Abstract
Hypophosphatemia due to isolated renal phosphate wasting results from a heterogeneous group of disorders. Hereditary hypophosphatemic rickets with hypercalciuria (HHRH) is an autosomal recessive form that is characterized by reduced renal phosphate reabsorption, hypophosphatemia, and rickets. It can be distinguished from other forms of hypophosphatemia by increased serum levels of 1,25-dihydroxyvitamin D resulting in hypercalciuria. Using SNP array genotyping, we mapped the disease locus in two consanguineous families to the end of the long arm of chromosome 9. The candidate region contained a sodium-phosphate cotransporter gene, SLC34A3, which has been shown to be expressed in proximal tubulus cells. Sequencing of this gene revealed disease-associated mutations in five families, including two frameshift and one splice-site mutation. Loss of function of the SLC34A3 protein presumably results in a primary renal tubular defect and is compatible with the HHRH phenotype. We also show that the phosphaturic factor FGF23 (fibroblast growth factor 23), which is increased in X-linked hypophosphatemic rickets and carries activating mutations in autosomal dominant hypophosphatemic rickets, is at normal or low-normal serum levels in the patients with HHRH, further supporting a primary renal defect. Identification of the gene mutated in a further form of hypophosphatemia adds to the understanding of phosphate homeostasis and may help to elucidate the interaction of the proteins involved in this pathway.
Hypophosphatemia due to isolated renal phosphate wasting can be caused by three well-defined Mendelian disorders: (1) X-linked hypophosphatemia (XLH [MIM 307800]), (2) autosomal dominant hypophosphatemic rickets (ADHR [MIM 193100]), and (3) hereditary hypophosphatemic rickets with hypercalciuria (HHRH [MIM 241530]). The genes mutated in XLH and ADHR have been identified as PHEX (HYP Consortium 1995) and FGF23 (ADHR Consortium 2000), respectively, whereas the gene mutated in HHRH has remained elusive. Phosphate wasting is also a prominent feature of tumor-induced osteomalacia (TIO), which is caused by rare, mostly benign mesenchymal tumors that overexpress fibroblast growth factor 23 (FGF23) (Shimada et al. 2001; White et al. 2001). Besides these well-defined conditions, there may be further Mendelian hypophosphatemias, such as hypophosphatemic bone disease and autosomal recessive hypophosphatemia without hypercalciuria.
HHRH was first described in a Bedouin tribe (Tieder et al. 1985). Since that article, one other Bedouin kindred and several sporadic cases have been described (Nishiyama et al. 1986; Chen et al. 1989; Tieder et al. 1992; Schnabel et al. 1996; van den Heuvel et al. 2001). Like other phosphate wasting disorders, HHRH is characterized by reduced tubular reabsorption of phosphate (TRP) and hypophosphatemia. However, in contrast to the characteristics of XLH and ADHR, serum levels of 1,25-dihydroxyvitamin D (1,25(OH)2D) in HHRH are appropriately elevated for low serum phosphorus levels despite suppressed parathyroid function, and there is an increase in urinary calcium excretion. It has been suggested that the pivotal defect consists of renal phosphate depletion, which stimulates renal 25-hydroxyvitamin D 1α-hydroxylase followed by an increase in 1,25(OH)2D (Tieder et al. 1985), enhanced intestinal absorption of calcium, and, subsequently, hypercalcemia. The accurate diagnosis of HHRH has important therapeutic implications. Unlike for XLH and ADHR, phosphate supplementation alone can cause a complete remission of HHRH, whereas the addition of vitamin D can create complications, such as hypercalcemia, nephrocalcinosis, and renal damage.
HHRH is inherited in an autosomal recessive mode. Obligate carriers of a single mutant allele have been described to have borderline hypercalciuria but do not have hypophosphatemia or rickets. Here, we describe the isolation of the gene mutated in HHRH by genomewide linkage analysis and mutation screening in a candidate gene.
Material and Methods
Patients
A total of seven patients with HHRH and their relatives from five families were included in this study. The patients were admitted to the hospital with rickets at the age of 2 to 12 years. All showed low serum phosphate levels and reduced or low-normal renal phosphate reabsorption. Alkaline phosphatase levels and urinary calcium excretion were clearly elevated. 1,25(OH)2D levels were elevated or highly normal, whereas 25-hydroxyvitamin D was in the normal range. Table 1 shows representative values for five patients. We also included 22 patients with hypophosphatemic rickets who were referred to us for molecular genetic diagnostics and in whom PHEX and FGF23 mutations have been excluded. We used 360 European individuals and 93 Israeli Arabic individuals as controls.
Table 1.
Chemical Findings
|
Patient |
|||||||
| Characteristic | 23724 | 23727 | 27018 | 5669 | 5919 | HeterozygousIndividualsa | ReferenceValue |
| Age (years) | 2 | 12 | 10 | 3.7 | 6 | 2–49 | … |
| Calcium (mmol/liter) | 2.45 | 2.42 | 2.35 | 2.26 | 2.54 | 2.2–2.3 | 2.15–2.64 |
| Phosphate (mmol/liter)b | .87 (1.23–1.94) | .74 (1.03–1.70) | .81 (1.10–1.87) | .87 (1.22–2.13) | .77c (1.16–1.91) | 1.00–1.39 | … |
| Alkaline phosphatase (U/liter)b | 736 (60–500) | 1,133 (80–224) | 689 (60–500) | 908 (182–446) | 890 (110–500) | 88–189 | … |
| PTH (pg/ml)d | 36 | 8.8 | 8.6 | 4.5 | …e | 18–68 | 12–65 |
| 25-hydroxyvitamin D (ng/ml) | 8 | 25 | 29 | 14 | 24 | 16–31 | 12–48 |
| 1,25(OH)2D (pg/ml) | 73 | 174 | 208 | 129 | 93 | 44–83 | 36–96 |
| Urine calcium/creatine (mg/mg)b | 1.24 (<.28) | .55 (<.25) | .66 (<.27) | .89 (<.17) | .26 (<.25) | .23–.55 | … |
| TRP (%) | 91 | 78 | 82 | 74 | 87.6 | 83–88 | 83–95 |
| TmP/GFRf | 2.1 | .9 | 2.0 | 2.0 | … | 2.9–4.1 | 2.5–4.2 |
Heterozygous individuals are six parents and seven siblings.
Given in parentheses are age- and sex-dependent reference values (Hochberg and Tiosano 2004).
The patient had several phosphate levels within the low-normal range (4.1–4.9 mg/dl) during the first months of evaluation.
PTH = parathyroid hormone.
At the time of diagnosis (before 1986), only PTH fragments could be measured. The value for C-terminal PTH (aa 35–84) was 0.4 ng/ml (normal 0.4–1.5). Measurement of C-terminal PTH (aa 44–68) was 159 pg/ml (normal 100–300). Actual PTH level was <3 pg/ml.
TmP/GFR = maximum tubular phosphate reabsorption per glomerular filtration rate.
Genetic Studies
Genomewide genotyping was performed using the Linkage IV Panel from Illumina, which contains ∼5,600 SNP markers. In addition, microsatellite markers were used in the candidate region: D9S164, D9S1826, D9S158, D9S905, and D9S1838. For the genomewide scan, frequencies of marker alleles were derived from dbSNP as provided by Illumina. For microsatellite analysis, the frequencies were based on the individuals genotyped in this project. The map positions of these markers were obtained by calculating the arithmetic mean of the sex-specific distances taken from the Genetic Location Database (Collins et al. 1996), by use of MAP-O-MAT version 1.1 (Lander and Green 1987; Matise and Gitlin 1999). Data for linkage analysis were prepared with a modified version of Alohomora (Rueschendorf and Nurnberg 2005). This tool uses PedCheck1.1 to detect Mendelian or genotyping errors (O’Connell and Weeks 1998) and uses Merlin to avoid unlikely genotypes (Abecasis et al. 2002). Multipoint linkage analysis was performed using Allegro (version 1.1d) (Gudbjartsson et al. 2000).
Mutation Analysis
SLC34A3 exons were amplified with intronic primers (table A1) and were directly sequenced using a BigDye Cycle sequencing kit (Applied Biosystems). Genomic DNA (∼50 ng) was subjected to PCR amplification performed in a 25-μl volume containing 1× PCR buffer/1.5 mM MgCl2, dNTPs (200 μM of each dNTP), 1× Q-Solution, HotStar Taq DNA Polymerase (2.5 U/reaction [Qiagen]), and 0.25 μM of each forward and reverse primer under the following cycle conditions: initial step at 95°C for 15 min; 10 cycles at 95°C for 30 s, 70°C for 30 s, and 72°C minus 1°C/cycle for 30 s; 25 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s; and final extension at 72°C for 10 min. Primers were designed with ExonPrimer.
Circulating FGF23 Levels
Plasma samples were isolated within 30 min after blood withdrawal by centrifugation and were stored at −70°C before biochemical analysis. FGF23 levels were measured using a commercial C-terminal ELISA (Immutopics) and a full-length FGF23 ELISA (Kainos Laboratories). The intra-assay coefficient of variation (CV) for the C-terminal assay is 5%, and the interassay CV is 7.3%.
Results
Linkage Analysis
We identified three families of Arab origin and two families of German origin with HHRH (fig. 1). Seven individuals in these families were classified as affected. The parents of the Arab families were first-degree cousins. Diagnosis was based on rickets, reduced TRP, hypophosphatemia, and hypercalciuria. Furthermore, mutations in the coding regions of PHEX and FGF23 were excluded by sequencing.
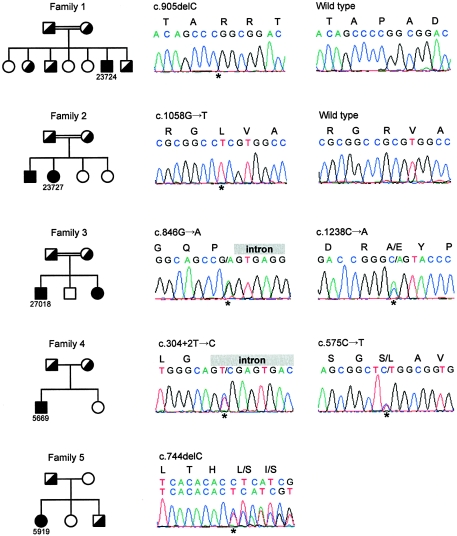
Figure 1.
Mutation analysis of SLC34A3 in five families (left) and electropherograms of the affected individuals (right). The mutations are indicated above the electropherograms. Families 1 and 2 presented homozygous mutations. Affected individuals from families 3 and 4 were compound heterozygous. In the affected individual from family 5, we found only a heterozygous deletion. Affected individuals are indicated by fully blackened symbols, and unaffected heterozygous individuals by half-blackened symbols.
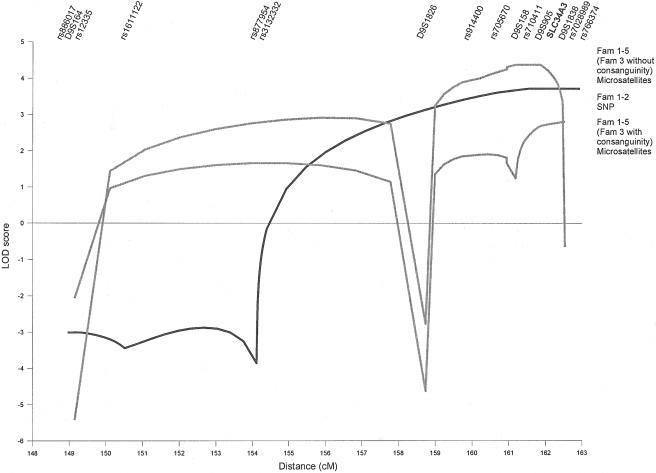
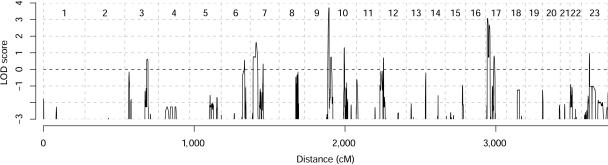
To identify the HHRH locus, we performed a genomewide linkage analysis by SNP genotyping, using the Linkage IV Panel (Illumina) for families 1 and 2. We assumed an autosomal recessive model. The frequency of the deleterious allele was set to 0.001, and the penetrance to 99% (q=0.001; f1=0.0; f2=0.0; f3=0.99). Data analysis revealed a single candidate region, with a maximum multipoint LOD score of 3.7 at the tip of chromosome 9 and distal of marker rs3132332 (fig. 2). Linkage was supported by microsatellite marker analysis in all five families. The maximum multipoint LOD score with allowance for heterogeneity was 2.8 distal of marker D9S158. Most of the LOD score came from families 1 and 2 (fig. 3). Families 4 and 5 were too small to give prominent LOD scores but were in agreement with linkage to this locus. Family 3 showed LOD scores of −1 in this region. Ultimately, it was discovered that the affected individuals in this family were compound heterozygous, although the family was consanguineous. Data analysis without the assumption of consanguinity in family 3 resulted in a LOD score of 4.4 distal of marker D9S1826.
Figure 2.
Multipoint linkage analysis of 15 cM of the telomeric end of the long arm of chromosome 9. The analyses were performed using ALLEGRO (q=0.001; f1=0.0; f2=0.0; f3=0.99) and included affected and unaffected family members. Depicted are summary LOD scores under the assumption of homogeneity. Analysis of SNP markers was performed in families (Fam) 1 and 2 (black line), and analyses of microsatellite markers were performed in all families, once with (bottom gray line) and once without (top gray line) consanguinity in family 3. The SLC34A3 gene maps between D9S905 and D9S1838.
Figure 3.
Genomewide linkage analysis of families 1 and 2. Analysis was performed using ALLEGRO (q=0.001; f1=0.0; f2=0.0; f3=0.99) and included affected and unaffected family members. Depicted are summary LOD scores under the assumption of homogeneity.
Mutation Analysis
The candidate region between marker D9S1826 and the chromosome end extends across 2.7 Mbp. According to Ensembl release 31.35d, the region contained 85 genes. Among these was an obvious candidate gene, the sodium-phosphate cotransporter type IIc gene (SLC34A3). SLC34A3 consists of 13 exons, with the initiation codon located in exon 2. Sequencing of the entire exon sequence and the flanking intronic regions revealed one heterozygous, two compound heterozygous, and two homozygous mutations in the affected individuals (fig. 1). Furthermore, the parents were shown to be heterozygous. Three of the mutations are predicted to result in a transcript with a prematurely terminated translation product: the proband in family 1 had a homozygous 1-bp deletion in exon 9 (c.905delC), one mutation in family 4 disrupts an essential residue at the intron 4 donor splice site (c.304+2T→C), and one mutation in family 5 was a 1-bp deletion in exon 7 (c.744delC). The other mutations were missense mutations and a silent mutation at the last nucleotide position of exon 8 (c.846G→A), which might lead to an aberrant splicing product. Attempts to confirm this consequence in peripheral blood leukocyte mRNA have been unsuccessful because of low abundance of SLC34A3 mRNA in this tissue. However, the splice-site prediction score (NNSPLICE version 0.9; see Splice Site Prediction by Neural Network Web site) decreased from 0.78 to 0.11. We found only a heterozygous mutation in family 5. Attempts to find a second mutation by searching for deletions with quantitative PCR failed. All mutations from the five families were not found in 350 European and 93 Israeli-Arabic control individuals.
We then sequenced DNA from another 22 individuals with hypophosphatemic rickets, for whom mutations in the PHEX and FGF23 genes had been excluded by sequencing and for whom we had received incomplete clinical reports. We did not find homozygous or compound heterozygous SLC34A3 mutations in these patients. During sequencing of the families with HHRH and the additional 22 individuals with hypophosphatemic rickets, we found five nonsynonymous and four synonymous sequence variations (table 2). One of the nonsynonymous changes (p.D237N) was found in a single control chromosome; all others were present in at least six control chromosomes. We concluded that these variants, with the possible exception of p.D237N, are polymorphisms because, under the assumption of random mating, a disease-allele frequency of 0.014 (6/440) would result in a recessive-disease frequency of 0.00019, which is much higher than the incidence of HHRH.
Table 2.
SLC34A3 Sequence Variations
|
Control Individualsb |
|||||||
| Variation Groupand Sequence Change | Patienta | dbSNP AccessionNumber | Exonor UTR | n | Genotype 11 | Genotype 12 | Genotype 22 |
| Mutations: | |||||||
| c.304+2T→C | 5669 (comp. het.) | … | Exon 4 | 353 (93) | TT: 353 (93) | TC: 0 (0) | CC: 0 (0) |
| c.575C→T (p.S192L) | … | … | Exon 7 | 350 (93) | CC: 350 (93) | CT: 0 (0) | TT: 0 (0) |
| c.744delC | 5919 (het.) | … | Exon 7 | 354 | CC: 354 | CdelC: 0 | delCdelC: 0 |
| c.905delC | 23724c (hom.) | … | Exon 9 | 352 | CC: 352 | CdelC: 0 | delCdelC: 0 |
| c.1058G→T (p.R353L) | 23727c (hom.) | … | Exon 10 | 353 (93) | GG: 353 (93) | GT: 0 (0) | TT: 0 (0) |
| c.1238C→A (p.A413E) | 27018c (comp. het.) | … | Exon 12 | 338 (91) | CC: 338 (91) | CA: 0 (0) | AA: 0 (0) |
| c.846G→A (p.P282P) | … | … | Exon 8 | 338 (91) | GG: 338 (91) | GA: 0 (0) | AA: 0 (0) |
| Nonsynonymous variations: | |||||||
| c.200G→A (p.R67H) | … | ss48531127 | Exon 4 | 366 | GG: 306 | GA: 57 | AA: 3 |
| c.539G→C (p.G180A) | … | ss48531129 | Exon 6 | 337 (92) | GG: 330 (88) | GC: 7 (4) | CC: 0 (0) |
| c.709G→A (p.D237N) | … | … | Exon 7 | 334 (92) | GG: 334 (91) | GA: 0 (1) | AA: 0 (0) |
| c.1009G→A (p.G337S) | … | ss48531134 | Exon 10 | 338 (93) | GG: 331 (86) | GA: 7 (7) | AA: 0 (0) |
| c.1538T→A (p.V513E) | … | ss48531135 | Exon 13 | 353 (93) | TT: 265 (83) | TA: 85 (10) | AA: 3 (0) |
| Synonymous variations: | |||||||
| c.204C→T (p.R68R) | … | ss48531128 | Exon 4 | 354 (93) | CC: 348 (92) | CT: 6 (1) | TT: 0 (0) |
| c.625C→T (p.L209L) | … | ss48531130 | Exon 7 | 338 (93) | CC: 332 (93) | CT: 6 (0) | TT: 0 (0) |
| c.757T→C (p.L249L) | … | ss48531131 | Exon 8 | 353 (93) | TT: 124 (30) | TC: 176 (44) | CC: 53 (18) |
| c.942G→C (p.A314A) | … | ss48531133 | Exon 10 | 338 | GG: 332 | GC: 6 | CC: 0 |
| Noncoding variations: | |||||||
| c.1094−10T→A | … | ss48531132 | Exon 11 | 351 (92) | TT: 282 (55) | TA: 67 (33) | AA: 2 (4) |
| c.1800+14A→C | … | ss48531136 | 3′ UTR | 338 (92) | AA: 104 (39) | AC: 171 (44) | CC: 63 (9) |
comp. het. = compound heterozygous; het. = heterozygous; hom. = homozygous.
n = number of control individuals tested for a particular variation. Genotypes are followed by the number of control individuals tested who have that genotype. The number of Arabic controls is given in parentheses.
Arabic index patient.
Phylogeny
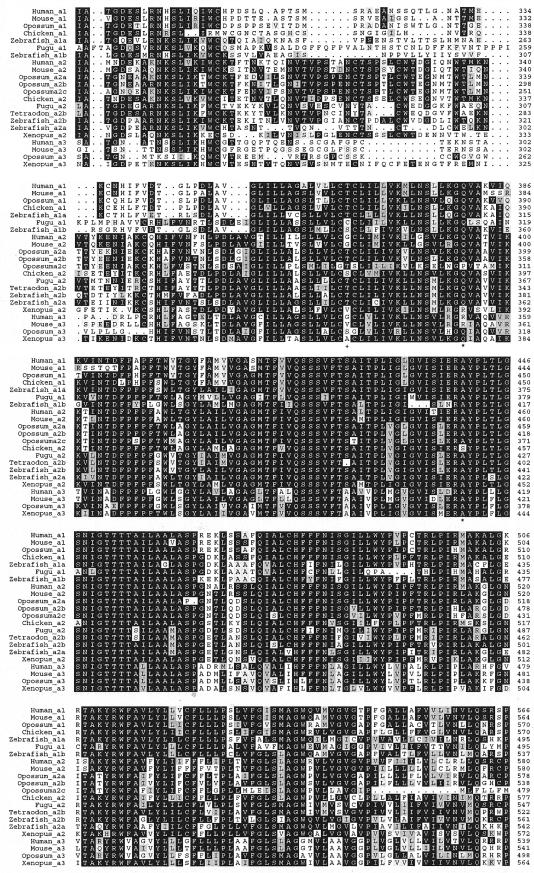
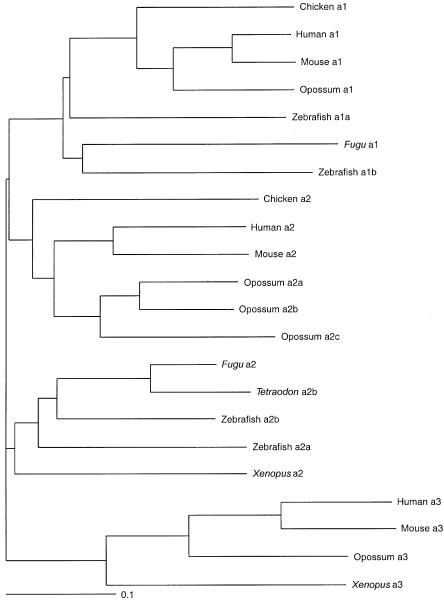
SLC34A3 has recently been identified as the third member of the sodium-phosphate cotransporter family type II, also called “NaPi-IIc” or “Npt2c.” In rats, it has been reported to be expressed only in kidney and at high levels at weaning age (Segawa et al. 2002). The protein has been shown to be localized at the apical membrane of proximal tubular cells (Segawa et al. 2002) and to be reduced in Hyp mice (Tenenhouse et al. 2003), which suggests that it is involved in renal phosphate reabsorption. To study whether the missense mutations are evolutionarily conserved, we undertook database searches and protein predictions from the genomic sequence of several vertebrate species to construct a phylogenetic tree of all type II cotransporters (fig. 4). SLC34A3 is present in eutherians, marsupials, and amphibians, because it can be found in human, mouse, rat, opossum, and Xenopus laevis, whereas it cannot be detected in fish genomes (fig. 5). Instead, zebrafish, having the most complete fish genome sequence, has four NaPi cotransporter genes, two each belonging to the type IIa family and the more divergent type IIb family. These four genes seem to be transcribed, since all sequences are supported by expressed sequence tags. One of the mutated amino acids (p.A413E) is conserved in all but one of the investigated cotransporters, one (p.S192L) in all type IIc cotransporters, and one (p.R353L) in only human and mouse type IIc cotransporters.
Figure 4.
Amino acid alignment of SLC34A1, SLC34A2, and SLC34A3 proteins from different vertebrate species. The proteins were obtained from GenBank, the UCSC Genome Browser, or Ensembl or were predicted by tblastn and Genewise from the corresponding genome sequence. Part of the predicted sequences is supplemented with partial cDNA sequences. The alignment was generated using ClustalW and Prettybox. Polymorphisms and variations of uncertain effect are indicated by plus signs (+), and missense mutations are indicated by asterisks (*). GenBank sequences (and accession numbers) include Human_a1 (NP_003043.2), Human_a2 (NP_006415.1), Human_a3 (NP_543153.1), Mouse_a1 (NP_035522.1), Mouse_a2 (NP_035532.1), Mouse_a3 (NP_543130.1), Chicken_a2 (NP_989805), Xenopus_a2 (AAH67316.1), Zebrafish_a2a (NP_571699.1), and Zebrafish_a2b (AAG35356.2). Ensembl proteins (and accession numbers) include Chicken_a1 (ENSGALP00000004850 and BU122185), Xenopus_a3 (ENSXETP00000050672 and CX972852), Zebrafish_a1a (ENSDARP017391 and C0920054), Zebrafish_a1b (ENSDARP00000041987 and CR929642), Fugu_a1 (SINFRUP00000132898), and Fugu_a2 (SINFRUP00000149494). Predicted proteins include Opossum_a1, Opossum_a2a, Opossum_a2b, Opossum_a2c, Opossum_a3, and Tetraodon_a2b.
Figure 5.
Dendrogram of SLC34A1, SLC34A2, and SLC34A3 proteins from different vertebrate species. The sequences were obtained from GenBank or Ensembl or were predicted by tblastn and Genewise from the corresponding genome sequence and were aligned using ClustalW (fig. 4).
FGF23 Plasma Levels
FGF23 is a phosphaturic factor (Shimada et al. 2001; White et al. 2001) that is elevated in most patients with X-linked hypophosphatemia and in patients with TIO (Jonsson et al. 2003). It is thought that FGF23 directly or indirectly inhibits renal phosphate reabsorption through SLC34A1 and SLC34A3 sodium-phosphate cotransporters. If this were the case, one would expect normal or low FGF23 plasma levels in individuals with HHRH. We measured FGF23 plasma levels with two ELISAs, detecting C-terminal/intact and only intact FGF23 in four of the affected individuals and in their relatives and controls. The C-terminal assays showed normal levels in the affected persons, whereas the intact assay revealed low-normal and undetectable FGF23 plasma levels in three of four patients (table 3).
Table 3.
FGF23 Plasma Levels
| Patient | C-Terminal/Intact FGF23a(RU/ml) | Intact FGF23b(pg/ml) |
| 23727 | 28 | 5 |
| 27018 | 33 | ND |
| 5669 | 53 | 35 |
| 5919 | 30 | 9 |
Normal <150 reference units (RU)/ml.
Normal 8–54 pg/ml. ND = not detectable.
Discussion
We identified the gene mutated in HHRH by a positional candidate-gene approach. Several observations substantiate the causal role of SLC34A3 mutations in HHRH. We identified two different homozygous, one heterozygous, and two compound heterozygous SLC34A3 mutations in five families that were not present in 440 control individuals. At least three of seven mutations are predicted to result in loss of function of the sodium-phosphate cotransporter. Final proof of the causal role of the missense mutations awaits functional studies. We also found five nonsynonymous sequence variations. All but one are considered to be polymorphisms because the minor-allele frequency was at least 8%. That HHRH is caused by loss-of-function mutations is compatible with the HHRH phenotype and the current view of renal phosphate regulation. Long-term phosphate supplementation alone results in reversal of the clinical and biochemical abnormalities in HHRH, with the exception of decreased renal phosphate reabsorption. Thus, it is likely that HHRH is a primary disorder of renal phosphate reabsorption.
A key process in phosphate homeostasis is the transport of inorganic phosphate from the primary urine across the renal proximal tubule, which involves secondary active sodium-phosphate (NaPi) cotransport at the apical brush border membrane of the proximal renal tubular epithelium. The closely related type II NaPi cotransporter genes, SLC34A1 (Magagnin et al. 1993) and SLC34A3 (Segawa et al. 2002), are predominantly expressed in the kidney and are thought to play a key role in renal phosphate handling. The importance of Slc34a1 in renal handling of Pi was demonstrated in Npt2a knockout mice (Beck et al. 1998). Mice homozygous for the disrupted Npt2a gene exhibit increased urinary Pi excretion, hypophosphatemia, an adaptive increase in serum 1,25(OH)2D, and hypercalciuria. These biochemical features are typical for patients with HHRH, yet Npt2a−/− mice do not show rickets.
The Slc34a3 protein is maximally upregulated in Npt2a−/− mice and may therefore be also involved in physiological phosphate reabsorption, although upregulation is obviously not sufficient to compensate for the phosphate loss through Slc34a1 (Tenenhouse et al. 2003). It has been estimated that SLC34A1 accounts for ∼70%–80% of NaPi-cotransport activity (Murer et al. 2004). A significant part of the remaining activity might be related to SLC34A3 (Segawa et al. 2002; Tenenhouse et al. 2003).
Both transporters are likely to be regulated by the endopeptidase homolog PHEX and the phosphaturic factor FGF23, since decreased renal Slc34a1 and Slc34a3 mRNA or protein expression has been reported in FGF23 transgenic mice and in Hyp mice (Tenenhouse et al. 1994; Larsson et al. 2004; Shimada et al. 2004), which harbor a Phex deletion (Strom et al. 1997) and serve as a model for X-linked hypophosphatemia. It is further likely that PHEX acts upstream of FGF23, since double-mutant Phex−/−Hyp−/− and FGF23−/− mice do not rescue the FGF23−/− phenotype (Sitara et al. 2004), although there is evidence against a direct interaction between PHEX and FGF23 (Liu et al. 2003; Benet-Pages et al. 2004). Hypophosphatemia caused by both PHEX and FGF23 mutations affects not only renal phosphate reabsorption but also the synthesis of 1,25(OH)2D due to the lack of upregulation of 25-hydroxyvitamin D 1α-hydroxylase (Scriver et al. 1978; Lobaugh and Drezner 1983). This upregulation in response to low serum phosphate is intact in HHRH, which further argues for a primary defect in renal phosphate reabsorption in this condition.
Heterozygous mutations in the SCL34A1 gene have been described in two individuals with nephrolithiasis, renal phosphate loss, and hypophosphatemia (Prié et al. 2002), but that report has not been confirmed yet. Hypophosphatemia has also been described in heterozygous Npt2a+/− mice, although they do not develop nephrolithiasis (Beck et al. 1998). Of note, borderline hypercalciuria and hypophosphatemia have been described in individuals presumably carrying a heterozygous SLC34A3 mutation (Tieder et al. 1987). Hypercalciuria is also documented in heterozygous mutation carriers in the present study and may predispose these individuals to nephrolithiasis and skeletal abnormalities.
In summary, the identification of SLC34A3 mutations in HHRH adds the protein SLC34A3 to the group of proteins—PHEX, FGF23, SLC34A1, and GALNT3—that are all involved in renal phosphate reabsorption. The interaction and regulation of these proteins remain largely elusive. The identification of mutated genes in additional, rare forms of hypophosphatemia may help to unravel these questions.
Acknowledgments
We thank the patients and their families for participation in this study. We also thank K. E. von Mühlendahl, for providing DNA samples, and Jack Favor, for critical discussions. This work was supported by Deutsche Forschungsgemeinschaft grant STR304/2–1.
Appendix A
Table A1.
Primers for the Amplification of SLC34A3 Exons
|
Primer Sequence(5′→3′) |
||||
| Exon(s) | Forward | Reverse | Annealing Temperature(°C) | Length(bp) |
| 1 | TCTTTGACCTGCAACTGCTC | CACCTGGACCCCTCCAC | 60 | 265 |
| 2 and 3 | TGTGAATCCAGCTTGTGAGG | CTGTCTCCCCAGTCCCG | 60 | 637 |
| 4 and 5 | AGAGAATGAGGGGCCTGG | GACACCCGGACAGTCAGC | 60 | 523 |
| 6 and 7 | CAGCATGGTGGCTGCTAAG | CAGACCCTCCGCTGACC | 60 | 595 |
| 8 and 9 | CCCGGGTGAGTCCTGAG | GGCTGTGCTGTTCCTCTCC | 60 | 448 |
| 10 and 11 | GACAGGCTGCCCTGTGAG | CACCACAGGACCCCAGC | 60 | 560 |
| 12 | GACAGAGGCCTCGGGAAC | CTGTCTGTGGGGCTGAGAG | 60 | 247 |
| 13 | GTAGGGTGGAGGAGGGCAG | GTGTCCACCAGGAGGCAG | 60 | 768 |
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Ensembl, http://www.ensembl.org/ (for proteins from chicken [accession numbers ENSGALP00000004850 and BU122185], Xenopus [accession numbers ENSXETP00000050672 and CX972852], zebrafish [accession numbers ENSDARP017391, C0920054, ENSDARP00000041987, and CR929642], and Fugu [accession numbers SINFRUP00000132898 and SINFRUP00000149494])
- ExonPrimer, http://ihg.gsf.de/ihg/ExonPrimer.html
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for sequences from human [accession numbers NP_003043.2, NP_006415.1, and NP_543153.1], mouse [accession numbers NP_035522.1, NP_035532.1, and NP_543130.1], chicken [accession number NP_989805], Xenopus [accession number AAH67316.1], and zebrafish [accession numbers NP_571699.1 and AAG35356.2])
- MAP-O-MAT, http://compgen.rutgers.edu/mapomat/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HHRH, ADHR, and XLH)
- Splice Site Prediction by Neural Network, http://www.fruitfly.org/seq_tools/splice.html
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- ADHR Consortium (2000) Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet 26:345–348 10.1038/81664 [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Beck L, Karaplis AC, Amizuka N, Hewson AS, Ozawa H, Tenenhouse HS (1998) Targeted inactivation of Npt2 in mice leads to severe renal phosphate wasting, hypercalciuria, and skeletal abnormalities. Proc Natl Acad Sci USA 95:5372–5377 10.1073/pnas.95.9.5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benet-Pages A, Lorenz-Depiereux B, Zischka H, White KE, Econs MJ, Strom TM (2004) FGF23 is processed by proprotein convertases but not by PHEX. Bone 35:455–462 10.1016/j.bone.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Chen C, Carpenter T, Steg N, Baron R, Anast C (1989) Hypercalciuric hypophosphatemic rickets, mineral balance, bone histomorphometry, and therapeutic implications of hypercalciuria. Pediatrics 84:276–280 [PubMed] [Google Scholar]
- Collins A, Frezal J, Teague J, Morton NE (1996) A metric map of humans: 23,500 loci in 850 bands. Proc Natl Acad Sci USA 93:14771–14775 10.1073/pnas.93.25.14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 10.1038/75514 [DOI] [PubMed] [Google Scholar]
- Hochberg Z, Tiosano D (2004) Disorders of mineral metabolism. In: Pescovitz O, Eugster E (eds) Pediatric endocrinology: mechanisms, manifestations and managment. Lippincott Williams and Wilkins, Philadelphia, pp 614–640 [Google Scholar]
- HYP Consortium (1995) A gene (PEX) with homologies to endopeptidases is mutated in patients with X-linked hypophosphatemic rickets. Nat Genet 11:130–136 10.1038/ng1095-130 [DOI] [PubMed] [Google Scholar]
- Jonsson KB, Zahradnik R, Larsson T, White KE, Sugimoto T, Imanishi Y, Yamamoto T, Hampson G, Koshiyama H, Ljunggren O, Oba K, Yang IM, Miyauchi A, Econs MJ, Lavigne J, Jüppner H (2003) Fibroblast growth factor 23 in oncogenic osteomalacia and X-linked hypophosphatemia. N Engl J Med 348:1656–1663 10.1056/NEJMoa020881 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson T, Marsell R, Schipani E, Ohlsson C, Ljunggren O, Tenenhouse HS, Jüppner H, Jonsson KB (2004) Transgenic mice expressing fibroblast growth factor 23 under the control of the α1(I) collagen promoter exhibit growth retardation, osteomalacia, and disturbed phosphate homeostasis. Endocrinology 145:3087–3094 10.1210/en.2003-1768 [DOI] [PubMed] [Google Scholar]
- Liu S, Guo R, Simpson LG, Xiao ZS, Burnham CE, Quarles LD (2003) Regulation of fibroblastic growth factor 23 expression but not degradation by PHEX. J Biol Chem 278:37419–37426 10.1074/jbc.M304544200 [DOI] [PubMed] [Google Scholar]
- Lobaugh B, Drezner MK (1983) Abnormal regulation of renal 25-hydroxyvitamin D-1α-hydroxylase activity in the X-linked hypophosphatemic mouse. J Clin Invest 71:400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magagnin S, Werner A, Markovich D, Sorribas V, Stange G, Biber J, Murer H (1993) Expression cloning of human and rat renal cortex Na/Pi cotransport. Proc Natl Acad Sci USA 90:5979–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise TC, Gitlin JA (1999) MAP-O-MAT: marker-based linkage mapping on the World Wide Web. Am J Hum Genet 65:A435 [Google Scholar]
- Murer H, Forster I, Biber J (2004) The sodium phosphate cotransporter family SLC34. Pflugers Arch 447:763–767 10.1007/s00424-003-1072-5 [DOI] [PubMed] [Google Scholar]
- Nishiyama S, Inoue F, Makuda I (1986) A single case of hypophosphatemic rickets with hypercalciuria. J Pediatr Gastroenterol Nutr 5:826–829 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prié D, Huart V, Bakouh N, Planelles G, Dellis O, Gerard B, Hulin P, Benque-Blanchet F, Silve C, Grandchamp B, Friedlander G (2002) Nephrolithiasis and osteoporosis associated with hypophosphatemia caused by mutations in the type 2a sodium-phosphate cotransporter. N Engl J Med 347:983–991 10.1056/NEJMoa020028 [DOI] [PubMed] [Google Scholar]
- Rueschendorf F, Nurnberg P (2005) ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics 21:2123–2125 10.1093/bioinformatics/bti264 [DOI] [PubMed] [Google Scholar]
- Schnabel D, von Mühlendahl KE, Morlot M, Wassmann A, Grüters A, Kruse K (1996) The hereditary syndrome of hypophosphatemic rickets and hypercalciuria (HHRH): possible diagnostic pitfalls and clinical follow-up [abstract]. Horm Res Suppl 2 46:84 [Google Scholar]
- Scriver CR, Reade TM, DeLuca HF, Hamstra AJ (1978) Serum 1,25-dihydroxyvitamin D levels in normal subjects and in patients with hereditary rickets or bone disease. N Engl J Med 299:976–979 [DOI] [PubMed] [Google Scholar]
- Segawa H, Kaneko I, Takahashi A, Kuwahata M, Ito M, Ohkido I, Tatsumi S, Miyamoto K (2002) Growth-related renal type II Na/Pi cotransporter. J Biol Chem 277:19665–19672 10.1074/jbc.M200943200 [DOI] [PubMed] [Google Scholar]
- Shimada T, Mizutani S, Muto T, Yoneya T, Hino R, Takeda S, Takeuchi Y, Fujita T, Fukumoto S, Yamashita T (2001) Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proc Natl Acad Sci USA 98:6500–6505 10.1073/pnas.101545198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Urakawa I, Yamazaki Y, Hasegawa H, Hino R, Yoneya T, Takeuchi Y, Fujita T, Fukumoto S, Yamashita T (2004) FGF-23 transgenic mice demonstrate hypophosphatemic rickets with reduced expression of sodium phosphate cotransporter type IIa. Biochem Biophys Res Commun 314:409–414 10.1016/j.bbrc.2003.12.102 [DOI] [PubMed] [Google Scholar]
- Sitara D, Razzaque MS, Hesse M, Yoganathan S, Taguchi T, Erben RG, Jüppner H, Lanske B (2004) Homozygous ablation of fibroblast growth factor-23 results in hyperphosphatemia and impaired skeletogenesis, and reverses hypophosphatemia in Phex-deficient mice. Matrix Biol 23:421–432 10.1016/j.matbio.2004.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom TM, Francis F, Lorenz B, Böddrich A, Econs MJ, Lehrach H, Meitinger T (1997) Pex gene deletions in Gy and Hyp mice provide mouse models for X-linked hypophosphatemia. Hum Mol Genet 6:165–171 10.1093/hmg/6.2.165 [DOI] [PubMed] [Google Scholar]
- Tenenhouse HS, Martel J, Gauthier C, Segawa H, Miyamoto K (2003) Differential effects of Npt2a gene ablation and X-linked Hyp mutation on renal expression of Npt2c. Am J Physiol Renal Physiol 285:F1271–F1278 [DOI] [PubMed] [Google Scholar]
- Tenenhouse HS, Werner A, Biber J, Ma S, Martel J, Roy S, Murer H (1994) Renal Na+-phosphate cotransport in murine X-linked hypophosphatemic rickets: molecular characterization. J Clin Invest 93:671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieder M, Arie R, Bab I, Maor J, Liberman UA (1992) A new kindred with hereditary hypophosphatemic rickets with hypercalciuria: implications for correct diagnosis and treatment. Nephron 62:176–181 [DOI] [PubMed] [Google Scholar]
- Tieder M, Modai D, Samuel R, Arie R, Halabe A, Bab I, Gabizon D, Liberman UA (1985) Hereditary hypophosphatemic rickets with hypercalciuria. N Engl J Med 312:611–617 [DOI] [PubMed] [Google Scholar]
- Tieder M, Modai D, Shaked U, Samuel R, Arie R, Halabe A, Maor J, Weissgarten J, Averbukh Z, Cohen N, Edelstein S, Liberman UA (1987) “Idiopathic” hypercalciuria and hereditary hypophosphatemic rickets: two phenotypical expressions of a common genetic defect. N Engl J Med 316:125–129 [DOI] [PubMed] [Google Scholar]
- van den Heuvel L, Op de Koul K, Knots E, Knoers N, Monnens L (2001) Autosomal recessive hypophosphataemic rickets with hypercalciuria is not caused by mutations in the type II renal sodium/phosphate cotransporter gene. Nephrol Dial Transplant 16:48–51 10.1093/ndt/16.1.48 [DOI] [PubMed] [Google Scholar]
- White KE, Jonsson KB, Carn G, Hampson G, Spector TD, Mannstadt M, Lorenz-Depiereux B, Miyauchi A, Yang IM, Ljunggren O, Meitinger T, Strom TM, Jüppner H, Econs MJ (2001) The autosomal dominant hypophosphatemic rickets (ADHR) gene is a secreted polypeptide overexpressed by tumors that cause phosphate wasting. J Clin Endocrinol Metab 86:497–500 10.1210/jc.86.2.497 [DOI] [PubMed] [Google Scholar]