATGCACACAACACG | 136085575 |
| CB-4 | GACAG | TTGCACCACTGTACCTGTA | CGTTGGGGGGAAGTATTTG | 136179906 |
| CB-9 | AAAT | GGGCAACAAGAGTGAAACTAC | GTGGTTGTTAGGAGTCTGGA | 137208898 |
| CB-11 | AAT | CCAGGAGGTGGAGATTGGA | CCTACCACCCTAGCCAACAA | 137771033 |
| CB-14 | ATAGA | GGTGACAGAGCAAGACTTGACTAA | GTTTGAACCCAGGAATTGGA | 136257774 |
| CB-18 | AC | GTCACCCCAACCTCAAGCTA | TCCATACTCACCCCCATACAA | 136214185 |
| CB-26 | CAAA | ATTAGTCGGATGTGGTGACG | CAAATTAGGCCCCATTCTCA | 135762525 |
Note.— For PCR in a final volume of 50 μl, the thermal cycler conditions (Mastercycler gradient) after an initial denaturation at 94°C for 5 min were 40 cycles at 94°C for 50 s, 58°C for 50 s, and 72°C for 50 s, with a final extension at 72°C for 10 min.
Physical map position on chromosome 9, UCSC Genome Browser, May 2004 release.
Nucleotide Sequence Analysis
SLC34A3 spans ∼5 kb of genomic DNA. The entire SLC34A3 gene was amplified by PCR (Qiagen PCR kit) in four overlapping fragments by use of primer pairs 17 and 18, 19 and 20, 21 and 22, and 23 and 24 (see table A1 for primers and thermal cycler conditions). The PCR products were purified using spin columns (Qiagen), and nucleotide sequence analysis was performed using 20 ng/100 bp DNA and nested forward or reverse primers (100 ng each) at the Massachusetts General Hospital DNA Sequencing Core Facility.
SNP Analysis in Controls
DNA samples were obtained from the Harvard Partners Center for Genetics and Genomics (Cambridge, MA) (84% white, 8% black, 5% Asian, and 3% Hispanic) (n=95). Bedouin, Turkish, and Australian Aboriginal controls were not available to us; thus, we did not have the most-appropriate controls for all kindreds. When possible, sequence variations were analyzed in these control samples by restriction endonuclease digestion and subsequent 2% agarose/TAE gel electrophoresis, by use of appropriate PCR primers that amplify across the SNP. The region encompassing the 101-bp deletion (g.2259_2359del) was amplified with primers 21 and 22, followed by nested PCR with primers 173 and 174 to generate a wild-type 480-bp product and a mutant 380-bp product (see tables 2 and A1 for primers and thermal cycler conditions).
Table 2.
Amplification and Restriction Endonuclease Digestion to Confirm Mutations/SNPs[Note]
|
Digestion Fragmentsa(bp) |
||||||||
| Mutation/SNP | Exon | Primers | AmpliconSize(bp) | Enzyme | No. ofControlDNAs | Genotype 11 | Genotype 12 | Genotype 22 |
| c.1402C→T (p.R468W) | 13 | 52 and 53 | 336 | BtsI | 95 | CC (95): 283, 53 | CT (0): 336, 283, 53 | TT (0): 336 |
| c.1579_1581del (p.L527del) | 13 | 52 and 53 | 336 | BseRI | 86 | Wt (86): 223, 113 | Het (0): 336, 223, 113 | Mut (0): 336 |
| c.228delC | 4 | 32 and 33 | 451 | StuI | 94 | Wt (94): 359, 92 | Het (0): 451, 359, 92 | Mut (0): 451 |
| c.413C→T (p.S138F) | 5 | 32 and 33 | 451 | BmgBI | 95 | CC (95): 352, 99 | CT (0): 451, 352, 99 | TT (0): 451 |
| c.1538T→A (p.V513E) | 13 | 52 and 53 | 336 | AluI | 48 | TT (29): 196, 140 | TA (19): 336, 196, 140 | AA (0): 336 |
Note.— The region of SLC34A3 flanking the mutation/SNP was amplified by PCR in a final volume of 20 μl by use of the indicated primers (table A1) (Qiagen PCR kit); the thermal cycler conditions after an initial denaturation at 94°C for 5 min were 35 cycles at 94°C for 1 min, 60°C or 65°C for 1 min, and 72°C for 3 min, followed by a final extension at 72°C for 10 min. PCR products were then subjected to restriction endonuclease digestion to obtain the indicated genotype-specific fragments on 2% agarose/TAE gel electrophoresis.
Genotypes are followed by the respective fragment(s), given in bp, and the number of controls with a particular genotype is in parentheses. Het = heterozygote; Mut = mutant; Wt = wild type.
Multiplex PCR assays for the analysis of c.586G→A (p.G196R) and c.575C→T (p.S192L) were designed and performed at the Harvard Partners Center for Genetics and Genomics by use of Sequenom SpectroDESIGNER software and sequence containing the SNP and 100 bp on each side of the SNP (Lazarus et al. 2002). Single-primer extension across the SNP was performed as described elsewhere (Lazarus et al. 2002), and extension products were then spotted onto a 384-well spectroCHIP before being flown in the matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) mass spectrometer for analysis (see table A2 for primer information and experimental details).
Statistical Analysis
Since the mode of inheritance of HHRH was not definitively established, we used the maximized maximum likelihood score (MMLS) screening method of Greenberg et al. (1998) for analysis of the genomewide scan data and 13 additional markers on chromosome 9. In brief, two models were analyzed: a recessive model with 50% penetrance and a dominant model with 50% penetrance. The MMLS is the maximum LOD score obtained using one of the two models. Singlepoint parametric LOD scores at different recombination fractions were calculated using a pedigree of 294 individuals from the Bedouin kindred that connected all 10 patients with HHRH and contained 14 inbreeding/marriage loops. Linkage analyses were performed using the MLINK-like option of Superlink version 1.4 (Fishelson and Geiger 2002). Marker allele frequencies were estimated by counting all genotyped individuals. The disease-allele frequency was arbitrarily set at 0.1 for the recessive model and at 0.01 for the dominant model (Pal et al. 2001), since no estimates of the disease prevalence were available. To adjust for multiple testing with these two models, the LOD scores should be compared with significance thresholds that are 0.3 LOD units higher than the standard ones.
Results
Clinical Information
The Bedouin kindred, a subset of which is shown in figure 1A, has been well characterized elsewhere (Tieder et al. 1985, 1987; Gazit et al. 1991).
The parents of the affected child in kindred A (of Turkish origin) were initially reported to be second cousins (Jones et al. 2001; Sermet-Gaudelus et al. 2001); however, additional genealogic information did not confirm this. The affected female (V-1) was first evaluated at age 7 years and 7 mo because of genua vara—which ultimately required surgical correction—growth delay, and radiographical signs of rickets, but she had no fractures. At the time of presentation, the patient had nephrocalcinosis, as determined by renal ultrasound, in the absence of glycosuria, proteinuria, and acidosis; her remaining serum and urinary chemistries revealed hypophosphatemia due to renal phosphate wasting, hypercalciuria, and elevated 1,25(OH)2D levels (table 3). Bone mineral density (BMD) evaluated at age 13 years was reduced (total hip z score −3.8; lumbar spine z score −2.4). Since the initial reports describing family A (Jones et al. 2001; Sermet-Gaudelus et al. 2001), two additional younger siblings of V-1 have been evaluated: V-2 had normal serum and urine chemistries, and V-3 also had normal serum chemistry but had an increased calcium-to-creatinine ratio of 1.44 mmol/mmol (upper limit of normal for children younger than 2 years old is 1.0 mmol/mmol) and slightly reduced tubular reabsorption of phosphate (TRP) (see table 3). Their father (IV-4) and mother (IV-3) had, when first evaluated, hypophosphatemia and elevated 1,25(OH)2D levels in the setting of vitamin D deficiency, which did not persist after vitamin D repletion. However, the mean urinary calcium-tocreatinine ratio remained above the normal range throughout the observation period of 3 years for both parents (0.23–0.74 mmol/mmol; upper limit of normal for adults 0.4). Intact PTH was in the low- to mid-normal range for all individuals of kindred A (data not shown).
Table 3.
Clinical Data for Kindreds A, C, and D[Note]
| Kindredand Individual | Age atMeasurement(years) | SerumPhosphatea(mmol/liter) | TRPb(%) | UrineCalcium/Creatininea(mmol/mmol) | Serum1,25(OH)2Da |
| Kindred A: | |||||
| IV-3 | 32–35 | .72–1.04 (.85–1.20) | 91–96 | .23–.64 (<.4) | 46–140 (15–60) pg/ml |
| IV-4 | 31–34 | .68–.94 (.85–1.20) | 81–88 | .38–.74 (<.4) | 22–66 (15–60) pg/ml |
| V-1 | 8–13 | .71–.86 (1.20–1.70) | 70–75 | .95–1.4 (<.4) | 73–103 (30–70) pg/ml |
| V-2 | 6 | 1.35 (1.20–1.70) | 91 | .33 (<.4) | 40 (30–70) pg/ml |
| V-3 | 1.6 | 1.85 (1.50–1.80) | 80 | 1.44 (<1.0) | 38 (30–120) pg/ml |
| Kindred C: | |||||
| I-1 | 48 | .42 (.81–1.45) | 78 | .14 (<.4) | 72 (20–65) pg/ml |
| I-2 | 45 | 1.03 (.81–1.45) | 88 | .51 (<.4) | 68 (20–65) pg/ml |
| II-1 | 15 | 1.00 (.97–1.61) | 93 | .11 (<.4) | ND |
| II-2 | 14 | .77 (.97–1.61) | 82 | .64 (<.4) | 97 (20–65) pg/ml |
| Kindred D: | |||||
| I-2 | 48 | .74–.89 (.79–1.37) | ND | .50 (<.4) | 96 (38–162) pmol/liter |
| I-3 | 36 | .97 (.79–1.37) | ND | 1.00 (<.4) | 79 (38–162) pmol/liter |
| II-1 | 19 | .90 (.79–1.37) | ND | .66 (<.4) | 68 (38–162) pmol/liter |
| II-2 | 17 | .80 (.79–1.37) | ND | .61 (<.4) | 94 (38–162) pmol/liter |
| II-3 | 14 | .75–.93 (.95–1.85) | 80 | .57 (<.4) | 240 (38–162) pmol/liter |
| II-4 | 11 | .75–.91 (.95–1.85) | 89 | 1.03 (<.4) | 344 (38–162) pmol/liter |
| II-5 | 9 | .77–1.13 (.95–1.85) | 87 | .79 (<.4) | >566 (38–162) pmol/liter |
| II-6 | 16 | .96 (.79–1.37) | ND | .24 (<.4) | 68 (38–162) pmol/liter |
Note.— The IDs of individuals with compound heterozygous SLC34A3 mutations are indicated in bold italics. ND = not determined.
Given in parentheses are age-dependent normal values.
Calculated as 100 × [1 − (urine phosphate × serum creatinine)/serum phosphate × urine creatinine)]; normal range is 85%–95%.
In kindred C, the affected individual II-2 presented at age 14 years with gradual onset of bilateral knee and ankle pain. At age 13 years, he passed a calcium oxalate kidney stone. There were bilateral genua valga on initial examination, and he walked with a limp. He was found to have hypophosphatemia, an elevated urinary calcium-to-creatinine ratio, and an increased 1,25(OH)2D level (see table 3), whereas the concentration of 25-hydroxyvitamin D and midmolecule PTH were within normal limits (data not shown). His bone turnover was accelerated, as indicated by an increased urinary excretion of N-telopeptide (805 nmol bovine collagen equivalents/mmol creatinine; normal <86) and pyridinoline (451 nmol/mmol creatinine; normal 17–410), a urinary deoxypyridinoline excretion at the upper end of normal (115 nmol/mmol creatinine; normal 2–118), and an increased serum alkaline phosphatase (910 U/liter; normal <350). There was no evidence of hyperoxaluria, proteinuria, or acidosis. Radiographic studies were remarkable for osteopenia, an osteochondroma emanating from the left fibula, and subtle rachitic changes—that is, mild fraying of the metaphyses and epiphyses. The patient’s BMD was reduced to 0.367 g/cm2 at the hip (z score −4.7; t score −4.41) and to 0.447 g/cm2 at the lumbar spine (z score −3.7; t score −5.26). With oral phosphate supplements, his lumbar spine BMD improved to 0.977 g/cm2 (z score –1.0; t score −1.0) by age 20 years. A renal ultrasound at age 21 years showed no abnormalities. His mother (I-2) is of Irish, French, and German descent and had normal biochemistry, with the exception of slightly elevated 1,25(OH)2D levels and an increased urinary calcium-to-creatinine ratio. His father (I-1) is of Scottish descent and had hypophosphatemia and elevated 1,25(OH)2D levels but normal urinary calcium excretion (see table 3). There was no history of consanguinity in kindred C.
In kindred D, which is of mixed white and Aboriginal origin from Australia, three individuals are affected by HHRH (II-3, II-4, and II-5). II-5, the index patient in the report by Jones et al. (2001), had severe rickets with bowing, which required stapling of the lower femoral epiphyses. Her bone histology showed severe osteomalacia with increased cellularity. The sisters (II-3 and II-4) had milder bone disease. All three siblings met biochemical criteria for HHRH, including hypophosphatemia, hypercalciuria, and elevated 1,25(OH)2D levels (see table 3). Thus, the diagnoses for these individuals have changed since the initial publication (Jones et al. 2001). Lumbar BMD was 0.846 g/cm2 for II-4 (z score −3.0) and 0.617 g/cm2 for II-5 (z score −1.0). Their father (I-2) had abnormal bone histology, with increased osteoid surfaces and increased rate of bone formation. He had hypercalciuria and hypophosphatemia but normal 1,25(OH)2D levels. Their mother (I-3) also had abnormal bone histology, with an increased rate of bone formation, but normal osteoid surfaces. She had mild genua valga and hypercalciuria but normal serum phosphate and 1,25(OH)2D levels. One half-brother (II-1) died from causes unrelated to bone disease. The half-sister (II-2) had hypercalciuria but no other abnormalities. II-6 was phenotypically normal. There is no report of kidney stones in kindred D, nor were abnormalities seen on renal ultrasonography for II-3, II-4, or II-5. There was no known consanguinity in kindred D.
Genetic Analysis
An MMLS screening model was used to analyze the genomewide scan data on 63 individuals in the Bedouin kindred who were connected in a pedigree of 294 individuals with 14 inbreeding/marriage loops. The highest LOD score of 3.34 was obtained at marker D9S1838 by use of the recessive model (table 4). D9S1838 was the most telomeric marker on chromosome 9q in the initial screen. Additional markers that flank D9S1838 were identified (D9S158, D9S905, and D9S2168), but these markers did not change the evidence for or against linkage and could not narrow the candidate region. However, examination of the genotypes for the 10 patients with documented HHRH showed that all patients, with the exception of V-89 and VI-75, were homozygous for D9S1838 (fig. 1B). At marker D9S1826, V-89 and VI-75 were homozygous, but V-29, VI-64, and V-83 were heterozygous. Thus, under the assumption that there existed one disease-causing mutation in a homozygous chromosomal segment inherited identically by descent, the HHRH locus was predicted to be between D9S1826 and D9S1838.
Table 4.
Maximal LOD Scores for Each Chromosome Derived from the Genomewide Scan with Recessive and Dominant Disease Models[Note]
|
Dominant Model |
Recessive Model |
||||
| Chromosomeand Position (cM) | Marker | LOD | θ | LOD | θ |
| 1: | |||||
| 179.98 | D1S1625 | .918 | 0 | 1.160 | 0 |
| 218.46 | D1S1678 | 1.667 | 0 | .513 | .1 |
| 2: | |||||
| 47.97 | D2S405 | .127 | .2 | .218 | .15 |
| 99.41 | D2S1777 | .708 | 0 | −.172 | .2 |
| 3: | |||||
| 89.91 | D3S4542 | .986 | 0 | 1.702 | 0 |
| 4: | |||||
| 33.96 | D4S3340 | −.412 | .2 | .655 | 0 |
| 60.16 | D4S1627 | .155 | .2 | −.515 | .2 |
| 5: | |||||
| 168.49 | D5S1475 | .840 | 0 | .887 | .1 |
| 6: | |||||
| 92.85 | D6S1270 | 1.313 | 0 | .040 | .2 |
| 102.81 | D6S1056 | −.063 | 0 | 1.480 | 0 |
| 7: | |||||
| 17.17 | D7S2200 | .901 | 0 | .112 | .2 |
| 103.63 | D7S1813 | .562 | 0 | .273 | .1 |
| 8: | |||||
| 26.43 | D8S1106 | .400 | 0 | −.484 | .2 |
| 94.17 | D8S1123 | −.159 | .2 | .005 | .15 |
| 9: | |||||
| 163.84 | D9S1838 | 1.834 | 0 | 3.341 | 0 |
| 10: | |||||
| 109.98 | D10S1739 | 1.322 | 0 | .538 | .075 |
| 117.42 | D10S677 | 1.489 | 0 | .136 | .15 |
| 11: | |||||
| 33.02 | ATA34E08 | −.141 | .2 | .409 | .2 |
| 123.00 | D11S4464 | .975 | 0 | .267 | .15 |
| 12: | |||||
| 56.38 | D12S1663 | 1.150 | 0 | .247 | .15 |
| 13: | |||||
| 63.90 | D13S317 | .261 | .2 | −.089 | .2 |
| 14: | |||||
| 55.82 | D14S587 | .753 | 0 | −.059 | .2 |
| 105.53 | D14S617 | −.092 | .2 | .724 | .025 |
| 15: | |||||
| 52.33 | D15S643 | .202 | .1 | −.003 | .2 |
| 60.17 | D15S1507 | −.213 | .1 | .631 | .1 |
| 16: | |||||
| 8.16 | D16S2622 | .225 | .2 | .692 | .025 |
| 17: | |||||
| .63 | D17S1308 | −.172 | .2 | .097 | .15 |
| 82.00 | D17S1290 | .013 | .2 | .010 | .2 |
| 18: | |||||
| 96.48 | D18S1270 | .728 | 0 | .773 | .05 |
| 19: | |||||
| 68.08 | D19S178 | .417 | .15 | .329 | .1 |
| 92.56 | D19S418 | .591 | .05 | .115 | .2 |
| 20: | |||||
| 21.15 | D20S115 | .628 | 0 | −.115 | .2 |
| 42.28 | D20S471 | .089 | .2 | .123 | .2 |
| 21: | |||||
| 36.77 | D21S1440 | .419 | 0 | .703 | .075 |
| 22: | |||||
| 24.74 | D22S1167 | .503 | .1 | −.047 | .2 |
| 32.39 | D22S685 | −.155 | .2 | .173 | .2 |
| X: | |||||
| 52.63 | DXS1068 | .379 | .075 | 1.027 | .001 |
| 165.11 | DXS9908 | .419 | .1 | −.120 | .2 |
Note.— See the “Methods” section for a detailed description of the search for the genetic locus of HHRH. LOD scores are not corrected for multiple testing.
To refine the boundaries of this locus, publicly available sequence databases were searched to identify seven new microsatellite markers, which were used to genotype a subset of DNA samples (fig. 1B and table 1). For marker CB-9, all 10 patients were homozygous for the same allele, whereas all genotyped parents and grandparents were heterozygous. Patient V-83 was heterozygous for three markers (CB-3, CB-4, and CB-18) centromeric to CB-9, and patients V-89 and VI-75 were heterozygous for a marker (CB-26) telomeric to CB-9. Thus, under the assumption that heterozygosity represents ancestral recombinants, the candidate interval could be reduced to a 1.6-Mbp region between CB-18 and CB-26. The March 2004 release of the human genome sequence from the National Center for Biotechnology Information (NCBI) revealed 70 known or predicted genes in the linked interval, including a likely candidate gene, SLC34A3. This gene encodes the renal sodium-dependent phosphate cotransporter NaPi-IIc, which is involved in renal phosphate handling (Segawa et al. 2002) and spans ∼5 kb on contig NT_024000.16, comprising 13 exons, which give rise to a 2.1-kb mRNA.
The entire SLC34A3 gene was PCR-amplified in four overlapping fragments to search for mutations in the DNA from patient VI-75 (see table A1); these fragments were submitted for sequence analysis. A homozygous single-nucleotide deletion was identified that affects cytosine at position 228 of the cDNA sequence (c.228delC; numbering is according to cDNA [GenBank accession number NM_080877.1] and starts at the initiator ATG). In addition, three homozygous nucleotide changes were identified in patient VI-75. Two of these SNPs, c.558G→A and c.757T→C, are silent substitutions at codons Q186 and L253, respectively, whereas the third represents a nonsynonymous common polymorphism, c.1538T→A (p.V513E) (tables 2 and 5; the SNPs presented in table 5 were submitted to the NCBI dbSNP database.)
Table 5.
SLC34A3 Variations[Note]
| Region and Change inNucleotide (Amino Acid) | Region | Primers |
| Coding region: | ||
| c.73C/T (p.L25) | Exon 2 | 17 and 18 |
| c.399G/A (p.Q133) | Exon 5 | 32 and 33 |
| c.558G/A (p.Q186) | Exon 6 | 57 and 87 |
| c.756G/A (p.Q252) | Exon 7 | 20 and 34 |
| c.757T/C (p.L253) | Exon 8 | 20 and 34 |
| c.1538T→A (p.V513E) | Exon 13 | 52 and 53 |
| Noncoding region: | ||
| g.−804G/A | 5′ UTR | 17 and 18 |
| g.2704A/T | Intron 10 | 21 and 22 |
| g.3701G/C | Intron 12 | 23 and 24 |
| g.3736C/T | Intron 12 | 23 and 24 |
Note.— SLC34A3 was amplified in four overlapping fragments by PCR (for primers, see table A1) by use of DNA from VI-75, an affected individual from the previously published Bedouin kindred (Tieder et al. 1985, 1987) or DNA from three additional kindreds (A, C, and D) (Jones et al. 2001). The PCR products were then subjected to nucleotide sequence analysis; the listed polymorphisms were found in at least two independent sequencing reactions and were confirmed by restriction endonuclease digestion. Note that c.756G/A (p.Q252) (heterozygous for adenine in individuals IV-4 and V-1 of kindred A) affects a splice donor site and thus may lead to abnormal splicing.
The c.228delC deletion causes a shift in the ORF after codon 76, which is predicted to result in the translation of a novel sequence of 74 aa that is unrelated to NaPi-IIc and is then followed by a termination codon at codon 151. The predicted protein thus lacks all eight membrane-spanning domains as well as the C-terminal intracellular tail of NaPi-IIc, predicting a complete loss of function of this sodium-dependent phosphate cotransporter (fig. 3). The c.228delC mutation removes a recognition site for the endonuclease StuI, making it possible to readily assay the presence or absence of the identified mutation (see table 2). Using this assay, DNA from all 22 individuals was analyzed (fig. 1C). All patients affected by HHRH were homozygous for c.228delC, whereas all available parents and grandparents were heterozygous for this deletion. The c.228delC deletion was not found in 188 alleles from 94 controls (table 2).
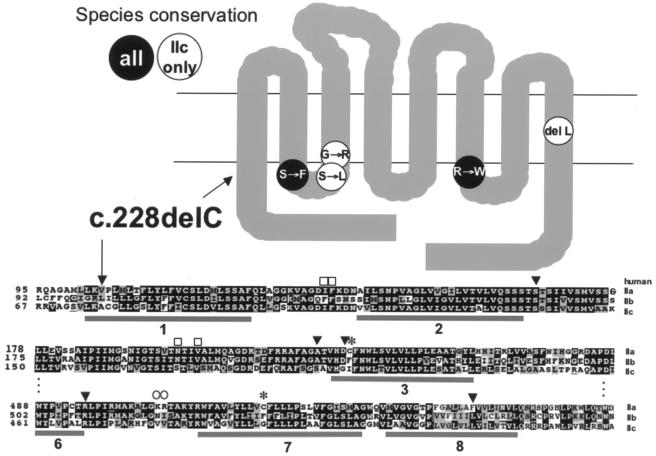
Figure 3.
Location and type of SLC34A3 mutations identified in four kindreds with HHRH. Upper panel, Predicted NaPi-IIc, with eight membrane-spanning domains. The approximate locations of point mutations are shown in black letters on a white background if they affect identical amino acid residues in the NaPi-IIc molecules of human, mouse, rat, and dog; they are depicted in white letters on a black background if they affect amino acid residues that are identical in all type II sodium-phosphate cotransporters. The c.228delC deletion (black arrow) is predicted to lead to truncation of NaPi-IIc within the first membrane-spanning domain. Lower panel, Partial sequence of human NaPi types IIa, IIb, and IIc. Species alignments were performed using Clustal W1.8 (Higgins et al. 1996). Amino acid residues are shown in white letters on black if they are identical in several sodium-phosphate cotransporter types (type IIa: human, rat, mouse, dog, chimpanzee, and chicken; type IIb: human, rat, mouse, dog, chimpanzee, and chicken; type IIc: human, rat, mouse, and dog). Amino acid residues are shown in black letters on gray if they are identical in all listed species for one sodium-phosphate cotransporter type. Amino acid residues not conserved among species for one sodium-phosphate cotransporter type are shown in black letters on white. Gray horizontal bars indicate the predicted location of membrane-spanning domains in NaPi-IIc (see Segawa et al. 2002). Squares above the IIa sequence indicate amino acid residues identified by site-directed mutagenesis in mouse and rat NaPi-IIa that are important for transport function, circles indicate residues important for PTH-mediated internalization, and asterisks (*) indicate cysteine residues thought to form intramolecular cysteine bonds. Black arrowheads indicate residues affected by point mutations. Two of these residues, S192 and G196, when mutated in the corresponding position of mouse Npt2c to the respective residues of mouse Npt2a, confer 3:1 sodium:phosphate stoichiometry and electrogenicity to Npt2c (Bacconi et al. 2005). See Web Resources for GenBank accession numbers of sequences.
We next searched for SLC34A3 mutations in three unrelated, previously reported kindreds with at least one member affected by HHRH (Jones et al. 2001; Sermet-Gaudelus et al. 2001). As shown in figure 2, compound heterozygous missense and frameshift mutations were identified in the affected female V-1 of kindred A. Missense mutation c.586G→A (p.G196R) was paternally inherited, whereas the other missense mutation, c.1402C→T (p.R468W), was maternally inherited. These two nucleotide changes were absent in the unaffected sibling and were not identified in 160 and 190 control alleles. The hypercalciuric brother (V-3) had inherited the maternal allele with the missense mutation c.1402C→T (p.R468W). In kindred C, the affected male (II-2) inherited one of the missense mutations identified in kindred A, c.586G→A (p.G196R), whereas a 101-bp deletion within intron 9, g.2259_2359del, was inherited from his mother. Neither mutation was found in 160 control alleles. In kindred D, the female who had first received the diagnosis of HHRH (II-5) has one maternally inherited mutation, c.413C→T (p.S138F), and one paternally inherited mutation, c.575C→T (p.S192L). Parental origin of a third mutation, c.1579_1581del (p.L527del), could not be confidently assigned. These three mutations were not found in 190, 160, and 172 control chromosomes. The sisters II-3 and II-4, who were classified as having HHRH on the basis of more-recent findings, were genotypically identical to II-5 with respect to the SLC34A3 mutations and flanking markers (see fig. 2 and table 3).
Discussion
Evidence That NaPi-IIc Mutations Cause HHRH
A genomewide linkage scan combined with a homozygosity mapping approach in an extended Bedouin kindred was used to map the genetic defect responsible for HHRH to a 1.6-Mbp interval in chromosome region 9q34. SLC34A3, a plausible candidate gene in this region, was sequenced, and all individuals with HHRH were found to be homozygous for a 1-nt deletion, c.228delC. This putative mutation is predicted to result in a shift in the ORF after amino acid residue 76 and to produce a presumably nonfunctional NaPi-IIc protein that is truncated within the first membrane-spanning domain (Segawa et al. 2002). Consistent with an important role for NaPi-IIc in the pathogenesis of HHRH, several compound heterozygous missense mutations or deletions were found in affected individuals from three additional unrelated kindreds but were not present in healthy controls. In kindred D, three different SLC34A3 variations were identified in the patients with HHRH. It remains to be determined, however, whether the identified amino acid changes contribute to the phenotype in each of these patients, since ethnically matched controls were not always available to exclude the possibility that the nucleotide changes represent rare polymorphisms. We would, however, conclude from the findings above that the mutations affecting both SLC34A3 alleles, which presumably lead to complete or partial loss of a functional NaPi-IIc protein, cause HHRH in our kindreds.
Physiological Role for NaPi-IIc Defined by SLC34A3 Mutations in Patients with HHRH
SLC34A3 encodes NaPi-IIc, a type II electroneutral, sodium-dependent phosphate transporter (Segawa et al. 2002), which shares significant amino acid identity with NaPi-IIa (54% between the two human cotransporters). Both cotransporters are expressed predominantly in the brush border membrane of proximal renal tubular cells (Murer et al. 2004). NaPi-IIa was previously thought to be the most important determinant of renal phosphate handling and thus overall phosphate homeostasis. Consistent with this conclusion, it was shown that the relative expression of Npt2a is an order of magnitude higher than that of Npt2c in kidneys of wild-type mice and rats maintained on a normal-phosphate diet (Segawa et al. 2002; Ohkido et al. 2003). Furthermore, on the basis of hybrid depletion experiments of rat kidney poly-A+ RNA in Xenopus oocytes, Npt2c was thought to be rate-limiting only during weaning and to contribute little to total sodium-dependent phosphate cotransport in adult rats (Segawa et al. 2002). On the basis of these data, NaPi-IIc was predicted to contribute 15%–20% to total proximal TRP (Segawa et al. 2002; Ohkido et al. 2003). However, the phenotype of Npt2a-null mice improves with age (Beck et al. 1998), suggesting that Npt2c or other renal sodium-dependent phosphate cotransporters can rescue these mice from more-severe hypophosphatemia and thus more-prominent and lifelong bone changes.
Npt2c expression in the kidneys of Npt2a-null mice is three- to fourfold higher than that of wild-type littermates (Tenenhouse et al. 2003). Furthermore, normal animals exhibit a more robust increase in Npt2c than in Npt2a in response to phosphate deprivation (Ohkido et al. 2003; Madjdpour et al. 2004). Thus, it is conceivable that Npt2c can compensate, at least partially, for the lack of Npt2a (Tenenhouse et al. 2003). Npt2c expression is also hormonally regulated. For example, it is reduced by recombinant FGF23 in wild-type mice and in Hyp mice, which display elevated levels of endogenous FGF23 (Tenenhouse et al. 2003, Liu et al. 2005), and there is preliminary evidence to suggest that Npt2c is downregulated after injection of synthetic PTH in wild-type mice (Miyamoto et al. 2004). NaPi-IIc thus appears to be critically involved in regulating phosphate homeostasis. Our finding that mutations in SLC34A3 cause HHRH now provides further evidence for this conclusion and shows that NaPi-IIc, despite its low expression levels in the kidney, plays a critical and nonredundant role in renal phosphate conservation in humans. Significant but previously unappreciated differences in the temporospacial regulation of its expression and/or direct or indirect interaction or functional interdependence with other cotransporters and regulatory and/or scaffolding proteins could explain why the importance of NaPi-IIc in normal phosphate homeostasis has remained undetected so far.
Npt2a haploinsufficiency leads to a blunted adaptive response to a low-phosphate diet in mice (Tenenhouse 2005), and dominant-negative NaPi-IIa mutations have been implicated in osteoporosis and nephrolithiasis in humans (Prié et al. 2002). Hypercalciuria, furthermore, leads to the development of nephrocalcinosis in homozygous Npt2a-ablated mice (Chau et al. 2003; Tenenhouse et al. 2004; Tenenhouse 2005), and both hypercalciuria and nephrocalcinosis can be prevented by concomitant ablation of the 25-hydroxyvitamin D3 1α-hydroxylase gene (Cyp27b1) or by supplementing Npt2adeficient animals with oral phosphate (Tenenhouse et al. 2004). Despite similar biochemical changes seen in heterozygous and homozygous carriers of SLC34A3 mutations, it is surprising that hypercalciuria does not lead to nephrolithiasis or nephrocalcinosis in most patients with HHRH, unless the affected individuals are inappropriately treated with vitamin D analogs (Chen et al. 1989; Tieder et al. 1993). The index patient of kindred A, however, presented with nephrocalcinosis, which resolved after treatment with phosphate supplements, and the index patient of kindred C passed a calcium oxalate stone 1 year before the diagnosis of HHRH was established. Thus, the involvement of NaPi-IIc in the pathogenesis of nephrolithiasis and nephrocalcinosis remains to be clarified. Furthermore, analysis of samples from the general population is needed to determine whether common variants of SLC34A3 can affect urinary calcium excretion.
Mice homozygous for the ablation of Npt2a do not develop rickets (Beck et al. 1998). In fact, these animals have an increased BMD compared with that of wild-type littermates (Tenenhouse 2005), which is thought to be related to increased trabecular thickness and decreased number and activity of osteoclasts, which express Npt2a (Gupta et al. 2001). This is in contrast to patients with HHRH, who usually develop bone changes consistent with rickets. Thus, NaPi-IIc may affect—either directly or indirectly via hypophosphatemia and elevated 1,25(OH)2D levels—skeletal function in a manner that is fundamentally different from that of NaPi-IIa. The basis for these differences, however, is not understood at this time.
Presumed Mechanism of the Identified SLC34A3 Mutations
Although the homozygous deletion of nucleotide 228 is predicted to result in a truncated and presumably nonfunctional protein (see fig. 3), the mechanism(s) through which the identified compound heterozygous mutations lead to a complete or partial loss of function is less apparent. However, the five identified missense and in-frame deletion mutations were not found in healthy controls, and all involve conserved amino acid residues that are located either in intracellular loops (ICLs) 1 or 3 or their adjacent membrane-spanning domains or within the eighth membrane-spanning domain (see fig. 3). If structure-function studies performed for NaPi-IIa can predict functionally important domains or residues in the closely related NaPi-IIc, the identified SLC34A3 mutations are likely to have significant functional consequences.
The capacity of the proximal tubule to reabsorb phosphate is determined by the amount of NaPi-II cotransporters residing in the brush border membrane. Site-directed mutagenesis and functional analysis of NaPi-IIa through the substituted cysteine accessibility method identified a dibasic motif K/R-K in the third ICL, which appears to be important for NaPi-IIa internalization induced by PTH (Karim-Jimenez et al. 2000). By use of the same experimental approach, several residues identified in the first extracellular loop (ECL), first ICL, and third ECL appear to be important for transport function, and the PRE motif in the third ECL determines pH-dependence of NaPi-IIa (Forster et al. 2002). Furthermore, the terminal three amino acid residues TRL and the PR motif in the C-terminal intracellular tail confer binding to PDZ-domain–containing proteins such as NHERF-1, thus permitting apical membrane insertion of this cotransporter (Karim-Jimenez et al. 2001). On the basis of these studies, it is possible that the missense mutations located within the first ICL or adjacent transmembrane domains affect transporter function, whereas mutations within the third ICL could potentially affect hormonal regulation of NaPi-IIc expression (Hernando et al. 2005; Virkki et al. 2005). Interestingly, 2 aa in NaPi-IIc (S192 and G196) that are mutated in some of our patients with HHRH were recently shown to be important determinants of stoichiometry and electroneutrality in the mouse ortholog Npt2c (Bacconi et al. 2005). When one of these residues was changed to alanine, or the other to asparagine (i.e., the residues found at these positions in Npt2a), mouse Npt2c became electrogenic, with a 3:1 sodium:phosphate stoichiometry, which is identical to that of mouse Npt2a. Mutations in these residues may thus lead to loss of function of NaPi-IIc in our patients with HHRH.
The intronic 101-bp deletion g.2259_2359del, identified on one allele in a single patient with HHRH (kindred C), may affect transcription or splicing of pre-mRNA between exons 9 and 10. Since this change in nucleotide sequence was not found in control samples, it also could provide a plausible explanation, if associated with a mutation on the second allele, for the development of HHRH.
It is likely that the laboratory abnormalities associated with some of the identified point mutations are the result of haploinsufficiency. However, a dominant-negative impact on wild-type NaPi-IIc derived from the intact SLC34A3 allele, or possibly on other sodium-phosphate cotransporters, such as NaPi-IIa, could also be considered. Functional in vitro studies are therefore needed to explore the impact of some of the mutations reported here.
Expressivity of SLC34A3 Mutations
On the basis of the findings in kindred D, it is conceivable that the identified SLC34A3 mutations do not lead to full expression of the clinical phenotype. In fact, in this kindred, not all individuals with compound heterozygous NaPi-IIc mutations developed HHRH; instead, some showed a milder phenotype (table 3). Furthermore, we found a homozygous c.228delC mutation in five individuals of the Bedouin kindred who were initially classified as having idiopathic hypercalciuria (data not shown), although subtle bone changes may have been missed. Consistent with these genetic findings, Tieder et al. (1985, 1987) demonstrated, in the Bedouin kindred, a significant overlap of the tubular maximum for phosphate reabsorption per glomerular filtration rate (TmP/GFR) and serum phosphate levels between healthy controls and individuals with hypercalciuria and between hypercalciuric individuals and patients with HHRH.
Similarly, heterozygous SLC34A3 mutations may lead to additional biochemical abnormalities besides hypercalciuria. For example, several members of the Bedouin kindred, who were shown here to be heterozygous for the c.228delC mutation, displayed mild hypophosphatemia, reduced TmP/GFR, and elevations in 1,25(OH)2D levels in addition to increased urinary calcium excretion (Tieder et al. 1985, 1987). Furthermore, heterozygosity for one of the missense mutations in the other kindreds was associated with increased calcium excretion and other laboratory abnormalities, such as hypophosphatemia and elevated serum 1,25(OH)2D levels; these mutations included c.586G→A (p.G196R) (IV-4 in kindred A), c.1402C→T (p.R468W) (V-3 and his mother IV-3 in kindred A), and c.1579_1581del (p.L527del) (II-1 and II-2, and possibly I-2 and I-3, in kindred D).
However, loss of one SLC34A3 allele does not always lead to laboratory abnormalities. For example, in the Bedouin kindred, heterozygosity for the c.228delC mutation did not lead to hypercalciuria in individuals V-10 and V-75, which indicates that additional factors contribute to the presence or absence of these urinary changes. Further investigations are thus required to identify environmental and/or genetic factors that affect expressivity of SLC34A3 mutations.
In summary, homozygous or compound heterozygous mutations in SLC34A3 were identified as the most plausible cause of HHRH in affected individuals from several different kindreds. Our data indicate that NaPi-IIc has a more prominent role in renal phosphate handling than previously thought.
Acknowledgments
We thank Dr. Thomas Hudson and Andrei Verner for facilitating the genomewide scan. We also thank Dr. Martin Epstein, from John Hunter Hospital, Newcastle, Australia, who was much involved in the investigation of kindred D. This study was supported by National Institutes of Health grants RO1 DK 46718 (to H.J.) and K24-HD-01288 (to T.O.C.), Canadian Institutes of Health Research grant GR-13297 (to H.S.T.), and a grant from the Canadian Networks of Centres of Excellence program–the Canadian Genetics Diseases Network (to K.M.).
Appendix A
Table A1.
Primers for PCR Amplification and Nucleotide Sequence Analysis[Note]
| Primer | Primer Sequence(5′→3′) | Orientation |
| 17 | TGGAGGTCCTAGCCACAGAG | Forward |
| 18 | GTCCCATTTCCTCCTCTTCC | Reverse |
| 19 | CCTCCAGTTCTGCTCCAGTC | Forward |
| 20 | CGCACCAGTGCTTAATGAGA | Reverse |
| 21 | CCTGCTGGAGAGGCTAAGTG | Forward |
| 22 | AGGAGGTCTCAAGGGAGGAGA | Reverse |
| 23 | CACAGGTCCTGCACACATTTC | Forward |
| 24 | CCATTCCTTGGGAGCTTC | Reverse |
| 25 | TGACAGGATGGAGACAAG | Forward |
| 26 | AGGAGCCTTGAAGTGGTG | Forward |
| 27 | CGTCTCGGCTTGATAAGG | Reverse |
| 28 | TCTCTGACACGCGCGTCC | Forward |
| 29 | TGGTCAGACTGGTGAAGG | Reverse |
| 30 | TTGGAGGAAGGGGACACA | Forward |
| 31 | GGCCTGCCAGCCTGCCAC | Reverse |
| 32 | GAGGGCCAGCCAGGGACA | Forward |
| 33 | AGTCAGCCTGCGGCCCAC | Reverse |
| 34 | CGTGGGTGCACACTCCCT | Forward |
| 35 | TGCTGTTAGTGGCGTTGC | Reverse |
| 36 | AGCTAGCCCTGGGTGCCG | Forward |
| 37 | TCACACCCTCCCTGCGTC | Forward |
| 38 | GACGCAGGGAGGGTGTGA | Reverse |
| 39 | CCGAAACTCCGGGTCATG | Forward |
| 43 | TCCCCAGCGGCTGCCGTG | Reverse |
| 44 | GCATGGAGCTGGCCGCTG | Forward |
| 45 | CTCTGACCTCTGTCTGCC | Forward |
| 46 | CAGGGCTGACCCAGCATC | Forward |
| 47 | GGAAGGGGAAGTCTATGG | Reverse |
| 52 | CATCCACTTCTTCTTCAACCTG | Forward |
| 53 | TCCAGAGAATGGAGCCAGAC | Reverse |
| 58 | AATGTGTGCAGGACCTGT | Reverse |
| 102 | CCTCCCCGGGTGGTGG | Forward |
| 103 | CACTTAGCCTCTCCAGCAGG | Reverse |
| 173 | CGTGCACAGAGAAGAACAGC | Forward |
| 174 | GGAAGTCTATGGGGGATGCT | Reverse |
Note.— For PCR, thermal cycler conditions (Mastercycler gradient) after an initial denaturation at 94°C for 5 min were 35 cycles at 94°C for 1 min, 60°C or 65°C for 1 min, and 72°C for 3 min, followed by a final extension at 72°C for 10 min.
Table A2.
Multiplex PCR Assays[Note]
|
Nucleotide Sequence(5′→3′) |
|||
| Polymorphism | Forward Primer | Reverse Primer | Probe |
| c.586G→A | ACGTTGGATGCAGCACTGTGAGCCAGTTGA | ACGTTGGATGTGACCCTGCCCACTCTCTG | GAGCCAGTTGAAGATCC |
| c.575C→T | ACGTTGGATGTGTGAGCCAGTTGAAGATCC | ACGTTGGATGTGACCCTGCCCACTCTCTG | AGATCCCGTGCACCGCC |
Note.— Please note that the PCR primers all have a 5′ tail of 10 bases (ACGTTGGATG), which can be used for nucleotide sequence analysis. PCR reactions were performed in a 384-well format. In brief, 2.5 ng of genomic DNA was amplified in a 5-μl reaction containing 1× HotStar Taq PCR buffer (Qiagen), 2.5 mM MgCl2, 200 μM of each dNTP, 50 nM of each PCR primer, and 0.1 U HotStar Taq (Qiagen). The reaction was incubated at 95°C for 15 min, followed by 45 cycles at 95°C for 20 s, 56°C for 30 s, and 72°C for 1 min, with a final step at 72°C for 3 min. Excess dNTPs were then removed from the reaction by incubation with 0.3 U shrimp alkaline phosphatase (USB) at 37°C for 20 min, followed by 5 min at 85°C to deactivate the enzyme. Single-primer extension over the SNP or insertion/deletion was performed in a final concentration of 600 nM of each extension primer (probe), 50 μM d/ddNTP, and 0.126 U Thermosequenase (USB) and was incubated at 94°C for 2 min, followed by 45 cycles at 94°C for 5 s, 52°C for 5 s, and 72°C for 5 s. The reaction was then desalted by addition of a cation exchange resin followed by mixing and centrifugation to precipitate the contents of the tube. The extension product was then spotted onto a 384-well spectroCHIP before being flown in the MALDI-TOF mass spectrometer.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SLC34A3 genomic contig [909158 in accession number NT_024000.16, or physical map nucleotide chr9:137401992 in human genome release March 2004], cDNA [187 in accession number NM_080877.1], and protein [accession number NP_543153.1]; SLC34A1 (NaPi-IIa) protein sequences for Homo sapiens [accession number NP_003043.2], Canis familiaris [accession number XP_536418.1], Pan troglodytes [accession number XP_518129.1], Rattus norvegicus [accession number NP_037162.1], Mus musculus [accession number NP_035522.1], and Gallus gallus [accession number XP_425204.1]; SLC34A2 (NaPi-IIb) protein sequences for H. sapiens [accession number NP_006415.1], C. familiaris [accession number XP_545968.1], P. troglodytes [accession number XP_526805.1], R. norvegicus [accession number XP_579555.1], M. musculus [accession number NP_035532.2], and G. gallus [accession number NP_989805.1]; and SLC34A3 (NaPi-IIc) protein sequences for C. familiaris [accession number XP_548353.1], R. norvegicus [accession number NP_647554.1], and M. musculus [accession number NP_543130.1])
- Marshfield Center for Medical Genetics, http://www2.marshfieldclinic.org/research/genetics/ (for Marshfield genetic maps)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HHRH, XLH, and ADHR)
References
- ADHR Consortium (2000) Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet 26:345–348 10.1038/81664 [DOI] [PubMed] [Google Scholar]
- Bacconi A, Virkki LV, Biber J, Murer H, Forster IC (2005) Renouncing electroneutrality is not free of charge: switching on electrogenicity in a Na+-coupled phosphate cotransporter. Proc Natl Acad Sci USA 102:12606–12611 10.1073/pnas.0505882102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Miao D, Li J, Goltzman D, Karaplis AC (2004) Transgenic mice overexpressing human fibroblast growth factor 23 (R176Q) delineate a putative role for parathyroid hormone in renal phosphate wasting disorders. Endocrinology 145:5269–5279 10.1210/en.2004-0233 [DOI] [PubMed] [Google Scholar]
- Bastepe M, Jüppner H (2000) Identification and characterization of two new, highly polymorphic loci adjacent to GNAS1 on chromosome 20q13.3. Mol Cell Probes 14:261–264 10.1006/mcpr.2000.0308 [DOI] [PubMed] [Google Scholar]
- Bastepe M, Lane AH, Jüppner H (2001) Paternal uniparental isodisomy of chromosome 20q—and the resulting changes in GNAS1 methylation—as a plausible cause of pseudohypoparathyroidism. Am J Hum Genet 68:1283–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastepe M, Pincus JE, Jüppner H (1999) Two frequent tetra-nucleotide repeat polymorphisms between VAPB and STX16 on chromosome 20q13. Mol Cell Probes 13:449–451 10.1006/mcpr.1999.0271 [DOI] [PubMed] [Google Scholar]
- Beck L, Karaplis AC, Amizuka N, Hewson AS, Ozawa H, Tenenhouse HS (1998) Targeted inactivation of Npt2 in mice leads to severe renal phosphate wasting, hypercalciuria, and skeletal abnormalities. Proc Natl Acad Sci USA 95:5372–5377 10.1073/pnas.95.9.5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau H, El-Maadawy S, McKee MD, Tenenhouse HS (2003) Renal calcification in mice homozygous for the disrupted type IIa Na/Pi cotransporter gene Npt2. J Bone Miner Res 18:644–657 [DOI] [PubMed] [Google Scholar]
- Chen C, Carpenter T, Steg N, Baron R, Anast C (1989) Hypercalciuric hypophosphatemic rickets, mineral balance, bone histomorphometry, and therapeutic implications of hypercalciuria. Pediatrics 84:276–280 [PubMed] [Google Scholar]
- Fishelson M, Geiger D (2002) Exact genetic linkage computations for general pedigrees. Bioinformatics Suppl 1 18:S189–S198 [DOI] [PubMed] [Google Scholar]
- Forster IC, Kohler K, Biber J, Murer H (2002) Forging the link between structure and function of electrogenic cotransporters: the renal type IIa Na+/Pi cotransporter as a case study. Prog Biophys Mol Biol 80:69–108 10.1016/S0079-6107(02)00015-9 [DOI] [PubMed] [Google Scholar]
- Gazit D, Tieder M, Liberman UA, Passi-Even L, Bab IA (1991) Osteomalacia in hereditary hypophosphatemic rickets with hypercalciuria: a correlative clinical-histomorphometric study. J Clin Endocrinol Metab 72:229–235 [DOI] [PubMed] [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Tenenhouse HS, Hoag HM, Wang D, Khadeer MA, Namba N, Feng X, Hruska KA (2001) Identification of the type II Na+-Pi cotransporter (Npt2) in the osteoclast and the skeletal phenotype of Npt2−/− mice. Bone 29:467–476 10.1016/S8756-3282(01)00601-9 [DOI] [PubMed] [Google Scholar]
- Hernando N, Gisler SM, Pribanic S, Deliot N, Capuano P, Wagner CA, Moe OW, Biber J, Murer H (2005) NaPi-IIa and interacting proteins. J Physiol 567:21–26 10.1113/jphysiol.2005.087049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266:383–402 [DOI] [PubMed] [Google Scholar]
- Hilfiker H, Kvietikova II, Hartmann CM, Stange G, Murer H (1998) Characterization of the human type II Na/Pi-cotransporter promoter. Pflugers Arch 436:591–598 10.1007/s004240050676 [DOI] [PubMed] [Google Scholar]
- Holm IA, Huang X, Kunkel LM (1997) Mutational analysis of the PEX gene in patients with X-linked hypophosphatemic rickets. Am J Hum Genet 60:790–797 [PMC free article] [PubMed] [Google Scholar]
- HYP Consortium (1995) A gene (PEX) with homologies to endopeptidases is mutated in patients with X-linked hypophosphatemic rickets. Nat Genet 11:130–136 10.1038/ng1095-130 [DOI] [PubMed] [Google Scholar]
- Jones A, Tzenova J, Frappier D, Crumley M, Roslin N, Kos C, Tieder M, Langman C, Proesmans W, Carpenter T, Rice A, Anderson D, Morgan K, Fujiwara T, Tenenhouse HS (2001) Hereditary hypophosphatemic rickets with hypercalciuria is not caused by mutations in the Na/Pi cotransporter NPT2 gene. J Am Soc Nephrol 12:507–514 [DOI] [PubMed] [Google Scholar]
- Karim-Jimenez Z, Hernando N, Biber J, Murer H (2000) A dibasic motif involved in parathyroid hormone-induced down-regulation of the type IIa NaPi cotransporter. Proc Natl Acad Sci USA 97:12896–12901 10.1073/pnas.220394197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2001) Molecular determinants for apical expression of the renal type IIa Na+/Pi-cotransporter. Pflugers Arch 442:782–790 10.1007/s004240100602 [DOI] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ (2003) The UCSC Genome Browser Database. Nucleic Acids Res 31:51–54 10.1093/nar/gkg129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh MP, Kabat D (1996) Identification and characterization of a widely expressed phosphate transporter/retrovirus receptor family. Kidney Int 49:959–963 [DOI] [PubMed] [Google Scholar]
- Kempson SA, Lotscher M, Kaissling B, Biber J, Murer H, Levi M (1995) Parathyroid hormone action on phosphate transporter mRNA and protein in rat renal proximal tubules. Am J Physiol 268:F784–F791 [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kos CH (1998) The role of the renal sodium-dependent phosphate cotransporter genes, NPT1 and NPT2, in inherited hypophosphatemias. PhD dissertation, McGill University, Montreal [Google Scholar]
- Larsson T, Marsell R, Schipani E, Ohlsson C, Ljunggren O, Tenenhouse HS, Jüppner H, Jonsson KB (2004) Transgenic mice expressing fibroblast growth factor 23 under the control of the α1(I) collagen promoter exhibit growth retardation, osteomalacia, and disturbed phosphate homeostasis. Endocrinology 145:3087–3094 10.1210/en.2003-1768 [DOI] [PubMed] [Google Scholar]
- Lazarus R, Klimecki WT, Palmer LJ, Kwiatkowski DJ, Silverman EK, Brown A, Martinez F, Weiss ST (2002) Single-nucleotide polymorphisms in the interleukin-10 gene: differences in frequencies, linkage disequilibrium patterns, and haplotypes in three United States ethnic groups. Genomics 80:223–228 10.1006/geno.2002.6820 [DOI] [PubMed] [Google Scholar]
- Liu S, Brown TA, Zhou J, Xiao ZS, Awad H, Guilak F, Quarles LD (2005) Role of matrix extracellular phosphoglycoprotein in the pathogenesis of X-linked hypophosphatemia. J Am Soc Nephrol 16:1645–1653 10.1681/ASN.2004121060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madjdpour C, Bacic D, Kaissling B, Murer H, Biber J (2004) Segment-specific expression of sodium-phosphate cotransporters NaPi-IIa and -IIc and interacting proteins in mouse renal proximal tubules. Pflugers Arch 448:402–410 [DOI] [PubMed] [Google Scholar]
- Mira MT, Alcaïs A, Van Thuc N, Thai VH, Huong NT, Ba NN, Verner A, Hudson TJ, Abel L, Schurr E (2003) Chromosome 6q25 is linked to susceptibility to leprosy in a Vietnamese population. Nat Genet 33:412–415 10.1038/ng1096 [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Ito M, Kuwahara M, Segawa H (2002) Regulation of a growth-related type IIc Na/Pi cotransporter by parathyroid hormone. J Am Soc Nephrol 13:284A [Google Scholar]
- Miyamoto K, Segawa H, Ito M, Kuwahata M (2004) Physiological regulation of renal sodium-dependent phosphate cotransporters. Jpn J Physiol 54:93–102 10.2170/jjphysiol.54.93 [DOI] [PubMed] [Google Scholar]
- Murer H, Forster I, Biber J (2004) The sodium phosphate cotransporter family SLC34. Pflugers Arch 447:763–767 10.1007/s00424-003-1072-5 [DOI] [PubMed] [Google Scholar]
- Murer H, Hernando N, Forster I, Biber J (2000) Proximal tubular phosphate reabsorption: molecular mechanisms. Physiol Rev 80:1373–140911015617 [Google Scholar]
- Ohkido I, Segawa H, Yanagida R, Nakamura M, Miyamoto K (2003) Cloning, gene structure and dietary regulation of the type-IIc Na/Pi cotransporter in the mouse kidney. Pflugers Arch 446:106–115 [DOI] [PubMed] [Google Scholar]
- Pal DK, Durner M, Greenberg DA (2001) Effect of misspecification of gene frequency on the two-point LOD score. Eur J Hum Genet 9:855–859 [DOI] [PubMed] [Google Scholar]
- Prié D, Huart V, Bakouh N, Planelles G, Dellis O, Gérard B, Hulin P, Benqué-Blanchet F, Silve C, Grandchamp B, Friedlander G (2002) Nephrolithiasis and osteoporosis associated with hypophosphatemia caused by mutations in the type 2a sodium-phosphate cotransporter. N Engl J Med 347:983–991 10.1056/NEJMoa020028 [DOI] [PubMed] [Google Scholar]
- Segawa H, Kaneko I, Takahashi A, Kuwahata M, Ito M, Ohkido I, Tatsumi S, Miyamoto K (2002) Growth-related renal type II Na/Pi cotransporter. J Biol Chem 277:19665–19672 10.1074/jbc.M200943200 [DOI] [PubMed] [Google Scholar]
- Sermet-Gaudelus I, Garabedian M, Dechaux M, Lenoir G, Rey J, Tieder M (2001) Hereditary hypophosphatemic rickets with hypercalciuria: report of a new kindred. Nephron 88:83–86 10.1159/000045964 [DOI] [PubMed] [Google Scholar]
- Shimada T, Hasegawa H, Yamazaki Y, Muto T, Hino R, Takeuchi Y, Fujita T, Nakahara K, Fukumoto S, Yamashita T (2004a) FGF-23 is a potent regulator of vitamin D metabolism and phosphate homeostasis. J Bone Miner Res 19:429–435 [DOI] [PubMed] [Google Scholar]
- Shimada T, Kakitani M, Yamazaki Y, Hasegawa H, Takeuchi Y, Fujita T, Fukumoto S, Tomizuka K, Yamashita T (2004b) Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J Clin Invest 113:561–568 10.1172/JCI200419081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Urakawa I, Yamazaki Y, Hasegawa H, Hino R, Yoneya T, Takeuchi Y, Fujita T, Fukumoto S, Yamashita T (2004c) FGF-23 transgenic mice demonstrate hypophosphatemic rickets with reduced expression of sodium phosphate cotransporter type IIa. Biochem Biophys Res Commun 314:409–414 10.1016/j.bbrc.2003.12.102 [DOI] [PubMed] [Google Scholar]
- Sitara D, Razzaque MS, Hesse M, Yoganathan S, Taguchi T, Erben RG, Jüppner H, Lanske B (2004) Homozygous ablation of fibroblast growth factor-23 results in hyperphosphatemia and impaired skeletogenesis, and reverses hypophosphatemia in Phex-deficient mice. Matrix Biol 23:421–432 10.1016/j.matbio.2004.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenhouse HS (2005) Regulation of phosphorus homeostasis by the type IIa Na/phosphate cotransporter. Annu Rev Nutr 25:197–214 10.1146/annurev.nutr.25.050304.092642 [DOI] [PubMed] [Google Scholar]
- Tenenhouse HS, Gauthier C, Chau H, St-Arnaud R (2004) 1αHydroxylase gene ablation and Pi supplementation inhibit renal calcification in mice homozygous for the disrupted Npt2a gene. Am J Physiol Renal Physiol 286:F675–F681 10.1152/ajprenal.00362.2003 [DOI] [PubMed] [Google Scholar]
- Tenenhouse HS, Martel J, Gauthier C, Segawa H, Miyamoto K (2003) Differential effects of Npt2a gene ablation and X-linked Hyp mutation on renal expression of Npt2c. Am J Physiol Renal Physiol 285:F1271–F1278 [DOI] [PubMed] [Google Scholar]
- Tenenhouse HS, Roy S, Martel J, Gauthier C (1998) Differential expression, abundance, and regulation of Na+-phosphate cotransporter genes in murine kidney. Am J Physiol 275:F527–F534 [DOI] [PubMed] [Google Scholar]
- Tieder M, Blonder J, Strauss S, Shaked U, Maor J, Gabizon D, Manor H, Sela BA (1993) Hyperoxaluria is not a cause of nephrocalcinosis in phosphate-treated patients with hereditary hypophosphatemic rickets. Nephron 64:526–531 [DOI] [PubMed] [Google Scholar]
- Tieder M, Modai D, Samuel R, Arie R, Halabe A, Bab I, Gabizon D, Liberman UA (1985) Hereditary hypophosphatemic rickets with hypercalciuria. N Engl J Med 312:611–617 [DOI] [PubMed] [Google Scholar]
- Tieder M, Modai D, Shaked U, Samuel R, Arie R, Halabe A, Maor J, Weissgarten J, Averbukh Z, Cohen N, Edelstein S, Liberman UA (1987) “Idiopathic” hypercalciuria and hereditary hypophosphatemic rickets: two phenotypical expressions of a common genetic defect. N Engl J Med 316:125–129 [DOI] [PubMed] [Google Scholar]
- van den Heuvel L, Op de Koul K, Knots E, Knoers N, Monnens L (2001) Autosomal recessive hypophosphataemic rickets with hypercalciuria is not caused by mutations in the type II renal sodium/phosphate cotransporter gene. Nephrol Dial Transplant 16:48–51 10.1093/ndt/16.1.48 [DOI] [PubMed] [Google Scholar]
- Virkki LV, Forster IC, Biber J, Murer H (2005) Substrate interactions in the human type IIa sodium-phosphate cotransporter (NaPi-IIa). Am J Physiol Renal Physiol 288:F969–F981 10.1152/ajprenal.00293.2004 [DOI] [PubMed] [Google Scholar]