Abstract
We report the clinical characteristics of a schizophrenia sample of 409 pedigrees—263 of European ancestry (EA) and 146 of African American ancestry (AA)—together with the results of a genome scan (with a simple tandem repeat polymorphism interval of 9 cM) and follow-up fine mapping. A family was required to have a proband with schizophrenia (SZ) and one or more siblings of the proband with SZ or schizoaffective disorder. Linkage analyses included 403 independent full-sibling affected sibling pairs (ASPs) (279 EA and 124 AA) and 100 all-possible half-sibling ASPs (15 EA and 85 AA). Nonparametric multipoint linkage analysis of all families detected two regions with suggestive evidence of linkage at 8p23.3-q12 and 11p11.2-q22.3 (empirical Z likelihood-ratio score [Zlr] threshold ⩾2.65) and, in exploratory analyses, two other regions at 4p16.1-p15.32 in AA families and at 5p14.3-q11.2 in EA families. The most significant linkage peak was in chromosome 8p; its signal was mainly driven by the EA families. Zlr scores >2.0 in 8p were observed from 30.7 cM to 61.7 cM (Center for Inherited Disease Research map locations). The maximum evidence in the full sample was a multipoint Zlr of 3.25 (equivalent Kong-Cox LOD of 2.30) near D8S1771 (at 52 cM); there appeared to be two peaks, both telomeric to neuregulin 1 (NRG1). There is a paracentric inversion common in EA individuals within this region, the effect of which on the linkage evidence remains unknown in this and in other previously analyzed samples. Fine mapping of 8p did not significantly alter the significance or length of the peak. We also performed fine mapping of 4p16.3-p15.2, 5p15.2-q13.3, 10p15.3-p14, 10q25.3-q26.3, and 11p13-q23.3. The highest increase in Zlr scores was observed for 5p14.1-q12.1, where the maximum Zlr increased from 2.77 initially to 3.80 after fine mapping in the EA families.
Schizophrenia (SZ [MIM 181500]) is a disorder (or group of disorders) with onset typically in adolescence or young adulthood and characterized by disruption of thinking (e.g., delusions or disorganization), perception (hallucinations), mood, and behavior. The symptoms, if untreated, tend to persist, and the course of the disease tends to be chronic, even with treatment. Although psychotic symptoms can be detected in the general population, and a continuum of severity from normality to psychosis has been proposed (Strauss 1969; van Os et al. 2000; Johns et al. 2004), SZ, as defined by the DSM-IV criteria of the American Psychiatric Association (1994), is widely viewed as a discrete illness. Heritability is ∼80%, on the basis of twin studies (Cardno et al. 1999). There is a 10-fold increase in risk to first-degree relatives (Gottesman and Shields 1982), as well as a familial coaggregation of SZ with other psychotic disorders and with “schizophrenia spectrum” personality disorders (Kendler et al. 1993b, 1993c, 1993d; Stompe et al. 1998). SZ risk is also increased in adopted-away children who have a biological parent with SZ (Heston 1966; Kety et al. 1971; Rosenthal et al. 1971) and in the offspring of an unaffected MZ cotwin of a proband with SZ (Gottesman and Bertelsen 1989). Complex inheritance is suggested by the high MZ:DZ risk ratio (∼4:1) and by the rapid decrease in risk as biological relatedness to a proband decreases (O’Rourke et al. 1982).
For complex disorders such as SZ, it has proven difficult to obtain replicable positive evidence of genetic linkage, but careful analysis of the SZ literature suggests that there is considerable support for a set of potential susceptibility loci. Three types of evidence are available: linkage statistics from published studies and from two recent meta-analyses of genomewide linkage scans (Badner and Gershon 2002; Lewis et al. 2003), evidence from a chromosomal deletion syndrome on chromosome 22q (Lindsay et al. 1995), and reported associations with positional susceptibility genes, some of which have gathered independent support (Owen et al. 2004) and convergent support from independent neuropharmacological findings (Cloninger 2002). We anticipate that many SZ susceptibility genes remain unidentified and that some of these can be characterized using genetic linkage and association methods. Therefore, we have recruited a large sample of affected sibling pairs (ASPs) to search for susceptibility loci by whole-genome linkage screening, to prioritize regions for more-intensive study. The families were ascertained through probands with SZ who have a sibling with SZ or schizoaffective disorder (SA). The sample is part of the National Institute of Mental Health (NIMH) Genetics Initiative for Schizophrenia, and the NIMH will make available the DNA specimens and blinded clinical data (see NIMH Center for Collaborative Genetics Studies of Mental Disorders Web site).
We report here the results of a genome scan of 409 pedigrees with SZ—263 of European ancestry (EA) and 146 of African American ancestry (AA), all collected by the Molecular Genetics of Schizophrenia (MGS1) Collaboration—with the use of a 9-cM map of STRPs. Using nonparametric multipoint linkage analysis, we detected two chromosomal regions with suggestive evidence of linkage (Lander and Kruglyak 1995) on chromosomes 8p23.3-p12 and 11p11.2-q22.3 in the full sample and, in exploratory analyses, two regions that reached similar thresholds on chromosomes 4p16.1-p15.32 and 5p14.3-q11.2 in the AA and EA samples, respectively. Chromosome 8p is one of the most consistently observed regions in SZ genome scans, as discussed below. There have been several reports of significant association between SZ and polymorphisms in neuregulin 1 (NRG1) in this region (Craddock et al. 2005), but it is not, in fact, clear to what extent this gene accounts for the linkage signals, and replication has not been uniform (Duan et al. 2005). Most such linkage signals have been observed closer to the p-telomere than to NRG1. In the present sample, there is evidence of linkage across a broad region, and the two peaks observed primarily in EA families are both telomeric to NRG1. Also within this region is a paracentric inversion that is common in Europeans (Broman et al. 2003). A large block of linkage disequilibrium (LD) was observed in this region in our EA sample, presumably because of the absence of crossing-over when a parent is heterozygous for the inversion. The inversion also creates uncertainty about the location and order of certain markers. Previous SZ linkage reports on this region have not specifically addressed the effects of this inversion on the results. We discuss below our efforts to accommodate this inversion.
Material and Methods
Family Ascertainment
The study participants were enrolled at nine sites in the United States and one in Australia: University of Chicago, Chicago; University of California, Irvine; University of Colorado Health Sciences Center, Denver; Baylor College of Medicine, Houston; University of Iowa, Iowa City; Washington University, St. Louis; Mount Sinai School of Medicine, New York; University of Pennsylvania, Philadelphia (this site included a subcontract with the Louisiana State University Health Sciences Center, New Orleans); and the University of Queensland, Brisbane. The focus of recruitment efforts was on EA and AA families. EA populations in the United States and Australia have similar ethnic characteristics (Cavalli-Sforza et al. 1994). Families were identified from a variety of sources, including local treatment facilities, physician referrals, the National Alliance for the Mentally Ill and other advocacy groups, Web sites, media announcements, and advertisements. Participants gave informed consent to an interview, provided a blood specimen for DNA and cell lines, and granted permission to obtain their psychiatric records and (usually) to contact a family informant. Available parents were asked to provide a blood specimen. If one or both parents were not available, as many as two unaffected sibs were asked to provide blood specimens. Local institutional review board approval was obtained for each site.
Clinical Assessment
Subjects (aged ⩾18 years) were interviewed by trained clinicians using the semistructured Diagnostic Interview for Genetic Studies (DIGS) 2.0 (Nurnberger et al. 1994), to elicit information required to determine diagnoses of psychotic, mood, and substance-use disorders in accordance with DSM-IV criteria (American Psychiatric Association 1994), the comorbidity of these disorders, medical history, and ratings of positive and negative symptoms of SZ. A family informant was also interviewed, when possible, about each patient’s history and about the family psychiatric history, by use of the Family Interview for Genetic Studies (FIGS) (Gershon et al. 1988; Maxwell 1992). Two experienced research clinicians independently reviewed the DIGS, FIGS, interviewer’s narrative report, and all available psychiatric records and then assigned all relevant diagnoses (with a confidence level and age at onset identified for each one), a judgment about presence or absence of each DSM-IV criterion for SZ and SA, and the estimated lifetime duration of SZ and of mood syndromes. For all these items, the diagnosticians resolved any disagreements by discussion (or occasionally asked a third diagnostician to serve as a tiebreaker). Thus, a primary best-estimate final diagnosis (BEFD) (Leckman et al. 1982) was assigned on the basis of this consensus procedure. All individuals (interviewed or not) who did not receive an SZ or SA diagnosis were considered to have “diagnosis unknown” for the linkage analyses. One of the two diagnosticians also completed the Lifetime Dimensions of Psychosis Scale (LDPS), a 21-item scale for rating the lifetime duration and severity of positive, bizarre positive, negative, disorganized, and mood symptoms of psychotic disorders, on the basis of all available information for each patient (Levinson et al. 2002). Analyses using LDPS ratings will be presented elsewhere.
Inclusion and Exclusion Criteria
To be eligible, a family was required to be multiplex—minimally, to have a proband with a BEFD of SZ and one or more siblings with SZ or SA, with a confidence level of “likely” or “definite.” Additional first-degree relatives with SZ and SA were recruited when possible. Both SZ-affected and SA-affected relatives have been included in this and most other SZ linkage studies, on the basis of evidence from multiple-family studies showing that both disorders cluster in families ascertained through a proband with SZ (Gershon et al. 1988; Kendler et al. 1993a; Maier et al. 1993; Taylor et al. 1993). For an SA diagnosis, DSM-IV requires that criteria for SZ and for manic, mixed, and/or major depressive episodes be met simultaneously at some point, with persistence of psychotic symptoms without prominent mood symptoms for at least 2 wk, and with mood episodes persisting for “a substantial portion of the total duration” (Cloninger et al. 1998, p. 278) of illness, defined here as 30% for consistency with the first NIMH Genetics Initiative for Schizophrenia study. Subjects were excluded from receiving a final diagnosis of SZ or SA if psychosis was limited to periods of likely substance intoxication or withdrawal, if persistent psychotic symptoms were considered likely to be related to substance use (e.g., increasing paranoia after years of amphetamine use or symptoms limited to visual hallucinations and “flashbacks” after hallucinogen use), if psychosis might have been caused by another disorder (e.g., epilepsy predating SZ onset) as determined by consensus judgment, or if the individual had moderate or severe mental retardation.
Diagnostic Reliability and Supplemental Clinical Measures
The cross-site reliability of diagnoses was measured by comparing the original site’s consensus BEFD with a new consensus BEFD produced blindly by a second site using blinded copies of all available case material for 32 cases from the first NIMH Genetics Initiative for Schizophrenia from Washington University (Cloninger et al. 1998) and 36 cases from the present study (total N=68). The 68 total cases included 40 with a diagnosis of SZ from the original site, 10 with an SA diagnosis, and 18 with mood, personality, and/or substance-use diagnoses, most with at least some symptoms suggestive of an SZ spectrum disorder. Cohen’s kappas (Cohen 1960) were 0.88 for SZ and 0.89 for SA, indicating no notable variation across sites.
STRP Genotyping
DNA was extracted from lymphoblastoid cell lines by the Rutgers University Cell and DNA Repository and was delivered in two “waves” to the Center for Inherited Disease Research (CIDR) for genotyping. CIDR’s methods and details of the CIDR map are available online (see CIDR Web site). The marker set consisted of ∼400 STRPs, with mean spacing of 9 cM (the largest gap was 18 cM) and a mean marker heterozygosity of 0.76. Family relationships were checked prior to the scan with a forensic panel of 12 markers. Samples with failed or very low amplification and contaminated samples were identified, and sex checks and a review of Mendelian discrepancies were performed when required. Failed wave I specimens were replaced when possible, and the entire family was regenotyped (for 23 families) in wave II. Each gel included four CEPH controls, blind duplicates, and blank PCR products. Over 5% of wave I specimens were genotyped a second time in wave II. Analyses with RELCHECK (Boehnke and Cox 1997; Broman and Weber 1998) to assess family relationships were performed with the full set of genotypes. Genotypes were removed in cases of Mendelian inconsistencies or unlikely genotypes (P<.01), as determined by MERLIN (multipoint engine for rapid likelihood inference) (Abecasis et al. 2002). Quality control analyses for waves I and II, respectively, showed missing data rates of 3.96% and 3.48%, genotypewise error rates of 0.1% and 0.03% based on blind duplicates, and Mendelian inconsistency rates of 0.72% and 0.67%.
Analysis of Population Substructure
Accurate estimation of marker-allele frequencies is critical when DNA is unavailable for some parents and when population substructure exists (Curtis and Sham 1996), and allele frequencies can be significantly different for EA and AA subjects (Goddard et al. 2000; Grigull et al. 2001). It was decided that separate allele-frequency estimates for these two ethnic groups would be established for all analyses. Therefore, we used the program STRUCTURE (Pritchard et al. 2000) to identify and exclude families that were outliers with respect to the two main ethnic clusters, because these families would otherwise have been analyzed with inappropriate frequency estimates. Self-reported ancestries were EA or AA, plus 10 families with Hispanic ancestry. STRUCTURE analyses were completed assuming K=2, 3, or 4 clusters by use of all autosomal STRP genotypes. Each run consisted of 10,000 burn-ins and 10,000 subsequent iterations. There were clear EA and AA clusters regardless of the value of K, with no additional clusters that reflected self-reported ethnicity. Eight of the 10 Hispanic families consistently clustered (posterior P>.85) with either the EA or AA group (or showed approximately equal EA or AA admixture). Thus, K=2 was assumed, and 10 runs of K=2 with different starting seeds produced no changes in family assignments to the clusters. Thirteen families with 1 or 2 ungenotyped parents were removed from linkage analyses because they did not attain the selected threshold of a posterior probability of group membership of at least 0.85. Two families with self-reported Asian ancestry (without genotyped parents) were also removed.
Confirmation of Biological Relationships
All participant-reported genetic relationships were verified via RELCHECK (Boehnke and Cox 1997; Broman and Weber 1998) for autosomal and X-linked STRPs from the forensic tests, and Mendelian consistency was verified via PEDCHECK (O’Connell and Weeks 1998) for autosomal and X-linked STRPs from the genome scan, respectively. On the basis of these checks, 26 families were excluded because they no longer contained an ASP (full or half) or because the “siblings” proved to be not related, and 6 families were excluded because the siblings had identical genotypes (because of either an unreported MZ twin relationship or an error during phlebotomy, blood submission, or processing). For the retained families, two nuclear pedigrees contained self-reported full-sibling ASPs that were proven to be half-sibling ASPs in the EA sample; 18 nuclear pedigrees contained self-reported full-sibling ASPs that were proven to be half-sibling ASPs in the AA sample. Finally, one large pedigree was split into subfamilies so that they could be analyzed by GENEHUNTER-PLUS (GH+) (Kong and Cox 1997), resulting in the creation of a “new” family; the subfamilies were treated as if they were unrelated to one another.
STRP Maps and Allele Renumbering
The STRP genotyping was performed in two waves. Briefly, in addition to three Y-linked markers and one pseudoautosomal marker not used in linkage analysis, 393 autosomal and X-linked STRPs were typed in wave I, and 400 in wave II. Genotypes for the 387 markers common to both waves were hand-checked for consistency of allele designations, and alleles were renumbered when necessary, including 114 for which renumbering was complex (see tables A1 and A2 in appendix A for detailed examples). For the genome-scan analysis, we assumed map positions provided by CIDR, which are similar to Marshfield map positions (Broman et al. 1998). For the linkage fine-mapping analysis, all positions were drawn or interpolated from the deCODE map (Kong et al. 2002), as discussed below.
STRP Allele-Frequency Estimation
Maximum-likelihood allele-frequency estimates were obtainedfrom the USERM13 subroutine of MENDEL (Lange et al. 1988; Boehnke 1991) separately for the EA and AA families. By likelihood-ratio testing, EA and AA allele frequencies differed at P<10-16 for 82.5% of the markers, at 10-16⩽P⩽.01 for 17% of the markers, and at P>.01 for only 0.5% (three) of the markers. Accordingly, for the linkage analysis of the combined EA and AA sample, we used separate allele-frequency estimates. For instance, if there were N alleles in the EA sample, we renumbered the AA alleles, starting with N+1. The sum of the EA and AA allele frequencies totaled 200% (table A2).
Linkage Analyses of STRPs and Determination of Empirical Significance Thresholds
Linkage analyses were performed using GH+ (Kong and Cox 1997) under the exponential-model option and the SALL scoring function, to compute Z likelihood-ratio (Zlr) scores for each map position. Zlr follows a normal distribution asymptotically under the null hypothesis of no linkage. All results are presented on the Zlr scale. The correspondence between Zlr and the log10 likelihood ratio of linkage is LOD=(Zlr)2/2ln(10). Information content (IC) was computed by GH+. The algorithm computes entropy for the probability distribution for the inheritance vectors of all pedigrees with or without genotypic data. IC is then computed by subtracting the ratio of these terms from 1. Map locations were drawn from the CIDR map (Broman et al. 1998) or were interpolated into that map.
The linkage analysis of all families combined was considered to be the primary analysis. Exploratory analyses of the EA and AA families separately were also performed. To determine empirical significance thresholds for Zlr scores, 5,000 replicates of the actual sample, map, and allele frequencies (including patterns of missing data) were generated using SIMULATE (Terwilliger et al. 1993) under the assumption of an absence of linkage. Each replicate was analyzed separately for all families (the planned primary analysis) and then for the AA and EA families separately. For the primary analysis, the Zlr threshold was 3.60 for “significant” linkage (expected in ⩽5% of genome scans) and 2.65 for “suggestive” linkage (expected ⩽1 times per genome scan) (Holmans et al. 2004). In comparison, the thresholds for EA and AA samples (without correction for multiple testing) were similar: 3.62 and 3.63, respectively, for significant linkage and 2.71 and 2.69, respectively, for suggestive linkage.
Also, as discussed below, a common paracentric inversion has been reported (Broman et al. 2003) in the region of chromosome 8p that produced the greatest evidence of linkage in the present study. One STRP in the CIDR map (D8S1469) is located in the middle of the typical inversion segment, and another STRP in the CIDR map (D8S1130) is close to the boundary of the typical inversion segment. The exact positions of these two STRPs, and the probability of recombination around them, would differ in individuals with and individuals without the inversion. The primary genome scan analysis results reported below excluded D8S1469, the less informative of the two inversion STRPs.
SNP Genotyping and Data Cleaning
For linkage fine mapping, genotyping of SNP markers was performed at Evanston Northwestern Healthcare’s Center for Psychiatric Genetics (Evanston, IL) by use of SNPlex (Applied Biosystems [ABI]). Using SNPbrowser software, version 1.0 (ABI), 585 SNPs (13 SNPlex pools) were selected to form a 0.5–0.6-cM map across 304 cM (255 Mb [Matise et al. 2003]) of chromosomes 4p16.3-p15.2 (44 cM; 27 Mb), 5p15.2-q13.3 (59 cM; 62 Mb), 8p23.3-q12.1 (78 cM; 59 Mb), 10p15.3-p14 (20 cM; 7 Mb), 10q25.3-q26.3 (36 cM; 18 Mb), and 11p13-q23.3 (67 cM; 82 Mb) from HapMap, Celera (Venter et al. 2001), or public databases (see National Center for Biotechnology Information [NCBI] dbSNP Web site, build 34) (table 1). Validated SNPs were selected if their minor-allele frequency (MAF) was reported to be >25% in databases for both EA and AA samples, and they were tested using bioinformatics for suitability for the SNPlex assay. Forty nanograms of fragmented DNA was dried down on each well of a 384-well plate. After phosphorylation of oligonucleotide ligation assay probes and universal linkers, allele-specific ligation and enzymatic clean-up were performed. PCR was performed with universal primers, and biotinylated amplicons were captured on streptavidin-coated plates. Single-stranded PCR products were hybridized with a set of fluorescently labeled, universal ZipChute probes that have a unique sequence corresponding to each SNP. ZipChute probes were eluted and separated on a 3730 DNA Analyzer (ABI), and genotypes were called by GeneMapper 3.5 (ABI), blind to diagnosis.
Table 1.
Fine-Mapping Summary
|
Chromosome |
||||||||
| Value | 4 | 5 | 8 | 10p | 10q | 11 | Total | Average |
| STRP Zlr peak group | AA | EA | EA+AA | EA | EA+AA | EA+AA | … | … |
| Peak STRP Zlr | 2.72 | 2.77 | 3.25 | 2.04 | 2.23 | 2.74 | … | … |
| Approximate 1-LOD linkage interval of STRP scan (cM)a: | ||||||||
| Start | 0 | 33.6 | 2.1 | 0 | 120.3 | 61.9 | … | … |
| End | 35.5 | 74.3 | 60.3 | 20.9 | 170.9 | 94.6 | … | … |
| Total approximate 1-LOD interval (cM)a | 35.5 | 40.7 | 58.2 | 20.9 | 50.6 | 32.7 | 238.6 | … |
| Fine-mapping region chosen (cM)b: | ||||||||
| Start | .0 | 24.9 | .0 | .0 | 134.7 | 43.2 | … | … |
| End | 43.6 | 84.0 | 77.9 | 20.0 | 170.9 | 110.1 | … | … |
| Total distance fine mapped (cM)b | 43.6 | 59.1 | 77.9 | 20.0 | 36.2 | 67.0 | 304 | … |
| Cytogenetic positionc | 4p16.3–4p15.2 | 5p15.2–5q13.3 | 8p23.3–8q12.1 | 10p15.3–10p14 | 10q25.3–10q26.3 | 11p13–11q23.3 | … | … |
| Physical distance (Mb)c: | ||||||||
| Start | .3 | 14.0 | .6 | .3 | 116.4 | 34.9 | … | … |
| End | 27.2 | 75.8 | 59.5 | 6.7 | 134.0 | 116.6 | … | … |
| Total distance fine mapped (Mb) | 26.8 | 61.8 | 58.9 | 6.4 | 17.7 | 81.8 | 253 | … |
| No. of genotyped SNPs | 91 | 120 | 131 | 33 | 71 | 139 | 585 | … |
| Initial SNP interval (cM)b | .48 | .49 | .59 | .61 | .51 | .48 | … | .52 |
| No. of SNPs failed for assay | 4 | 4 | 4 | 1 | 3 | 3 | 19 | … |
| No. of SNPs failed for low (<90%) genotyping rates | 6 | 5 | 5 | 3 | 0 | 4 | 23 | … |
| No. of SNPs with excessive Mendelian errors | 7 | 0 | 1 | 0 | 2 | 1 | 11 | … |
| No. of SNPs remaining with clean genotypes | 74 | 111 | 121 | 29 | 66 | 131 | 532 | … |
| Cleaned SNP interval (cM)b | .59 | .53 | .64 | .69 | .55 | .51 | … | .57 |
| Average genotyping rate of cleaned SNPs (%) | .987 | .988 | .980 | .988 | .988 | .988 | … | .986 |
| Mendelian error rate (per genotype) of cleaned SNPs (%) | .00015 | .00023 | .00012 | .00012 | .00017 | .00018 | … | .00017 |
| Unlikely-recombinants rate (per genotype) of cleaned SNPs (%) | .00048 | .00030 | .00042 | .00069 | .00075 | .00033 | … | .00044 |
| Average MAF of cleaned SNPs: | ||||||||
| EA | .37 | .35 | .36 | .34 | .35 | .38 | … | .36 |
| AA | .35 | .34 | .35 | .37 | .34 | .35 | … | .35 |
| No. of clean SNPs removed because of deviation from HWE: | ||||||||
| EA | 1 | 2 | 0 | 0 | 1 | 2 | 6 | … |
| AA | 0 | 1 | 3 | 1 | 0 | 1 | 6 | … |
| No. of clean SNPs removed because of adjacent SNP LD: | ||||||||
| EA | 9 | 21 | 12 | 6 | 11 | 13 | 72 | … |
| AA | 6 | 5 | 9 | 3 | 7 | 12 | 42 | … |
| No. of clean SNPs removed because of inversion (only chromosome 8): | ||||||||
| EA | 0 | 0 | 5 | 0 | 0 | 0 | 5 | … |
| AA | 0 | 0 | 5 | 0 | 0 | 0 | 5 | … |
| No. of final SNPs analyzed for linkage: | ||||||||
| EA | 64 | 88 | 104 | 23 | 54 | 116 | 449 | … |
| AA | 68 | 105 | 104 | 25 | 59 | 118 | 479 | … |
| Final SNP interval (cM)b: | ||||||||
| EA | .68 | .67 | .75 | .87 | .67 | .58 | … | .68 |
| AA | .64 | .56 | .75 | .80 | .61 | .57 | … | .63 |
| Final fine-map IC: | ||||||||
| EA | .83 | .85 | .85 | .80 | .83 | .85 | … | .84 |
| AA | .77 | .78 | .79 | .77 | .78 | .78 | … | .76 |
| Fine-mapping Zlr peak group | AA | EA | EA | EA | EA | EA+AA | … | … |
| Peak fine-mapping Zlr | 3.09 | 3.80 | 3.33 | 2.52 | 2.08 | 3.08 | … | … |
| SNP nearest to peak fine-mapping Zlr | rs7681266 | rs1027164 | rs7834209 | rs1155931 | rs1343418 | rs4275647 | … | … |
| Approximate 1-LOD interval after fine mapping (cM)b: | ||||||||
| Start | 1.0 | 44.9 | 5.1 | .0 | 134.1 | 69.7 | … | … |
| End | 32.0 | 76.7 | 46.0 | 23.2 | 153.5 | 89.2 | … | … |
| Total approximate 1-LOD interval (cM)b | 31.0 | 31.8 | 40.9 | 23.2 | 19.4 | 19.5 | 166 | … |
| Total narrowing of fine-mapping peak achieved (cM)b | 4.5 | 8.9 | 17.3 | −2.3 | 31.2 | 13.2 | 73 | … |
| Cytogenetic position of approximate 1-LOD interval after fine mappingc | 4p16.3–4p15.32 | 5p14.1–5q12.1 | 8p23.2–8p21.2 | 10p15.3–10p14 | 10q25.3–10q26.13 | 11q13.1–11q14.1 | … | … |
| Physical distance of approximate 1-LOD interval after fine mapping (Mb)c: | ||||||||
| Start | .00 | 25.22 | 2.54 | .00 | 115.78 | 63.69 | … | … |
| End | 16.65 | 61.62 | 25.93 | 9.27 | 126.53 | 82.59 | … | … |
| Total approximate 1-LOD interval (Mb)c | 16.7 | 36.4 | 23.4 | 9.3 | 10.8 | 18.9 | 115 | … |
cM values refer to the STRP map of CIDR (used for linkage scan).
cM values refer to the deCODE map (used for fine mapping).
Cytogenetic locations and Mb values are from the UCSC July 2003 freeze.
Mendelian errors and unlikely genotypes were evaluated using MERLIN (Abecasis et al. 2002), as described above. We first excluded 19 SNPs because of failed assays and 23 SNPs because of low call rates (<90%). Then, we excluded 11 SNPs because of excessive Mendelian errors (>1%). The remaining 532 SNPs had 0.017% Mendelian errors per SNP (range 0.00%–0.66%) and 0.044% unlikely recombinants (range 0.00%–0.58%) for all fine-mapping regions (table 1). Two families were excluded—one because of excessive Mendelian errors (due to a specimen swap) and one without genotyped parents, because of excessive unlikely recombinants. The final call rate for all 532 cleaned SNPs was 98.6% (range 92.1%–100.0%), with a total of 729,158 cleaned genotypes remaining for further analysis. The average interval between neighboring SNPs was 0.59 Mb for chromosome 4, 0.53 Mb for chromosome 5, 0.64 Mb for chromosome 8, 0.69 Mb for chromosome 10p, 0.55 Mb for chromosome 10q, and 0.51 Mb for chromosome 11. There were several sizable centromeric gaps where the genome sequence information was incomplete (a 3.9-Mb gap for chromosome 5, a 3.8-Mb gap for chromosome 8, and a 4.1-Mb gap for chromosome 11). The average MAF was 0.35 (range 0.04–0.50) for AA subjects and 0.36 (range 0.00–0.50) for EA subjects. We also evaluated the repeatability with 121 SNPs from the chromosome 8 fine mapping, in which we observed repeatability of 99.9% when 268 DNA samples were blindly regenotyped. Of 31,931 genotypes that were nonzero in both experiments, 31 were discrepant, with 3 changes involving both alleles in a homozygous subject and 28 others involving a change of only one of the two alleles in a heterozygous individual.
Deviations from Hardy-Weinberg equilibrium (HWE) were analyzed separately in EA and AA samples in a set of as many unrelated individuals as possible. Six SNPs were removed because of significant deviations at P<.01 in the EA sample, and 6 SNPs were likewise removed in the AA sample, including 2 SNPs with highly significant deviations in the AA sample (P=.00006 for rs1355305; P=.0002 for rs896044) (table 1).
Selection of a Map for Linkage Fine Mapping
We therefore undertook to select a subset of markers for fine-mapping analyses that had the highest heterozygosity (and hence the highest IC). Pairwise marker LD between the cleaned fine-mapping SNPs was analyzed with a set of unrelated individuals, separately for EA and AA samples, by use of ASSOCIATE (Ott 1985), which computes maximum-likelihood estimates of the LD parameter by the expectation-maximization algorithm (Dempster et al. 1977). For example, of 121 SNPs in the chromosome 8 fine-mapping region, significant (P<.05) pairwise LD was observed between 40 and 20 adjacent marker pairs in the EA and AA samples, respectively, which is consistent with other reports of greater LD in EA than in AA samples (Hinds et al. 2005).
A contiguous block of highly significant LD was observed in the EA sample across 3.31 Mb (∼6 cM), with six adjacent SNPs extending from rs2980438 (at 8.132 Mb) to rs7824640 (at 11.442 Mb) on chromosome 8. It appears likely that this LD block is due to the large polymorphic inversion that is observed with a frequency >20% in the European (Giglio et al. 2001; Broman et al. 2003) and Japanese (Sugawara et al. 2003) populations. The inversion typically spans 4.7 Mb between two low-copy-repeat regions, each of which contains several olfactory receptor genes (Giglio et al. 2001). Direct demonstration of the inversion by use of FISH is beyond the scope of the present study, but we inferred that inversions could be present in some unknown but substantial proportion of our EA subjects on the basis of the large LD block and the fact that recombination cannot occur between chromosomes that are heterozygous for an inversion, leading to LD if the inversion is common. We detected no significant LD in this region in AA subjects, which suggests that this inversion is infrequent in sub-Saharan Africa, similar to the 17q21.31 paracentric inversion reported recently (Stefansson et al. 2005).
In families with missing parental genotypes, linkage scores can be inflated if substantial LD is present between marker pairs (Huang et al. 2004; Dunn et al., in press). Recent work by one of us (Levinson and Holmans, in press) indicates, however, that there is no appreciable inflation of linkage scores if dense SNP maps are trimmed to eliminate marker pairs whose LD (as measured by r2) is >0.05. Accordingly, we used this criterion to trim our SNP maps separately for the EA and AA families by removing one of each adjacent pair of SNPs that showed this level of LD. Removal of a SNP will create a new adjacent pair, and these new pairs were also evaluated for LD, until all adjacent pairs had r2 estimates <0.05. For chromosome 8, a total of 17 SNPs were trimmed from both EA and AA families. In the EA families, 12 SNPs were removed because r2 was >0.05, and 5 were removed because they are located in the inversion (where there was strong LD). In the AA families, three SNPs were removed because they deviated from HWE (at P<.01), nine were removed because r2 was >0.05, and five were removed because they are located in the inversion. We retained a single SNP (hCV1965865) from the inversion region. However, we deleted this SNP in analyses that included both SNPs and STRPs, since an STRP in the inversion (D8S1130) is more polymorphic. Similarly, we had already deleted the less-informative inversion STRP (D8S1469) for the combined SNP and STRP analysis, leaving a total of 104 SNPs and 9 STRPs for the final chromosome 8p fine map. Fine-mapping linkage results are reported for the EA and AA families separately and combined.
For other fine-mapping regions, we followed similar rules to exclude SNPs that may be in LD with adjacent SNPs (table 1). The final numbers of SNPs used in fine-mapping analyses for the EA and AA samples, respectively, were 64 and 68 for chromosome 4, 88 and 105 for chromosome 5, 104 and 104 for chromosome 8, 23 and 25 for chromosome 10p, 54 and 59 for chromosome 10q, and 116 and 118 for chromosome 11, for a total of 449 and 479 cleaned fine-mapping SNPs without significant intermarker LD that proceeded to linkage analyses (table 1).
Correct specifications of marker-marker genetic distances are also critical for accurate multipoint linkage analysis. Genetic-mapping information was not available for most of our fine-mapping SNPs, and our sample did not include the kinds of complete and multigenerational pedigrees that ideally should be used to estimate genetic distances. The deCODE map includes genetic locations for a larger number of markers, on the basis of more meioses, than does the CIDR map. Therefore, we created a genetic and a physical map of the STRPs in the region, using the UniSTS database (see UniSTS Web site), deCODE map information for genetic locations, and the NCBI sequence map for physical positions. Genetic locations were assigned to SNPs by use of linear interpolation of physical positions in relation to the genetic locations of flanking STRPs.
Sharing of Biomaterials and Clinical Data
Biological materials, genotypes, and blinded clinical data will be made available to the scientific community by the NIMH Center for Collaborative Genetics Studies of Mental Disorders (see the Center's Web site).
Results
We report here results of the clinical characteristics of the sample, a genomewide linkage scan of SZ, and the fine mapping of 4p16.3-p15.2, 5p15.2-q13.3, 8p23.3-q12.1, 10p15.3-p14, 10q25.3-q26.3, and 11p13-q23.3.
Families and Individuals Included in Linkage Analyses
A total of 459 families met eligibility criteria and were included in the two waves of CIDR genotyping, although 4 of these families were removed because one of the siblings was no longer considered to be affected after further diagnostic review in preparation for dimensional analyses. After removal of families as described above, 409 families (408 independent families) were included in the linkage analysis: 263 (64.3%) EA and 146 (35.7%) AA. The 409 families resulted from the splitting of 1 family that was too large to be analyzed by GH+. A full description of the DNA specimens available from this data set, including those excluded from the present analysis, will be made available on the NIMH Human Genetics Initiative for Schizophrenia Web site. Table 2 describes the number of statistically independent affected full-sib pairs (AFSPs) and the number of all possible affected half-sib pairs (AHAPs) used in the linkage analysis. Statistically independent AFSPs were counted as S-1 for S affected full sibs in a sibship with only full sibs; all possible AHAPs were counted as S(S-1)/2 for S affected half sibs in a sibship with only half sibs. Because there is no optimal method for counting independent AHAPs when there are full and half sibs in the same family, all possible half-sib pairs have been counted in these cases (see fig. A1 in appendix A for an illustration and explanation). Linkage analyses included 403 independent AFSPs (279 EA and 124 AA), 100 AHAPs (15 EA and 85 AA), and 26 other genotyped affected relatives (parents, aunts, uncles, and offspring). Genotypes were available for 33% of all parents (45% EA and 17% AA), and 156 unaffected full or half sibs were genotyped (96 EA and 60 AA). Linkage analyses included genotypes from 1,380 subjects.
Table 2.
Counts of Families and Individuals Genotyped[Note]
|
No. of Genotyped Parents |
|||||||||
| Sample andNo. of AFSPs per Family | AFSPsa | AHAPsb | AffectedNonsibsc | UnaffectedSibsd | 0 | 1 | 2 | 3 | 4 |
| EA sample: | |||||||||
| 0 AFSPs | 0 | 15 | 2 | 5 | 5 | 8 | 1 | 0 | 0 |
| 1 AFSPs | 226 | 0 | 6 | 77 | 81 | 75 | 70 | 0 | 0 |
| 2 AFSPs | 18 | 0 | 1 | 5 | 7 | 7 | 4 | 0 | 0 |
| 3 AFSPs | 4 | 0 | 1 | 9 | 2 | 0 | 1 | 0 | 1 |
| 4 AFSPs | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 AFSPs | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| No. of families | 249 | NA | NA | NA | 96 | 90 | 76 | 0 | 1 |
| No. of individuals or pairs | 279 | 15 | 10 | 96 | 0 | 90 | 152 | 0 | 4 |
| AA sample: | |||||||||
| 0 AFSPs | 0 | 53 | 7 | 15 | 27 | 20 | 0 | 0 | 0 |
| 1 AFSPs | 80 | 17 | 8 | 29 | 57 | 18 | 5 | 0 | 0 |
| 2 AFSPs | 15 | 11 | 1 | 9 | 6 | 7 | 1 | 0 | 1 |
| 3 AFSPs | 2 | 4 | 1 | 2 | 0 | 2 | 0 | 0 | 0 |
| 4 AFSPs | 2 | 0 | 0 | 5 | 1 | 1 | 0 | 0 | 0 |
| 5 AFSPs | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| No. of families | 99 | NA | NA | NA | 91 | 48 | 6 | 0 | 1 |
| No. of individuals or pairs | 124 | 85 | 16 | 60 | 0 | 48 | 12 | 0 | 4 |
| Total sample: | |||||||||
| No. of families | 348 | NA | NA | NA | 187 | 138 | 82 | 0 | 2 |
| No. of individuals or pairs | 403 | 100 | 26 | 156 | 0 | 138 | 164 | 0 | 8 |
Note.— All counts reflect genotyped individuals only. A total of 1,380 individuals were genotyped in 409 families. The number of all (genotyped or not) parents was 920 (552 EA and 368 AA). NA = not applicable.
Genotyped independent AFSPs (n-1).
Genotyped AHAPs, all possible pairs; see main text for description of this counting method.
Genotyped affected nonsiblings; this includes persons who are not siblings or half siblings of affected individuals, so it includes parents, aunts, uncles, and offspring.
Genotyped unaffected siblings (and also unaffected half siblings) of affected individuals.
Descriptions of the 1,380 individuals who were genotyped are given in tables 3 and 4. Of the 409 families, 263 were EA, and 146 (from 145 independent families) were AA. The mean age at enrollment was 39.1 and 41.4 years for the EA and AA probands, respectively, with onset at ∼20 years of age for all groups of male and female probands, both EA and AA. As noted in table 3, 35% of fathers and 56% of mothers of the EA probands and 7% of fathers and 33% of mothers of the AA probands were genotyped. Overall, 4.9% of fathers and 6.6% of mothers received a diagnosis of SZ, and 1% of mothers received a diagnosis of SA. The “other” group consisted of 61 half sibs plus other non–first-degree relatives. In table 4, we display the characteristics of probands and siblings with SZ. The percentage of SZ-affected subjects with a mood disorder did not differ by ethnicity or between probands and relatives, but females had a higher percentage than males did (64.5% vs. 46.2%; χ21=24.8; P<.0001). All SA-affected subjects had mood episodes (manic, mixed, and/or major depressive), by definition. We used a general linear model to examine differences in age at enrollment, age at onset, duration of psychosis, and duration of mood episodes in the subjects with SZ and those with SA. The independent variables were ethnicity, sex, and familial relationship (probands vs. siblings). Both sex and ethnicity were significant predictors of duration of nonaffective psychosis (P<.01), with females and AA subjects having longer durations. No other variables were significant at the P<.01 level. There was a trend toward females having a longer duration of mood episodes (P=.03). Next, we compared the siblings with SZ and those with SA. The subjects with SA had a longer duration of mood episodes (P<.0001). The only other significant variable was ethnicity as a predictor of the duration of nonaffective psychosis (P=.02).
Table 3.
Clinical Characteristics of All Genotyped Individuals
|
Probands |
Siblings |
Fathers |
Mothers |
Othersa |
||||||
| Characteristic | EA | AA | EA | AA | EA | AA | EA | AA | EA | AA |
| No. of individuals | 263 | 145 | 368 | 193 | 92 | 10 | 148 | 48 | 38 | 78 |
| Percentage male | 68.8 | 53.4 | 57.7 | 48.2 | 100 | 100 | 0 | 0 | 60.5 | 44.3 |
| Mean (SD) age at enrollment (years) | 39.1 (10.8) | 41.4 (10.5) | 40.6 (10.8) | 43.3 (10.4) | 65.5 (10.2) | 69.6 (11.1) | 63.3 (11.1) | 60.8 (12.9) | 33.9 (17.5) | 38.5 (12.3) |
| Percentage with SZ | 100 | 100 | 62.7 | 63.2 | 4.4 | 10.0 | 2.7 | 18.8 | 50.0 | 64.6 |
| Percentage with SA | 0 | 0 | 12.1 | 10.4 | 0 | 0 | .7 | 2.1 | 13.2 | 6.3 |
| Percentage with substance dependence: | ||||||||||
| Any: | ||||||||||
| Males | 24.6 | 42.3 | 24.7 | 31.2 | 4.3 | 20.0 | … | … | 17.4 | 34.3 |
| Females | 16.9 | 23.5 | 15.3 | 20.0 | … | … | 4.1 | 8.3 | 13.3 | 25.0 |
| Alcohol | 15.8 | 26.7 | 12.4 | 15.5 | 4.3 | 20.0 | 2.7 | 4.2 | 5.3 | 16.5 |
| Cannabis | 7.5 | 8.2 | 7.0 | 7.3 | 0 | 0 | 0 | 2.1 | 5.3 | 6.3 |
| Cocaine | 2.3 | 12.3 | 1.6 | 10.9 | 0 | 0 | 0 | 0 | 0 | 8.8 |
| Amphetamine | 2.6 | 1.4 | 1.9 | 1.0 | 0 | 0 | 1.0 | 0 | 2.6 | 0 |
The “Others” group consists of half siblings and other non–first-degree relatives.
Table 4.
Additional Clinical Characteristics of Genotyped Affected Subjects
|
Probands |
Siblings |
|||||||
| EA |
AA |
EA |
AA |
|||||
| Affected Groupand Characteristic | Males | Females | Males | Females | Males | Females | Males | Females |
| SZ: | ||||||||
| No. of individuals | 182 | 81 | 78 | 67 | 155 | 77 | 66 | 54 |
| Mean age at enrollment (years) | 37.7 | 42.2 | 40.0 | 43.1 | 38.2 | 42.1 | 43.2 | 42.5 |
| Mean age at illness onset (years) | 20.3 | 20.3 | 19.6 | 22.2 | 20.1 | 21.1 | 19.3 | 20.3 |
| Mean duration of nonaffective psychosis (mo) | 207.5 | 258.9 | 238.1 | 253.9 | 215.4 | 246.6 | 276.4 | 253.9 |
| Percentage of subjects with mood episodes | 46.4 | 64.6 | 38.2 | 61.8 | 51.3 | 64.9 | 42.4 | 69.1 |
| Mean duration of mood episodes (mo) | 11.6 | 23.5 | 13.1 | 13.1 | 13.2 | 13.1 | 14.0 | 16.8 |
| SA: | ||||||||
| No. of individuals | 24 | 21 | 8 | 12 | ||||
| Mean age at enrollment (years) | 40.0 | 41.8 | 35.4 | 41.8 | ||||
| Mean age at illness onset (years) | 21.1 | 21.9 | 18.1 | 19.3 | ||||
| Mean duration of nonaffective psychosis (mo) | 218.4 | 234.3 | 182.6 | 222.4 | ||||
| Mean duration of mood episodesa (mo) | 94.8 | 123.6 | 89.3 | 115.7 | ||||
All subjects with SA had one or more mood episodes (manic, mixed, and/or major depressive), by definition.
Genome Scan
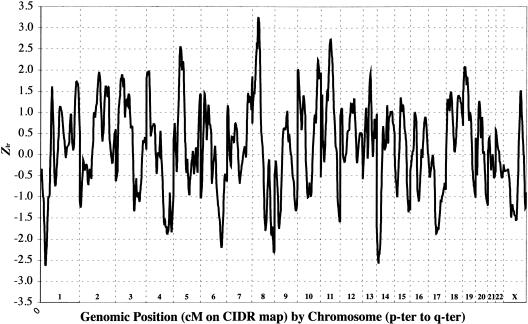
For the STRP genome scan, the CIDR map was used, although, for purposes of display alongside the fine-mapping SNPs, we converted to the deCODE map. Figure 1 displays the multipoint Zlr graph across the genome for all families, and table 5 lists the STRPs that attained a multipoint Zlr with nominal significance at P<.01 (results for all STRPs are found in table 6). We detected two chromosomal regions with suggestive evidence (Zlr⩾2.65) of linkage in the primary analyses of all families: 8p23.3-p12 and 11p11.2-q22.3. The mean IC for the STRP scan was 0.60 for the EA sample and 0.55 for the AA sample.
Figure 1.
Results of the STRP genome scan for SZ. Zlr scores for the entire sample are plotted on the Y-axis, and genomic position (cM on CIDR map) by chromosome (p-ter to q-ter) is plotted on the X-axis.
Table 5.
STRPs with a Multipoint Zlr of Nominal Significance at P < .01[Note]
|
Zlr |
|||||
| Chromosomeand Marker | Alias | Position(cM)a | EA Sample | AA Sample | Total Sample |
| 3: | |||||
| D3S2406 | GGAT2G03 | 103 | 2.33 | … | … |
| 4: | |||||
| D4S2366 | GATA22G05 | 13 | … | 2.66 | … |
| D4S403 | AFM157xg3 | 26 | … | 2.43 | … |
| 5: | |||||
| D5S2848 | GATA145D09 | 40 | 2.65 | … | … |
| D5S1470 | GATA7C06 | 45 | 2.72 | … | 2.52 |
| D5S2500 | GATA67D03 | 69 | 2.37 | … | … |
| 6: | |||||
| D6S2427 | GGAA15B08 | 54 | … | 2.54 | … |
| 8: | |||||
| D8S264 | AFM143xd8 | 1 | 2.56 | … | … |
| D8S262 | AFM127xh2 | 7 | 3.33 | … | … |
| D8S1130 | GATA25C10 | 22 | 3.06 | … | … |
| D8S1106 | GATA23D06 | 26 | 2.72 | … | … |
| D8S1145 | GATA72C10 | 37 | 3.09 | … | 2.47 |
| D8S560 | AFMa127ye5 | 43 | 2.96 | … | 2.51 |
| D8S1771 | AFMb320va5 | 50 | 2.95 | … | 3.24 |
| D8S1477 | GGAA20C10 | 60 | … | … | 2.64 |
| 11: | |||||
| D11S2371 | GATA90D07 | 76 | 2.66 | … | 2.74 |
| D11S2002 | GATA30G01 | 85 | … | … | 2.36 |
| 20: | |||||
| D20S851 | AFMa218yb5 | 25 | 2.34 | … | … |
Note.— A nominal P<.01 is a Zlr>2.326 here.
cM values refer to the STRP map of CIDR.
Table 6.
STRPs by Map Position and Multipoint Zlr for Linkage Scan
|
Zlr |
|||||
| Chromosomeand Marker | Alias | Position(cM)a | EA Sample | AA Sample | Total Sample |
| 1: | |||||
| D1S2845 | AFM344we9 | 9 | −.63 | −.18 | −.62 |
| D1S2660 | AFMa203yc1 | 11 | −.70 | −.05 | −.60 |
| D1S1612 | GGAA3A07 | 16 | −.26 | −.22 | −.34 |
| D1S1597 | GATA27E01 | 30 | −.46 | −1.14 | −1.05 |
| D1S3669 | GATA29A05 | 37 | −.72 | −1.88 | −1.70 |
| D1S552 | GGAT2A07 | 45 | −1.22 | −2.84 | −2.64 |
| D1S1622 | ATA20F08 | 57 | −.74 | −1.79 | −1.64 |
| D1S255 | AFM260zg5 | 65 | −.15 | −1.46 | −.99 |
| D1S3721 | GATA129H04 | 73 | −.23 | −1.34 | −.96 |
| D1S2134 | GATA72H07 | 76 | .09 | −.84 | −.41 |
| D1S3728 | GATA165C03 | 89 | 2.04 | −.06 | 1.62 |
| D1S1665 | GATA61A06 | 102 | .14 | .15 | .20 |
| D1S1728 | GATA109 | 109 | −.54 | −.36 | −.65 |
| D1S551 | GATA6A05 | 114 | −.71 | −.29 | −.75 |
| D1S1588 | ATA2E04 | 126 | −.02 | −.30 | −.19 |
| D1S1631 | ATA29D04 | 137 | 1.59 | −1.43 | .47 |
| D1S3723 | GATA176G01 | 140 | 2.13 | −1.37 | .96 |
| D1S534 | GATA12A07 | 152 | 1.28 | .04 | 1.06 |
| D1S1653 | GATA43A04 | 164 | 1.03 | −.03 | .83 |
| D1S1679 | GGAA5F09 | 171 | .70 | −.12 | .50 |
| D1S1677 | GGAA22G10 | 176 | .58 | −.17 | .37 |
| D1S1619 | ATA14D03 | 188 | .61 | −.98 | −.08 |
| D1S1589 | ATA4E02 | 192 | .76 | −1.00 | .03 |
| D1S518 | GATA7C01 | 202 | .67 | −.59 | .20 |
| D1S1660 | GATA48B01 | 212 | .87 | −.74 | .27 |
| D1S1647 | GATA25A11 | 216 | .96 | −.23 | .64 |
| C1S1248 | GATA124F08 | 226 | .86 | .41 | .94 |
| D1S2141 | GATA87F04 | 233 | .03 | .48 | .30 |
| D1S549 | GATA4H09 | 240 | .23 | −.14 | .10 |
| D1S3462 | ATA29C07 | 247 | .38 | −.12 | .23 |
| D1S235 | AFM203yg9 | 255 | 1.29 | .57 | 1.38 |
| D1S547 | GATA4A09 | 268 | 1.16 | 1.24 | 1.66 |
| D1S1609 | GATA50F11 | 275 | 1.10 | 1.30 | 1.65 |
| D1S2682 | AFMa272xc9 | 288 | −.25 | 1.22 | .45 |
| 2: | |||||
| D2S2976 | GATA165C07 | 4 | −.50 | −1.04 | −1.01 |
| D2S1780 | GATA72G11 | 10 | −.91 | −.88 | −1.25 |
| D2S2952 | GATA116B01 | 18 | −.61 | −.59 | −.84 |
| D2S1400 | GGAA20G10 | 28 | −.39 | .28 | −.13 |
| D2S1360 | GATA11H10 | 38 | −.98 | .68 | −.37 |
| D2S405 | GATA8F07 | 48 | −.57 | .42 | −.22 |
| D2S1788 | GATA86E02 | 56 | −.64 | .50 | −.25 |
| D2S1356 | ATA4F03 | 64 | −.60 | −.40 | −.72 |
| D2S1352 | ATA27D04 | 74 | −.14 | −.39 | −.33 |
| D2S441 | GATA8F03 | 87 | .25 | −1.35 | −.57 |
| D2S1394 | GATA69E12 | 91 | .69 | −1.09 | −.06 |
| D2S1777 | GATA71G04 | 99 | 1.34 | −1.38 | .25 |
| D2S1790 | GATA88G05 | 103 | 1.84 | −.91 | .95 |
| D2S2972 | GATA176C01 | 114 | 1.45 | −.51 | .87 |
| D2S410 | GATA4E11 | 125 | 1.66 | −.23 | 1.21 |
| D2S1328 | GATA27A12 | 133 | 1.71 | .43 | 1.64 |
| D2S442 | GATA8H05 | 147 | 1.70 | .77 | 1.82 |
| D2S1399 | GGAA20G04 | 152 | 1.45 | .24 | 1.32 |
| D2S1353 | ATA27H09 | 165 | 1.02 | −.95 | .31 |
| D2S1776 | GATA71D01 | 173 | 1.58 | −1.43 | .51 |
| D2S1391 | GATA65C03 | 186 | 2.25 | −.50 | 1.61 |
| D2S1384 | GATA52A04 | 200 | 1.38 | .40 | 1.36 |
| D2S2944 | GATA30E06 | 210 | 1.65 | .46 | 1.61 |
| D2S434 | GATA4G12 | 216 | .57 | −.12 | .40 |
| D2S1363 | GATA23D03 | 227 | .25 | −.38 | −.01 |
| D2S427 | GATA12H10 | 237 | −.19 | −.08 | −.20 |
| D2S2968 | GATA178G09 | 252 | −.44 | 1.38 | .47 |
| D2S125 | AFM112yd4 | 261 | −.22 | 1.32 | .60 |
| 3: | |||||
| D3S2387 | GATA22G12 | 6 | .15 | −1.23 | −.62 |
| D3S1560 | AFM217xd6 | 19 | .90 | −1.87 | −.37 |
| D3S4545 | GATA164B08 | 26 | 1.93 | −.87 | 1.04 |
| D3S1259 | AFM036yb8 | 37 | 1.35 | .55 | 1.43 |
| D3S3038 | GATA73D01 | 45 | 1.85 | .43 | 1.76 |
| D3S2432 | GATA27C08 | 58 | 1.91 | .08 | 1.60 |
| D3S1768 | GATA8B05 | 62 | 2.33 | −.10 | 1.82 |
| D3S2409 | ATA10H11 | 71 | 1.54 | −.65 | .88 |
| D3S1600 | AFM308xc9 | 86 | 1.77 | −.18 | 1.31 |
| D3S4542 | GATA148E04 | 90 | 1.73 | −.40 | 1.16 |
| D3S2406 | GGAT2G03 | 103 | 2.33 | −.86 | 1.37 |
| D3S4529 | GATA128C02 | 112 | .96 | −.25 | .62 |
| D3S2459 | GATA68D03 | 119 | .34 | .66 | .66 |
| D3S3045 | GATA84B12 | 124 | .21 | .86 | .67 |
| D3S2460 | GATA68F07 | 135 | −.98 | .03 | −.78 |
| D3S4523 | ATA34G06 | 138 | −1.47 | −.25 | −1.34 |
| D3S1764 | GATA4A10 | 153 | −.08 | −.83 | −.56 |
| D3S1744 | GATA3C02 | 161 | −.35 | −1.23 | −.99 |
| D3S1746 | GATA8F01 | 170 | −.41 | −.98 | −.89 |
| D3S1763 | GATA3H01 | 177 | −.87 | −.71 | −1.12 |
| D3S3053 | GATA92B06 | 182 | −.83 | −.38 | −.90 |
| D3S2427 | GATA22F11 | 188 | −.77 | −.10 | −.67 |
| D3S1262 | AFM059xa9 | 201 | .48 | −.11 | .32 |
| D3S2398 | GATA6G12 | 209 | .58 | −.33 | .27 |
| D3S2418 | ATA22E01 | 216 | .80 | −.37 | .42 |
| D3S1311 | AFM254ve1 | 225 | .35 | −.27 | .12 |
| 4: | |||||
| D4S2366 | GATA22G05 | 13 | .35 | 2.66 | 1.78 |
| D4S403 | AFM157xg3 | 26 | .57 | 2.43 | 1.88 |
| D4S2639 | GATA90B10 | 33 | .85 | 2.15 | 1.95 |
| D4S391 | AFM016xf3 | 44 | .16 | .52 | .44 |
| D4S2632 | GATA72G09 | 51 | .54 | .23 | .56 |
| D4S1627 | GATA7D01 | 60 | .32 | .71 | .67 |
| D4S3248 | GATA28F03 | 73 | .94 | −.17 | .67 |
| D4S2367 | GATA24H01 | 78 | .58 | −.49 | .20 |
| D4S3243 | GATA10G07 | 88 | .29 | −1.22 | −.46 |
| D4S2361 | ATA2A03 | 93 | .57 | −.75 | .02 |
| D4S1647 | GATA2F11 | 105 | −.06 | .06 | −.02 |
| D4S2623 | GATA62A12 | 114 | .16 | .72 | .56 |
| D4S2394 | ATA26B08 | 130 | −.30 | −.76 | −.67 |
| D4S1644 | GATA11E09 | 143 | −.73 | −1.87 | −1.67 |
| D4S1625 | GATA107 | 146 | −.50 | −1.93 | −1.52 |
| D4S1629 | GATA8A05 | 158 | −.36 | −2.47 | −1.69 |
| D4S2368 | GATA27G03 | 168 | −1.05 | −1.69 | −1.83 |
| D4S2431 | GGAA19H07 | 176 | −.89 | −1.59 | −1.64 |
| D4S2417 | GATA42H02 | 182 | −.08 | −1.46 | −.90 |
| D4S408 | AFM165xc11 | 195 | −1.39 | −1.22 | −1.84 |
| D4S1652 | GATA5B02 | 208 | .28 | −1.92 | −.85 |
| 5: | |||||
| D5S2488 | ATA20G07 | 0 | 1.17 | −1.09 | .32 |
| D5S2849 | GATA145D10 | 8 | 1.87 | −1.28 | .75 |
| D5S2505 | GATA84E11 | 14 | 1.33 | −1.45 | .23 |
| D5S817 | GATA3E10 | 23 | .39 | −1.28 | −.40 |
| D5S2845 | GATA134B03 | 36 | 1.97 | −.35 | 1.38 |
| D5S2848 | GATA145D09 | 40 | 2.65 | −.32 | 1.95 |
| D5S1470 | GATA7C06 | 45 | 2.72 | .52 | 2.52 |
| D5S1457 | GATA21D04 | 59 | 2.10 | .48 | 2.00 |
| D5S2500 | GATA67D03 | 69 | 2.37 | .33 | 2.14 |
| D5S424 | AFM214zg9 | 82 | .40 | .06 | .36 |
| D5S641 | AFM284VD1 | 92 | −.90 | .52 | −.43 |
| D5S1725 | GATA89G08 | 98 | −.67 | .76 | −.10 |
| D5S1503 | GATA61B11 | 108 | −1.14 | .27 | −.76 |
| D5S1453 | ATA4D10 | 115 | −1.75 | .82 | −.96 |
| D5S2501 | GATA68A03 | 117 | −1.30 | .76 | −.62 |
| D5S1505 | GATA62A04 | 130 | −1.00 | 1.14 | −.15 |
| D5S816 | GATA2H09 | 139 | .27 | −.23 | .09 |
| D5S1480 | ATA23A10 | 147 | −.27 | −.42 | −.46 |
| D5S820 | GATA6E05 | 160 | .85 | −.92 | .17 |
| D5S1471 | GATA7H10 | 172 | 1.44 | −2.69 | −.42 |
| D5S1456 | GATA11A11 | 175 | 1.82 | −2.17 | .25 |
| D5S211 | Mfd154 | 183 | 2.23 | −1.26 | 1.09 |
| D5S408 | AFM164xb8 | 195 | 2.01 | −.34 | 1.41 |
| 6: | |||||
| D6S942 | UT654 | 0 | −1.46 | .27 | −1.03 |
| F13A1 | SE30 | 9 | −.93 | .01 | −.74 |
| D6S2434 | ATA50C05 | 25 | .79 | 1.36 | 1.44 |
| D6S1660 | AFMb355wg5 | 40 | −.02 | 1.16 | .66 |
| D6S2439 | GATA163B10 | 42 | −.05 | .66 | .35 |
| D6S2427 | GGAA15B08 | 54 | −.45 | 2.54 | 1.18 |
| D6S1017 | GGAT3H10 | 63 | −.43 | 1.72 | .70 |
| D6S2410 | GATA11E02 | 73 | .04 | .91 | .55 |
| D6S1053 | GATA64D02 | 80 | −.74 | −.13 | −.68 |
| D6S1031 | ATA28B11 | 89 | −.89 | .87 | −.24 |
| D6S1056 | GATA68H04 | 103 | −.50 | 1.06 | .20 |
| D6S1021 | ATA11D10 | 112 | −1.04 | .35 | −.63 |
| D6S474 | GATA31 | 119 | −1.16 | −.08 | −1.00 |
| D6S1040 | GATA23F08 | 129 | −.45 | −1.71 | −1.30 |
| D6S1009 | GATA32B03 | 138 | −.57 | −1.98 | −1.58 |
| C6S1848 | GATA184A08 | 146 | −1.57 | −1.41 | −2.10 |
| D6S2436 | GATA165G02 | 155 | −1.79 | −.84 | −1.94 |
| D6S1035 | ATA6C09 | 165 | −1.05 | −.23 | −.98 |
| D6S1277 | GATA81B01 | 173 | −.09 | −.64 | −.45 |
| D6S1027 | ATA22G07 | 187 | −.82 | .08 | −.61 |
| 7: | |||||
| D7S2477 | AFMb035xb9 | 0 | −.23 | 1.52 | .75 |
| D7S3056 | GATA24F03 | 7 | .65 | 1.09 | 1.16 |
| D7S513 | AFM217yc5 | 18 | −.19 | .22 | −.02 |
| D7S3051 | GATA137H02 | 29 | −.28 | −.07 | −.26 |
| D7S1802 | GATA41G07 | 33 | .33 | .27 | .43 |
| D7S1808 | GGAA3F06 | 42 | .61 | −.16 | .40 |
| D7S817 | GATA13G11 | 50 | .58 | −.35 | .28 |
| D7S2846 | GATA31A10 | 58 | 1.00 | −.15 | .75 |
| D7S1818 | GATA24D12 | 70 | .73 | −.01 | .60 |
| D7S3046 | GATA118G10 | 79 | .48 | −.60 | .04 |
| D7S2204 | GATA73D10 | 91 | −.20 | −.90 | −.69 |
| D7S2212 | GATA87D11 | 95 | −.46 | −.29 | −.55 |
| D7S821 | GATA5D08 | 109 | −.42 | .26 | −.20 |
| D7S1799 | GATA23F05 | 114 | −.40 | .45 | −.06 |
| D7S3061 | GGAA6D03 | 128 | .13 | .16 | .20 |
| D7S1804 | GATA43C11 | 137 | −.32 | .33 | −.06 |
| D7S1824 | GATA32C12 | 150 | .94 | 1.20 | 1.47 |
| D7S2195 | GATA112F07 | 155 | .69 | 1.13 | 1.22 |
| D7S3070 | GATA189C06 | 163 | .37 | 1.80 | 1.37 |
| D7S3058 | GATA30D09 | 174 | −.29 | 2.20 | 1.05 |
| D7S559 | Mfd265 | 182 | 1.12 | 1.60 | 1.85 |
| 8: | |||||
| D8S264 | AFM143xd8 | 1 | 2.56 | −2.34 | .75 |
| D8S262 | AFM127xh2 | 7 | 3.33 | −2.61 | 1.19 |
| D8S1130 | GATA25C10 | 22 | 3.06 | −1.92 | 1.43 |
| D8S1106 | GATA23D06 | 26 | 2.72 | −.86 | 1.76 |
| D8S1145 | GATA72C10 | 37 | 3.09 | −.04 | 2.47 |
| D8S560 | AFMa127ye5 | 43 | 2.96 | .28 | 2.51 |
| D8S1771 | AFMb320va5 | 50 | 2.95 | 1.45 | 3.24 |
| D8S1477 | GGAA20C10 | 60 | 2.28 | 1.36 | 2.64 |
| D8S1110 | GATA8G10 | 67 | .51 | .27 | .57 |
| D8S1113 | GGAA8G07 | 78 | .08 | .09 | .12 |
| D8S1136 | GATA41A01 | 82 | −.53 | .57 | −.10 |
| D8S2324 | GATA14E09 | 94 | −1.81 | −.26 | −1.62 |
| D8S1119 | ATA19G07 | 101 | −1.91 | −.44 | −1.80 |
| C8S14 | GAAT1A4 | 110 | −1.26 | −.24 | −1.18 |
| D8S1132 | GATA26E03 | 119 | .49 | −1.12 | −.24 |
| D8S592 | GATA6B02 | 125 | .60 | −1.03 | −.11 |
| D8S1179 | GATA7G07 | 135 | −.51 | −1.60 | −1.34 |
| D8S1720 | AFMa197wg5 | 141 | −.40 | −2.64 | −1.88 |
| D8S256 | AFM073yb7 | 148 | −.37 | −2.35 | −1.71 |
| D8S1108 | GATA50D10 | 154 | −.86 | −2.28 | −2.06 |
| D8S373 | UT721 | 164 | −1.34 | −2.05 | −2.26 |
| 9: | |||||
| D9S1779 | AFM026tg9 | 0 | −.46 | −.21 | −.49 |
| D9S1871 | AFM345ta9 | 8 | −.68 | .67 | −.14 |
| D9S2169 | GATA62F03 | 14 | −.78 | .42 | −.37 |
| C9S1879 | GATA187D09 | 22 | −.97 | −.06 | −.83 |
| D9S925 | GATA27A11 | 32 | −1.47 | −.97 | −1.76 |
| D9S1121 | GATA87E02 | 44 | −1.31 | −.36 | −1.28 |
| D9S1118 | GATA71E08 | 58 | .44 | .21 | .48 |
| D9S301 | GATA7D12 | 66 | 1.22 | −.59 | .63 |
| D9S1122 | GATA89A11 | 76 | .71 | .02 | .59 |
| D9S922 | GATA21F05 | 80 | .64 | .21 | .64 |
| D9S1120 | GATA81C04 | 89 | .63 | .91 | 1.04 |
| D9S1796 | AFMa210ze5 | 98 | −.06 | .59 | .30 |
| D9S1786 | AFMa137yb9 | 104 | .03 | .79 | .49 |
| D9S938 | GGAA22E01 | 111 | −.39 | .77 | .13 |
| D9S930 | GATA48D07 | 120 | −.31 | .32 | −.07 |
| D9S934 | GATA64G07 | 128 | .04 | −.90 | −.48 |
| D9S1825 | AFMb029xg1 | 136 | .28 | −1.43 | −.60 |
| D9S2157 | ATA59H06 | 147 | −.26 | −1.70 | −1.20 |
| D9S1826 | AFMB030ZG9 | 160 | −.57 | −1.20 | −1.17 |
| D9S1838 | AFMb303zg9 | 164 | −.15 | −1.00 | −.71 |
| 10: | |||||
| D10S1435 | GATA88F09 | 4 | 2.01 | .68 | 2.02 |
| D10S189 | AFM063xf4 | 19 | 1.44 | .22 | 1.30 |
| D10S1412 | ATA31G11 | 28 | .84 | .31 | .87 |
| D10S2325 | GAAT5F06 | 33 | .88 | −.23 | .58 |
| D10S1423 | GATA70E11 | 46 | 1.14 | .83 | 1.41 |
| D10S1426 | GATA73E11 | 59 | .19 | 1.27 | .88 |
| D10S1208 | ATA5A04 | 63 | −.42 | .39 | −.11 |
| D10S1221 | ATA21A03 | 76 | −1.59 | .54 | −.99 |
| D10S1225 | ATA24F10 | 81 | −1.61 | .51 | −1.01 |
| C10S1218 | GATA121A08 | 88 | −1.58 | 1.07 | −.66 |
| D10S1432 | GATA87G01 | 94 | −1.37 | .49 | −.83 |
| D10S2327 | GGAT1A4 | 101 | −1.43 | .28 | −.99 |
| D10S2470 | GATA115E01 | 113 | −.35 | .60 | .06 |
| D10S1239 | GATA64A09 | 125 | 1.06 | .13 | .94 |
| D10S1237 | GATA48G07 | 135 | .94 | −.03 | .75 |
| D10S1656 | AFMa184xd9 | 149 | 1.55 | 1.66 | 2.23 |
| D10S217 | AFM212XD6 | 158 | .68 | 2.10 | 1.79 |
| D10S1248 | GGAA23C05 | 165 | 1.32 | 1.44 | 1.92 |
| D10S212 | AFM198zb4 | 171 | 1.48 | 1.18 | 1.89 |
| 11: | |||||
| D11S2362 | ATA33B03 | 9 | −.69 | .13 | −.49 |
| D11S1999 | GATA23F06 | 17 | 1.85 | −.13 | 1.42 |
| D11S1981 | GATA48E02 | 21 | .96 | −.69 | .38 |
| C11S348 | ATA34E08 | 33 | −.20 | −.04 | −.18 |
| D11S1392 | GATA6B09 | 43 | .69 | .14 | .64 |
| D11S1993 | ATA1B07 | 54 | 1.45 | .64 | 1.54 |
| D11S1344 | AFM298vc9 | 58 | 1.18 | .56 | 1.28 |
| D11S2371 | GATA90D07 | 76 | 2.66 | .98 | 2.74 |
| D11S2002 | GATA30G01 | 85 | 1.94 | 1.34 | 2.36 |
| D11S2000 | GATA28D01 | 101 | .20 | 1.39 | .99 |
| D11S1391 | GATA4E01 | 105 | −.12 | 1.61 | .86 |
| D11S1998 | GATA23E06 | 113 | −.51 | 1.02 | .20 |
| D11S4464 | GATA64D03 | 123 | −.63 | .92 | .04 |
| D11S912 | AFM157xh6 | 131 | −1.47 | .14 | −1.11 |
| D11S968 | AFM109xc3 | 148 | −1.58 | −.54 | −1.60 |
| 12: | |||||
| D12S372 | GATA4H03 | 6 | .26 | 1.21 | .90 |
| C12S4912 | GATA49D12 | 18 | .48 | 1.15 | 1.06 |
| D12S391 | GATA11H08 | 26 | −.22 | .21 | −.05 |
| D12S373 | GATA6C01 | 36 | −.25 | .03 | −.19 |
| D12S1042 | ATA27A06 | 49 | −.06 | .16 | .04 |
| C12S916 | GATA91H06 | 56 | .45 | .25 | .51 |
| D12S398 | GGAT2G06 | 68 | .24 | .63 | .57 |
| D12S1294 | GATA73H09 | 78 | 1.01 | .37 | 1.04 |
| D12S375 | GATA3F02 | 81 | 1.25 | .33 | 1.21 |
| D12S1052 | GATA26D02 | 83 | 1.31 | .25 | 1.20 |
| D12S1064 | GATA63D12 | 95 | .99 | .28 | .97 |
| D12S1300 | GATA85A04 | 104 | 2.08 | −.62 | 1.33 |
| PAH | PAH.PCR9 | 109 | 1.52 | −.49 | .97 |
| D12S2070 | ATA25F09 | 125 | .24 | −.66 | −.19 |
| D12S395 | GATA4H01 | 137 | .12 | .34 | .29 |
| D12S2078 | GATA32F05 | 150 | 1.11 | −.70 | .51 |
| D12S1045 | ATA29A06 | 161 | 1.05 | −.86 | .38 |
| D12S392 | GATA13D05 | 166 | .57 | −1.34 | −.30 |
| 13: | |||||
| D13S1243 | AFMa217yb5 | 10 | −.62 | −.12 | −.58 |
| D13S217 | AFM205XH12 | 17 | −.55 | .70 | −.03 |
| D13S1493 | GGAA29H03 | 26 | −.03 | 1.03 | .59 |
| D13S894 | GATA86H01 | 33 | .42 | 1.32 | 1.11 |
| D13S325 | GATA6B07 | 39 | .42 | .37 | .56 |
| D13S788 | GATA29A09 | 46 | .61 | −.97 | −.09 |
| D13S800 | GATA64F08 | 56 | 1.82 | .09 | 1.53 |
| D13S317 | GATA7G10 | 64 | 1.78 | .79 | 1.92 |
| D13S793 | GATA43H03 | 76 | .47 | −.06 | .36 |
| D13S779 | ATA26D07 | 83 | −.08 | .03 | −.05 |
| D13S796 | GATA51B02 | 94 | .09 | 1.30 | .85 |
| D13S1265 | AFMb318xh5 | 99 | −.39 | 1.14 | .37 |
| D13S285 | AFM309va9 | 111 | .11 | 1.44 | .96 |
| 14: | |||||
| D14S742 | GATA74E02 | 12 | −.99 | −1.35 | −1.58 |
| D14S1280 | GATA31B09 | 26 | −1.89 | −1.80 | −2.58 |
| D14S608 | GATA43H01 | 28 | −1.65 | −1.74 | −2.35 |
| D14S599 | ATA29G03 | 41 | −1.40 | −.94 | −1.68 |
| D14S306 | GATA4B04 | 44 | −.42 | −1.07 | −.96 |
| D14S587 | GGAA10C09 | 56 | .95 | −.17 | .67 |
| D14S592 | ATA19H08 | 67 | .85 | −.99 | .10 |
| D14S588 | GGAA4A12 | 76 | .86 | −.76 | .23 |
| D14S53 | Mfd190 | 86 | 1.01 | .61 | 1.17 |
| D14S606 | GATA30A03 | 92 | −.21 | .72 | .26 |
| C14S1937 | GATA193A07 | 96 | −.35 | 1.84 | .81 |
| D14S617 | GGAA21G11 | 106 | .33 | 1.09 | .92 |
| D14S1434 | GATA168F06 | 113 | .23 | 1.36 | 1.00 |
| D14S1426 | GATA136B01 | 126 | .87 | .05 | .74 |
| D14S1007 | AFMb002zf1 | 138 | .22 | .52 | .48 |
| 15: | |||||
| D15S128 | AFM273yf9 | 6 | 1.57 | −2.18 | .02 |
| D15S822 | GATA88H02 | 12 | .88 | −1.85 | −.38 |
| D15S165 | AFM248vc5 | 20 | .79 | −2.12 | −.64 |
| C15S503 | GATA50C03 | 35 | −.48 | −1.03 | −1.00 |
| D15S659 | GATA63A03 | 43 | 1.11 | −1.19 | .21 |
| D15S643 | GATA50G06 | 52 | 1.50 | .20 | 1.35 |
| D15S1507 | GATA151F03 | 60 | .91 | .48 | 1.02 |
| D15S131 | AFM262xb1 | 71 | .90 | .76 | 1.18 |
| D15S655 | ATA28G05 | 83 | .54 | .28 | .61 |
| D15S652 | ATA24A08 | 90 | .26 | .13 | .29 |
| D15S816 | GATA73F01 | 101 | −.62 | −.37 | −.72 |
| D15S657 | GATA22F01 | 105 | −1.28 | −.56 | −1.36 |
| D15S966 | AFMa140ye5 | 112 | −.55 | −1.08 | −1.08 |
| D15S642 | GATA27A03 | 122 | −.72 | −1.27 | −1.32 |
| 16: | |||||
| D16S3401 | 16PTEL06 | 0 | .65 | −1.54 | −.39 |
| D16S2616 | ATA41E04 | 11 | 1.02 | −1.57 | −.10 |
| D16S748 | ATA3A07 | 23 | 1.26 | −1.66 | .08 |
| D16S3103 | AFMb337zc9 | 32 | 1.51 | −.38 | 1.00 |
| D16S403 | AFM049xd2 | 44 | 1.17 | −.40 | .74 |
| D16S769 | GATA71H05 | 51 | .08 | −1.32 | −.70 |
| D16S540 | GATA7B02 | 58 | −.58 | −.89 | −.99 |
| D16S3396 | ATA55A11 | 64 | −.77 | −.82 | −1.10 |
| D16S3253 | GATA22F09 | 72 | −.67 | −.15 | −.63 |
| C16S1385 | GATA138C05 | 81 | .13 | .04 | .13 |
| D16S2624 | GATA81D12 | 88 | .27 | −.52 | −.07 |
| D16S3096 | AFMb322wb9 | 99 | .98 | −.16 | .70 |
| D16S3091 | AFMB297ZC1 | 111 | 1.52 | −.57 | .89 |
| D16S539 | GATA11C06 | 125 | .54 | −.65 | .07 |
| D16S2621 | GATA71F09 | 130 | .31 | −.51 | −.03 |
| 17: | |||||
| D17S1308 | GTAT1A05 | 1 | −.02 | .00 | −.01 |
| D17S1298 | GAAT2C03 | 11 | −.23 | −.59 | −.54 |
| D17S974 | GATA8C04 | 22 | .10 | −.64 | −.29 |
| D17S1303 | GATA64B04 | 24 | .46 | −.93 | −.17 |
| D17S799 | AFM192yh2 | 32 | 1.00 | −1.38 | −.01 |
| D17S2196 | GATA185H04 | 45 | −.03 | −.67 | −.41 |
| D17S975 | GGAT2C07 | 51 | −.68 | −1.50 | −1.43 |
| D17S1880 | AFMa072zh9 | 53 | −.66 | −1.85 | −1.61 |
| D17S1299 | GATA25A04 | 62 | −1.25 | −1.46 | −1.85 |
| D17S2180 | ATC6A06 | 67 | −1.41 | −.97 | −1.71 |
| D17S1290 | GATA49C09 | 82 | −1.42 | −.54 | −1.46 |
| D17S2193 | ATA43A10 | 89 | −1.12 | −.29 | −1.07 |
| D17S1301 | GATA28D11 | 100 | −1.20 | −.01 | −.99 |
| D17S784 | AFM044xg3 | 117 | −.12 | −.95 | −.66 |
| D17S928 | AFM217yd10 | 126 | −.41 | −.84 | −.83 |
| 18: | |||||
| C18S1781 | GATA178F11 | 3 | −.13 | −.57 | −.43 |
| D18S976 | GATA88A12 | 13 | 1.82 | −.34 | 1.28 |
| D18S843 | ACT1A01 | 28 | .97 | .42 | 1.03 |
| D18S542 | GATA11A06 | 41 | .58 | 1.77 | 1.49 |
| D18S877 | GATA64H04 | 54 | .65 | .30 | .70 |
| D18S535 | GATA13 | 64 | .19 | −.15 | .07 |
| D18S851 | GATA6D09 | 75 | .42 | −.04 | .32 |
| D18S858 | ATA23G05 | 80 | .81 | .03 | .68 |
| D18S862 | ATA7D07 | 89 | 1.74 | −.06 | 1.37 |
| D18S1364 | GATA7E12 | 99 | 1.26 | .52 | 1.33 |
| C18S822 | ATA82B02 | 107 | 1.39 | .20 | 1.24 |
| D18S1371 | GATA177C03 | 116 | 1.32 | −.02 | 1.06 |
| D18S1390 | 18QTEL11 | 126 | .79 | .09 | .70 |
| 19: | |||||
| D19S591 | GATA44F10 | 10 | .86 | 1.24 | 1.41 |
| D19S1034 | GATA21G05 | 21 | 1.35 | 1.67 | 2.07 |
| D19S586 | GATA23B01 | 33 | 1.31 | 1.00 | 1.65 |
| D19S714 | GATA66B04 | 42 | 1.28 | 1.30 | 1.80 |
| D19S433 | GGAA2A03 | 52 | 1.01 | −.02 | .81 |
| D19S245 | Mfd235 | 59 | 1.23 | .00 | 1.01 |
| D19S178 | Mfd139 | 68 | 1.20 | −.39 | .73 |
| D19S246 | Mfd232 | 78 | −.52 | .20 | −.32 |
| D19S589 | GATA29B01 | 88 | −1.09 | .26 | −.75 |
| D19S254 | Mfd238 | 101 | −.75 | −.70 | −1.01 |
| 20: | |||||
| D20S103 | AFM077xd3 | 2 | 1.09 | −.86 | .38 |
| D20S482 | GATA51D03 | 12 | .42 | −1.48 | −.48 |
| D20S851 | AFMa218yb5 | 25 | 2.34 | −1.12 | 1.26 |
| D20S604 | GATA81E09 | 33 | 2.21 | −1.19 | 1.10 |
| D20S470 | GGAA7E02 | 39 | 1.55 | −1.49 | .39 |
| D20S477 | GATA29F06 | 48 | 1.56 | −.65 | .89 |
| D20S478 | GATA42A03 | 54 | .71 | −.95 | .02 |
| D20S481 | GATA47F05 | 62 | 1.30 | −1.72 | .08 |
| D20S480 | GATA45B10 | 80 | −.17 | −1.65 | −1.07 |
| D20S451 | UT254 | 90 | −.32 | −1.61 | −1.20 |
| 21: | |||||
| D21S1432 | GATA11C12 | 3 | .20 | −.18 | .05 |
| D21S1437 | GGAA3C07 | 13 | .93 | −.60 | .41 |
| D21S2052 | GATA129D11 | 25 | .49 | −1.38 | −.37 |
| D21S1440 | ATA27F01 | 37 | .25 | −.33 | .01 |
| D21S266 | AFM234xg9 | 46 | −.18 | −.70 | −.56 |
| D21S1446 | GATA70B08 | 58 | −.52 | .08 | −.38 |
| 22: | |||||
| D22S420 | AFM217xf4 | 4 | 1.22 | −.70 | .59 |
| D22S345 | MFD313 | 19 | 1.10 | −1.23 | .16 |
| D22S689 | GATA21F03 | 29 | 1.29 | −1.65 | .07 |
| D22S685 | GATA6F05 | 32 | 1.19 | −1.88 | −.15 |
| D22S683 | GATA11B12 | 36 | 1.11 | −1.82 | −.16 |
| D22S683 | GGAT3C10 | 46 | .34 | −1.34 | −.53 |
| D22S1169 | AFMb337zh9 | 61 | −.31 | .00 | −.26 |
| 23: | |||||
| DXS9895 | GATA124B04 | 9 | −.39 | .20 | −.22 |
| DXS9902 | GATA175D03 | 22 | −.52 | .14 | −.37 |
| DXS9896 | GATA124E07 | 40 | .02 | −.55 | −.34 |
| DXS1068 | AFM238yc11 | 53 | −.68 | −.20 | −.68 |
| DXS6810 | GATA69C12 | 64 | −1.63 | −.22 | −1.49 |
| CXS1444 | GATA144D04 | 71 | −1.67 | .34 | −1.17 |
| DXS7132 | GATA72E05 | 83 | −1.47 | −.22 | −1.34 |
| DXS6800 | GATA31D10 | 93 | −1.33 | −.57 | −1.43 |
| DXS6789 | GATA31F01 | 104 | −.82 | −1.39 | −1.51 |
| DXS6797 | GATA10C11 | 112 | −.10 | −.98 | −.69 |
| CXS1725 | GATA172D05 | 116 | −.02 | .27 | .11 |
| DXS1001 | AFM248we5 | 130 | 1.53 | .55 | 1.53 |
| DXS1047 | AFM150xf10 | 143 | .49 | .48 | .65 |
| DXS8044 | AFM203ze11 | 143 | .48 | .47 | .64 |
| CXS318 | GATA31E08 | 154 | −.56 | .66 | −.08 |
| DXS9908 | GATA182E04 | 165 | −1.73 | .31 | −1.26 |
| DXS998 | AFM224zg11 | 173 | −1.23 | .25 | −.90 |
cM values refer to the STRP map of CIDR.
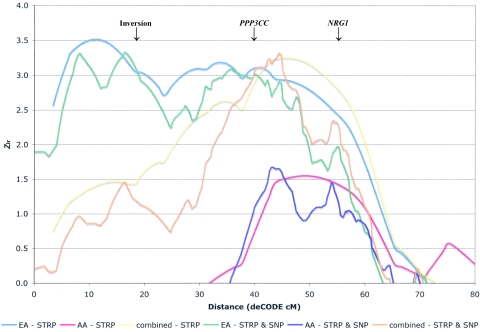
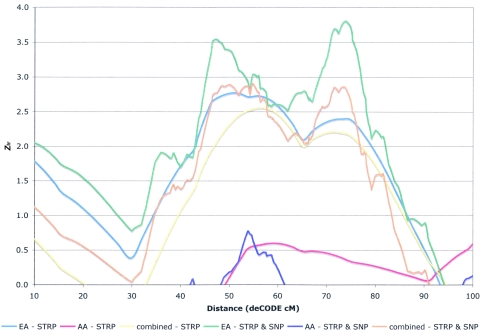
On chromosome 8p23.3-p12 (fig. 2), Zlr scores >2.0 were observed across 31 cM, from 30.7 to 61.7 cM on the CIDR map (∼26.6–58.5 cM on the deCODE map used for fine-mapping analyses; see below). The maximum multipoint evidence of linkage was a Zlr score of 3.25 (equivalent Kong-Cox LOD of 2.30) near D8S1771 (at 52 cM). Although the meaning of a 1-LOD interval is not clear for complex disorders, we note that here the 1-LOD interval (on the CIDR map) extended from 37.0 cM (18.36 Mb, at D8S1145) to 60.3 cM (32.12 Mb, at D8S1477). The linkage signal in this region was observed primarily in EA families, in which two peaks of similar magnitude were observed, at 11.5 cM (Zlr=3.52) and at 39.6 cM (Zlr=3.19), with a 1-LOD interval extending from 2.15 cM (∼3.03 Mb, centromeric to D8S264) to 58.3 cM (∼30.11 Mb, telomeric to D8S1477). In AA families, negative Zlr scores were observed from 8pter to 37.0 cM, with a maximum Zlr of 1.55 at 54.2 cM.
Figure 2.
Results of the STRP genome scan for SZ on chromosome 8 and the subsequent SNP fine mapping. Positive Zlr scores for the STRP scan for the entire sample (yellow line), EA subsample (blue line), and AA subsample (red line) are plotted on the Y-axis, and genomic position (CIDR cM converted to deCODE cM from p-ter, to allow plotting of the fine-mapping data) on chromosome 8 is plotted on the X-axis (0–80 cM). Similarly, positive Zlr scores for the scanning STRPs plus the fine-mapping SNPs for the entire sample (orange line), EA sample (green line), and AA sample (purple line) are plotted on the Y-axis, and genomic position (deCODE cM from p-ter) on chromosome 8 is plotted on the X-axis (0–80 cM). The PPP3CC gene is located at 39 deCODE cM, and the NRG1 (GGF2 isoform) gene spans 53–54 deCODE cM. The common inversion on chromosome 8p spans ∼18–22 deCODE cM and is marked.
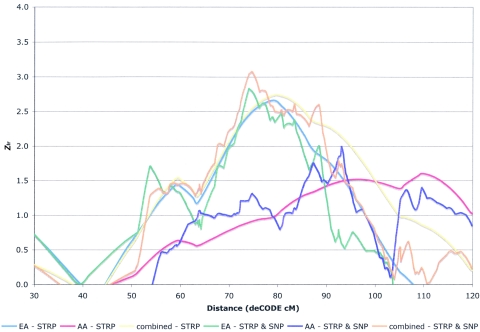
Suggestive linkage was also observed in the entire sample at 11p11.2-q22.3 (fig. 3), with a Zlr of 2.74 at D11S2371 (76.1 cM; equivalent LOD of 1.63), with a 1-LOD interval from 61.9 cM (∼60.55 Mb, centromeric to D11S1344) to 94.6 cM (∼94.39 Mb, centromeric to D11S2000). Again, greater evidence of linkage was observed in EA families (Zlr=2.66 at D11S2371, 76.1 cM) than in AA families (Zlr=1.61 at D11S1391, 104.6 cM).
Figure 3.
Results of the STRP genome scan for SZ on chromosome 11 and the subsequent SNP fine mapping. Positive Zlr scores for the STRP scan for the entire sample (yellow line), EA subsample (blue line), and AA subsample (red line) are plotted on the Y-axis, and genomic position (CIDR cM converted to deCODE cM from p-ter, to allow plotting of the fine-mapping data) on chromosome 11 is plotted on the X-axis (30–120 cM). Similarly, positive Zlr scores for the scanning STRPs plus the fine-mapping SNPs for the entire sample (orange line), EA subsample (green line), and AA subsample (purple line) are plotted on the Y-axis, and genomic position (deCODE cM from p-ter) on chromosome 11 is plotted on the X-axis (30–120 cM).
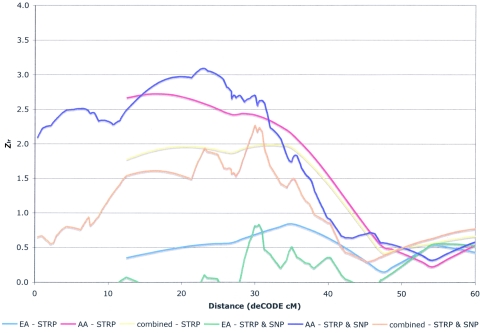
In the exploratory analysis, uncorrected suggestive evidence of linkage was observed in AA families on chromosome 4p16.1-p15.32 (fig. 4) and in EA families on chromosome 5p14.3-q11.2 (fig. 5). For chromosome 4, the maximum Zlr of 2.72 was at 15.5 cM (6.92 Mb, centromeric to D4S2366), with a 1-LOD interval from 4pter to 35.5 cM (∼22.65 Mb, centromeric to D4S2639) in AA families. Zlr did not exceed 1.0 in the EA families on chromosome 4. For chromosome 5, the maximum Zlr of 2.77 was at 43.2 cM (∼31.61 Mb, telomeric to D5S1470), with a 1-LOD interval from 33.6 cM (∼17.50 Mb, telomeric to D5S2845) to 74.3 cM (∼66.62 Mb, telomeric to D5S2500) in EA families. Zlr did not exceed 1.0 in the AA families in this region of chromosome 5.
Figure 4.
Results of the STRP genome scan for SZ on chromosome 4 and the subsequent SNP fine mapping. Positive Zlr scores for the STRP scan for the entire sample (yellow line), EA subsample (blue line), and AA subsample (red line) are plotted on the Y-axis, and genomic position (CIDR cM converted to deCODE cM from p-ter, to allow plotting of the fine-mapping data) on chromosome 4 is plotted on the X-axis (0–60 cM). Similarly, positive Zlr scores for the scanning STRPs plus the fine-mapping SNPs for the entire sample (orange line), EA subsample (green line), and AA subsample (purple line) are plotted on the Y-axis, and genomic position (deCODE cM from p-ter) on chromosome 4 is plotted on the X-axis (0– 60 cM).
Figure 5.
Results of the STRP genome scan for SZ on chromosome 5 and the subsequent SNP fine mapping. Positive Zlr scores for the STRP scan for the entire sample (yellow line), EA subsample (blue line), and AA subsample (red line) are plotted on the Y-axis, and genomic position (CIDR cM converted to deCODE cM from p-ter, to allow plotting of the fine-mapping data) on chromosome 5 is plotted on the X-axis (10–100 cM). Similarly, positive Zlr scores for the scanning STRPs plus the fine-mapping SNPs for the entire sample (orange line), EA subsample (green line), and AA subsample (purple line) are plotted on the Y-axis, and genomic position (deCODE cM from p-ter) on chromosome 5 is plotted on the X-axis (10–100 cM).
Linkage Fine-Mapping Analysis
For the fine-mapping analysis, the deCODE map was used. We performed fine mapping for the four suggestive regions (4p16.3-p15.2, 5p15.2-q13.3, 8p23.3-q12.1, and 11p13-q23.3) and two other regions that had multiple markers with a Zlr >2.0 (10p15.3-p14 and 10q25.3-q26.3) (table 1). The average IC across the fine-mapping region for EA and AA families was 0.64 and 0.59, respectively, for the STRP map and was 0.85 and 0.79, respectively, for the map of 104 SNPs and 9 STRPs on chromosome 8p. The average IC for other fine-mapping regions for EA and AA families, respectively, was 0.83 and 0.77 for chromosome 4, 0.85 and 0.78 for chromosome 5, 0.80 and 0.77 for chromosome 10p, 0.83 and 0.78 for chromosome 10q, and 0.85 and 0.78 for chromosome 11 (table 1).
Figure 2 shows Zlr scores for the full sample and for EA and AA families separately, with a combination of STRPs and SNPs on chromosome 8p. In the full sample, the maximum Zlr score of 3.32 was observed at 44.4 cM (deCODE), just telomeric to D8S1771, where a similar peak (Zlr = 3.24) was observed in the scan analysis. In EA families, two peaks were again observed, although at slightly different locations than in the scan analysis: a telomeric peak (Zlr = 3.33) at 16.4 cM (deCODE), near the boundary of the inversion region (D8S1130), and a more centromeric peak (Zlr=3.10) at 35.7 cM (deCODE), telomeric to D8S560. In AA families, a maximum Zlr score of 1.68 was observed at 43.1 cM (deCODE), contributing to the peak result for the full sample. Results were essentially identical for analyses with the SNPs alone.
Therefore, fine mapping did not significantly increase or decrease the overall significance or length of the linkage peak in chromosome 8, despite an increase in IC. We note that the effect of the inversion on the analysis remains unclear. The genome-scan map contains two STRPs in the inversion region, as discussed above. We removed D8S1469 from the linkage analysis of STRPs alone (and also in the fine mapping) so that only one STRP (D8S1130) remained in this region. In EA families, the peak Zlr in this analysis remains very close to the inversion region. Until a more comprehensive analysis of the presence and sequence of inversions in this region can be performed, their effect on linkage results cannot be clearly understood. We consider the results of our analysis of 9 STRPs and 104 SNPs, with only 1 STRP within the inversion region, to be the most conservative estimation possible from the present data, and we conclude that genomewide suggestive linkage has been observed in the full sample, with two somewhat distinct peaks observed in the EA families.
However, we observed a more noticeable increase in significance for fine mapping of 5p15.2-q13.3 (fig. 5). In EA families, the 1-LOD interval was narrowed from 40.7 cM to 31.8 cM (table 1). It became clear that the centromere separated the linkage region into two peaks, and the maximum Zlr score of the more significant peak increased from 2.77, telomeric to D5S1470 (43.2 cM and ∼31.6 Mb) (5p peak), to 3.80 at rs1027164 (73.8 cM and 57.3 Mb) (5q peak) (fig. 5). Zlr did not exceed 1 before or after fine mapping in the AA families (data not shown).
With regard to the fine mapping of chromosome 4p16.3-p15.2 that showed suggestive linkage in AA families and of chromosome 11p13-q23.3 that showed suggestive linkage in both the EA and the combined samples, the significance remained almost the same as before fine mapping (table 1 and figs. 3 and 4). However, we were able to narrow the 1-LOD interval for chromosome 11 from 32.7 cM to 19.5 cM in the full sample. For chromosome 10 regions, fine mapping did not improve any significance of linkage beyond a Zlr score of 3 (table 1).
Discussion
Genetic factors appear to predominate in the etiology of SZ, given a heritability estimated at 0.8. Most studies of the molecular genetics of SZ have investigated possible associations with functional polymorphisms in loci relevant to monoamine neurotransmission, which have been considered candidate genes on the basis of simple pharmacological models of SZ. Linkage studies have the advantage of not depending on any knowledge of the pathophysiology of the disorder. The most recent meta-analysis of SZ genome scans, including 20 analyses from 16 individual projects, showed significant evidence, across these studies, of linkage in a number of chromosomal regions (Lewis et al. 2003). In several of these regions, plausible candidate genes have been identified for which substantial evidence of association with SZ has been reported more than once (Craddock et al. 2005).
We have reported here one of the largest genome scans of SZ to date. The primary linkage analyses included predominantly EA or AA families. To take into account the possibility that these groups might differ in the effects of different SZ susceptibility loci, separate exploratory analyses of EA and AA families were also performed. Genomewide suggestive evidence of linkage was observed in the primary analysis on chromosomes 8p23.3-p12 and 11p11.2-q22.3, and, in the exploratory analyses, this threshold was also exceeded (without correction for multiple tests) on chromosome 4p16.1-p15.32 in AA families and on chromosome 5p14.3-q11.2 in EA families.
There are some limitations to our conclusions. For example, it is not known how to best define the phenotype for genetic studies of SZ. There is considerable support for the approach of combining the DSM-IV categories of SZ and SA, on the basis of their high relative risk in families of probands with SZ compared with the general population, their similar clinical characteristics in our (and other) samples, and the high reliability with which these diagnoses have been assigned. However, more-optimal phenotypes or endophenotypes might be identified in the future. Furthermore, despite the fact that this is a very large single sample of pedigrees with SZ, it lacks sufficient power to reliably detect loci with small effects.
The strongest evidence of linkage was at chromosome 8p23.3-p12. However, the presence of a common paracentric inversion in our strongest linkage region has complicated our analyses and the interpretation of results. It is possible, for example, that SZ is associated either with the presence of certain specific inversions or with one or more genes within the inversion segment. Also, failure to account for the presence of the inversion might have influenced the results of previous linkage analyses in this region (e.g., the location of the peak signals) in ways that are not yet clear. We evaluated the sensitivity of our linkage analyses to the genetic map assumptions, and linkage results remained basically unchanged whatever the assumed order of the SNPs located within the inverted region. Our results appeared to be more sensitive to the map length. Unsurprisingly, Zlr scores varied with the assumed length, but these variations remained relatively moderate. For instance, a twofold decrease in map length led to a 6% reduction in Zlr score. Conversely, the assumption of a twofold longer map led to a 5% increase in Zlr score. In spite of these perhaps modest limitations related to the map effects of the inversion, evidence of a susceptibility locus for SZ in 8p has been detected in other family samples (table A3), and two positional candidate genes—NRG1 and protein phosphatase 3 (formerly 2B), catalytic subunit, gamma isoform (calcineurin A gamma) (PPP3CC)—have been proposed to account for the linkages (Stefansson et al. 2002; Gerber et al. 2003), although neither can be considered definitively established (for example, see the detailed analysis of the reported association between NRG1 and SZ [Duan et al. 2005]). The linkage signal in the present sample was extremely broad: nine STRP markers across 60 cM produced linkage signals at nominal P<.01 (Zlr>2.326) in the combined sample and in the EA families separately (table 5). We continued to observe suggestive evidence of linkage in the fine-mapping analysis, and it is improbable that a denser SNP map would substantially increase the linkage signal. In the EA families, which are largely responsible for the positive signal in this region, there appear to be two peaks. Given this, plus the repeated observation of evidence of SZ linkage in this region and the diverse locations of the peak evidence of linkage across studies, there might be more than one SZ susceptibility gene in this region. The 8p23-p12 region contains 239 annotated genes (UCSC Genome Bioinformatics, May 2004 freeze); this includes 137 genes that are annotated as full length, with an initiating ATG and valid stop-codon, that can be translated from the genome without frameshifts, and that use consensus splice sites (Consensus Coding DNA Sequence [CCDS] database, March 2005 report; see CCDS Web site). In addition to NRG1 and PPP3CC in 8p, a substantial number of the genes in this region might be considered positional candidates because of their roles in brain function, and, given that the pathophysiology of SZ is largely unknown, there could be additional relevant genes. Therefore, large and systematic case-control and/or family-based LD mapping experiments should be undertaken in this region.
In summary, a genome scan of a large sample of pedigrees with SZ has produced suggestive evidence of linkage in two regions and evidence of two other suggestive regions in exploratory analyses of AA and EA families (see table 7 for results of fine mapping in our sample). Some of these regions have support from multiple other SZ linkage scans (see tables A3 and A4). The present study increases support for the hypothesis that one, or possibly more than one, SZ susceptibility gene is located on chromosome 8p. The SZ candidate regions observed in multiple samples should be studied systematically in large samples with the use of LD mapping methods.
Table 7.
STRPs and SNPs by Map Position and Multipoint Zlr for Fine-Mapping Regions
| Zlr |
|||||
| Chromosomeand Marker | Alias | Position(cM)a | EA Sample | AA Sample | Total Sample |
| 4: | |||||
| rs11727981 | hCV11282864 | .3 | −.71 | 2.09 | .65 |
| rs1135945 | hCV9657182 | .6 | −.73 | 2.15 | .66 |
| rs13138362 | hCV1884907 | 1.0 | −.77 | 2.22 | .67 |
| rs2290404 | hCV9039853 | 1.2 | −.81 | 2.24 | .65 |
| rs2236995 | hCV3253724 | 2.2 | −.97 | 2.26 | .55 |
| rs231200 | rs231200 | 2.5 | −.99 | 2.32 | .57 |
| rs4974665 | hCV3176043 | 2.9 | −.92 | 2.36 | .67 |
| rs445160 | hCV3243313 | 3.7 | −.89 | 2.45 | .77 |
| rs1475977 | hCV7565657 | 3.9 | −.88 | 2.46 | .78 |
| rs916171 | hCV7565369 | 4.3 | −.87 | 2.49 | .80 |
| rs1741553 | hCV1269876 | 5.2 | −.91 | 2.50 | .79 |
| rs910643 | hCV2523717 | 6.2 | −.93 | 2.51 | .76 |
| rs873924 | rs873924 | 7.2 | −.69 | 2.49 | .92 |
| rs4689174 | rs4689174 | 7.4 | −.65 | 2.46 | .94 |
| rs6833060 | hCV2876203 | 7.5 | −.79 | 2.44 | .82 |
| rs4361326 | hCV376560 | 8.0 | −.77 | 2.45 | .86 |
| rs1812310 | rs1812310 | 8.6 | −.58 | 2.33 | .94 |
| rs4689261 | hCV2881264 | 9.8 | −.32 | 2.33 | 1.15 |
| rs984576 | rs984576 | 10.7 | −.10 | 2.28 | 1.29 |
| rs7664611 | hCV2979334 | 11.5 | .01 | 2.34 | 1.41 |
| D4S2366 | GATA22G05 | 12.5 | .07 | 2.49 | 1.54 |
| rs2279195 | hCV1900723 | 21.3 | −.33 | 2.96 | 1.49 |
| rs938563 | hCV1216607 | 22.2 | −.08 | 3.06 | 1.76 |
| rs747357 | hCV2400825 | 22.4 | −.05 | 3.08 | 1.79 |
| rs2868942 | hCV2643298 | 22.7 | .02 | 3.09 | 1.85 |
| rs7681266 | hCV369676 | 23.1 | .11 | 3.09 | 1.94 |
| rs7662694 | rs7662694 | 23.5 | .06 | 3.06 | 1.89 |
| rs7674470 | hCV2976961 | 24.9 | .05 | 3.00 | 1.84 |
| rs1388875 | hCV11990265 | 25.4 | −.07 | 2.95 | 1.72 |
| rs717376 | hCV1895671 | 25.6 | −.10 | 2.94 | 1.69 |
| rs961996 | hCV8842641 | 25.7 | −.12 | 2.92 | 1.66 |
| rs763318 | hCV2308179 | 26.1 | −.08 | 2.82 | 1.64 |
| rs6838908 | hCV2967286 | 26.6 | −.02 | 2.78 | 1.65 |
| D4S403 | AFM157xg3 | 26.7 | −.04 | 2.77 | 1.63 |
| rs1019271 | hCV1192337 | 26.8 | −.03 | 2.65 | 1.57 |
| rs558473 | rs558473 | 27.1 | −.05 | 2.71 | 1.59 |
| rs6449004 | hCV2900115 | 27.4 | −.09 | 2.67 | 1.54 |
| rs1032732 | hCV7563531 | 27.9 | −.02 | 2.71 | 1.62 |
| rs11728802 | hCV2878633 | 28.2 | .17 | 2.66 | 1.75 |
| rs923985 | hCV9512533 | 28.5 | .32 | 2.61 | 1.82 |
| rs6449126 | rs6449126 | 29.3 | .59 | 2.66 | 2.06 |
| rs1861048 | hCV2672580 | 29.5 | .65 | 2.69 | 2.12 |
| rs6834674 | hCV1216865 | 30.0 | .81 | 2.71 | 2.27 |
| rs717176 | hCV343178 | 30.3 | .81 | 2.55 | 2.18 |
| rs10034719 | hCV8281272 | 30.5 | .83 | 2.62 | 2.24 |
| rs2036358 | hCV1485501 | 30.9 | .75 | 2.63 | 2.18 |
| rs1496747 | hCV3031687 | 31.2 | .48 | 2.59 | 1.94 |
| rs7671482 | hCV3142167 | 32.0 | .41 | 2.24 | 1.68 |
| rs7671329 | hCV1321122 | 33.5 | .21 | 2.07 | 1.43 |
| rs13104297 | hCV2801460 | 34.3 | .32 | 1.83 | 1.37 |
| rs7668417 | hCV1704075 | 34.5 | .39 | 1.79 | 1.41 |
| rs1861125 | hCV11989661 | 34.6 | .45 | 1.76 | 1.43 |
| D4S2639 | GATA90B10 | 34.7 | .46 | 1.75 | 1.44 |
| rs2643435 | rs2643435 | 34.8 | .47 | 1.75 | 1.46 |
| rs1452554 | rs1452554 | 35.0 | .52 | 1.74 | 1.49 |
| rs312364 | rs312364 | 35.0 | .52 | 1.74 | 1.49 |
| rs901123 | hCV1970471 | 35.1 | .49 | 1.80 | 1.50 |
| rs1323067 | hCV2946946 | 35.4 | .43 | 1.84 | 1.49 |
| rs525684 | hCV2946855 | 35.7 | .37 | 1.83 | 1.42 |
| rs1019997 | hCV8289078 | 36.3 | .33 | 1.65 | 1.27 |
| rs1364836 | hCV8847912 | 36.9 | .28 | 1.56 | 1.18 |
| rs925570 | hCV7462900 | 37.4 | .27 | 1.50 | 1.14 |
| rs2270851 | hCV11544446 | 37.6 | .23 | 1.47 | 1.09 |
| rs10030690 | rs10030690 | 38.0 | .21 | 1.38 | 1.01 |
| rs989631 | hCV9512541 | 38.1 | .21 | 1.32 | .98 |
| rs6857002 | hCV2645274 | 38.7 | .26 | 1.19 | .94 |
| rs4407488 | hCV1216773 | 39.8 | .36 | .93 | .87 |
| rs7684572 | hCV2668686 | 40.3 | .36 | .91 | .85 |
| rs1035091 | hCV336230 | 41.7 | .12 | .68 | .51 |
| rs6831932 | hCV3038799 | 42.3 | .05 | .64 | .44 |
| rs11731086 | hCV1222538 | 43.3 | .01 | .64 | .40 |
| rs2048506 | hCV1219482 | 44.9 | −.12 | .70 | .32 |
| rs13144326 | hCV1585404 | 45.2 | −.15 | .70 | .30 |
| rs260960 | hCV1228386 | 45.7 | −.15 | .72 | .31 |
| rs6833149 | hCV7672079 | 46.3 | −.11 | .70 | .33 |
| rs1947508 | hCV11938279 | 47.1 | .04 | .57 | .37 |
| D4S391 | AFM016xf3 | 47.3 | .04 | .57 | .38 |
| 5: | |||||
| D5S817 | GATA3E10 | 29.9 | .78 | −1.09 | .05 |
| rs1530495 | hCV7572662 | 31.9 | .87 | −.95 | .20 |
| rs30623 | hCV2936976 | 32.8 | 1.11 | −.86 | .45 |
| rs153931 | hCV3088146 | 33.8 | 1.44 | −.76 | .77 |
| rs983889 | hCV1401088 | 34.7 | 1.77 | −.74 | 1.04 |
| rs162859 | hCV2935668 | 35.9 | 1.91 | −.56 | 1.25 |
| rs31463 | hCV1162271 | 37.8 | 1.83 | −.29 | 1.33 |
| rs2036848 | hCV1454666 | 38.1 | 1.83 | −.18 | 1.40 |
| rs1443404 | hCV7580382 | 38.5 | 1.91 | −.19 | 1.46 |
| rs6870006 | rs6870006 | 38.9 | 1.84 | −.23 | 1.38 |
| rs10472818 | rs10472818 | 39.4 | 1.77 | −.04 | 1.43 |
| rs1911864 | hCV12086585 | 40.0 | 1.70 | −.03 | 1.38 |
| rs2434772 | hCV2941094 | 40.5 | 1.75 | −.05 | 1.42 |
| rs248186 | rs248186 | 40.9 | 1.82 | −.03 | 1.48 |
| rs7725275 | hCV198613 | 41.3 | 1.89 | −.06 | 1.52 |
| rs995178 | hCV9621778 | 41.9 | 1.85 | −.02 | 1.51 |
| D5S2845 | GATA134B03 | 42.5 | 1.84 | .09 | 1.55 |
| rs7716862 | hCV2939263 | 42.6 | 1.90 | .06 | 1.59 |
| rs1402426 | rs1402426 | 43.0 | 2.06 | −.03 | 1.67 |
| rs1366262 | hCV1399202 | 43.7 | 2.40 | −.06 | 1.93 |
| rs1505874 | hCV2935236 | 44.0 | 2.48 | −.11 | 1.97 |
| rs2014790 | hCV8796946 | 44.9 | 2.86 | −.16 | 2.23 |
| rs1566037 | hCV2708385 | 46.2 | 3.35 | −.19 | 2.61 |
| D5S2848 | GATA145D09 | 46.4 | 3.42 | −.19 | 2.66 |
| rs10942230 | hCV2940113 | 46.6 | 3.52 | −.19 | 2.75 |
| rs1501680 | hCV1399439 | 47.5 | 3.54 | −.19 | 2.78 |
| rs397425 | hCV1028719 | 47.9 | 3.51 | −.09 | 2.83 |
| rs9292323 | hCV2933709 | 48.2 | 3.50 | −.01 | 2.87 |
| rs4867029 | hCV28011100 | 48.9 | 3.41 | .11 | 2.86 |
| rs2133764 | rs2133764 | 50.0 | 3.38 | .18 | 2.89 |
| rs2068015 | hCV1428649 | 50.5 | 3.36 | .19 | 2.87 |
| rs500557 | hCV2940056 | 51.8 | 3.10 | .33 | 2.74 |
| rs641221 | hCV605081 | 53.8 | 2.92 | .78 | 2.87 |
| D5S1470 | GATA7C06 | 53.9 | 2.91 | .78 | 2.86 |
| rs6864434 | hCV1488765 | 54.2 | 2.80 | .74 | 2.75 |
| rs1981968 | hCV11324483 | 54.6 | 3.01 | .71 | 2.91 |
| rs7705177 | rs7705177 | 54.8 | 3.04 | .65 | 2.91 |
| hCV2838186 | hCV2838186 | 55.7 | 3.00 | .48 | 2.79 |
| rs213583 | hCV655159 | 56.0 | 3.03 | .44 | 2.79 |
| rs13189166 | hCV1114849 | 56.3 | 2.84 | .43 | 2.65 |
| rs460053 | hCV3172343 | 57.0 | 2.83 | .47 | 2.69 |
| hCV3020978 | hCV3020978 | 57.5 | 2.78 | .47 | 2.63 |
| rs7731139 | rs7731139 | 57.7 | 2.68 | .41 | 2.51 |
| rs10454863 | hCV1999272 | 57.9 | 2.59 | .42 | 2.44 |
| rs1549681 | hCV1400831 | 58.7 | 2.57 | .43 | 2.41 |
| rs158409 | hCV2935131 | 58.8 | 2.60 | .40 | 2.42 |
| rs11958226 | rs11958226 | 59.2 | 2.61 | .32 | 2.37 |
| rs2910797 | hCV1395027 | 59.6 | 2.61 | .29 | 2.35 |
| rs2289779 | hCV11360428 | 62.0 | 2.52 | −.09 | 2.09 |
| rs379707 | hCV646794 | 63.5 | 2.75 | −.22 | 2.20 |
| rs495237 | hCV1002294 | 64.6 | 2.78 | −.39 | 2.14 |
| rs1564269 | hCV7581372 | 64.7 | 2.78 | −.44 | 2.11 |
| rs1376178 | hCV1321333 | 65.0 | 2.79 | −.48 | 2.11 |
| rs1801033 | hCV115685 | 65.3 | 2.80 | −.58 | 2.05 |
| rs11740946 | hCV264064 | 65.5 | 2.80 | −.61 | 2.04 |
| rs2972408 | hCV1567451 | 66.2 | 2.70 | −.45 | 2.06 |
| D5S1457 | GATA21D04 | 66.3 | 2.67 | −.23 | 2.15 |
| rs2972756 | hCV2841551 | 66.3 | 2.68 | −.34 | 2.10 |
| rs6895327 | hCV450793 | 66.5 | 2.69 | −.34 | 2.12 |
| rs4285229 | hCV11553362 | 66.7 | 2.66 | −.33 | 2.12 |
| rs900379 | hCV8291179 | 66.8 | 2.66 | −.31 | 2.13 |
| rs10035564 | hCV11554044 | 67.1 | 2.64 | −.25 | 2.17 |
| rs2337954 | hCV2127591 | 67.3 | 2.71 | −.25 | 2.21 |
| rs166863 | hCV2994177 | 67.4 | 2.75 | −.20 | 2.29 |
| rs2288468 | hCV11371816 | 67.4 | 2.75 | −.19 | 2.30 |
| rs929819 | hCV8289991 | 67.4 | 2.75 | −.19 | 2.30 |
| rs1345981 | hCV2865963 | 67.7 | 2.82 | −.13 | 2.39 |
| rs12521153 | hCV2838776 | 68.7 | 3.03 | −.13 | 2.54 |
| rs10940347 | hCV3186696 | 68.9 | 3.09 | −.16 | 2.57 |
| rs250387 | hCV2386214 | 69.1 | 3.12 | −.17 | 2.58 |
| rs714459 | hCV2840288 | 69.5 | 3.16 | −.24 | 2.56 |
| rs1363864 | hCV3019869 | 70.1 | 3.29 | −.37 | 2.58 |
| rs3804264|s | hCV1401643 | 70.5 | 3.43 | −.46 | 2.63 |
| rs191205 | hCV1025536 | 70.9 | 3.59 | −.47 | 2.76 |
| rs30000 | hCV807035 | 71.5 | 3.70 | −.44 | 2.86 |
| rs252890 | hCV2840868 | 72.4 | 3.63 | −.47 | 2.79 |
| rs2972355 | hCV1238506 | 72.9 | 3.72 | −.49 | 2.85 |
| rs1027164 | hCV8886893 | 73.8 | 3.80 | −.56 | 2.85 |
| rs159729 | hCV3099052 | 74.8 | 3.72 | −.69 | 2.68 |
| D5S2500 | GATA67D03 | 75.2 | 3.62 | −.73 | 2.56 |
| rs27548 | hCV3248153 | 75.2 | 3.62 | −.73 | 2.56 |
| rs159195 | rs159195 | 75.2 | 3.53 | −.72 | 2.50 |
| rs1995780 | hCV11486548 | 75.4 | 3.51 | −.68 | 2.50 |
| rs153067 | hCV8963983 | 75.5 | 3.49 | −.68 | 2.48 |
| rs34634 | hCV1144877 | 75.7 | 3.45 | −.71 | 2.43 |
| rs4078970 | hCV7573130 | 76.2 | 3.33 | −.85 | 2.26 |
| rs2344396 | hCV1561870 | 76.7 | 3.16 | −1.00 | 2.07 |
| rs7715472 | hCV3248309 | 76.8 | 3.12 | −1.02 | 2.03 |
| rs346437 | hCV3045752 | 77.0 | 3.03 | −1.04 | 1.94 |
| rs4527551 | hCV1497859 | 77.1 | 2.97 | −1.04 | 1.89 |
| rs272629 | hCV2765438 | 77.5 | 2.79 | −.78 | 1.89 |
| rs12651884 | hCV7693739 | 77.7 | 2.75 | −.67 | 1.92 |
| rs10042323 | hCV1186571 | 77.9 | 2.69 | −.61 | 1.91 |
| rs37347 | hCV3166294 | 78.1 | 2.49 | −.62 | 1.74 |
| rs1317632 | hCV37054 | 78.6 | 2.33 | −.66 | 1.57 |
| rs257712 | hCV2841358 | 78.7 | 2.31 | −.67 | 1.55 |
| rs984091 | hCV7447368 | 79.2 | 2.11 | −.67 | 1.38 |
| rs7705123 | rs7705123 | 80.0 | 2.23 | −.49 | 1.58 |
| rs28155 | hCV946441 | 81.2 | 2.13 | −.36 | 1.59 |
| rs164578 | hCV918494 | 81.6 | 2.15 | −.34 | 1.62 |
| rs2561183 | hCV2934686 | 81.7 | 2.13 | −.36 | 1.59 |
| rs7448990 | hCV2837195 | 82.7 | 1.75 | −.49 | 1.22 |
| rs1531312 | hCV1698516 | 83.4 | 1.62 | −.67 | 1.01 |
| rs6886010 | rs6886010 | 83.7 | 1.55 | −.83 | .86 |
| rs4041421 | hCV1393623 | 84.0 | 1.49 | −.90 | .79 |
| rs1403728 | hCV3060555 | 84.4 | 1.46 | −.98 | .72 |
| rs347245 | hCV2828781 | 84.9 | 1.39 | −1.06 | .63 |
| rs282381 | hCV544172 | 85.5 | 1.21 | −1.05 | .49 |
| rs890995 | hCV2938484 | 86.6 | .94 | −1.22 | .21 |
| rs7734427 | hCV74896 | 87.8 | .95 | −1.40 | .17 |
| rs7717355 | rs7717355 | 88.9 | .90 | −1.39 | .19 |
| rs4374745 | hCV174414 | 89.5 | .86 | −1.33 | .16 |
| rs10045155 | rs10045155 | 90.2 | .90 | −1.25 | .22 |
| D5S424 | AFM214zg9 | 90.9 | .67 | −1.20 | .04 |
| 8: | |||||
| rs1979176 | hCV12003528 | .0 | 1.89 | −2.34 | .21 |
| rs6558451 | hCV2769705 | 1.8 | 1.90 | −2.25 | .25 |
| rs4354330 | hCV2062160 | 2.6 | 1.83 | −2.29 | .17 |
| D8S264 | AFM143xd8 | 4.0 | 1.99 | −2.46 | .19 |
| rs17515 | hCV12004129 | 5.1 | 2.51 | −2.59 | .55 |
| rs10088378 | hCV1423079 | 7.0 | 3.03 | −2.82 | .83 |
| D8S262 | AFM127xh2 | 7.9 | 3.28 | −2.93 | .95 |
| rs2740898 | hCV8849739 | 8.6 | 3.26 | −2.94 | .94 |
| rs1560428 | hCV7533717 | 9.6 | 3.17 | −2.95 | .86 |
| rs9314540 | rs9314540 | 11.0 | 3.00 | −2.78 | .87 |
| rs11779265 | hCV8315417 | 12.0 | 2.82 | −2.62 | .83 |
| rs10503309 | rs10503309 | 13.1 | 2.84 | −2.54 | .94 |
| rs6559058 | rs6559058 | 14.1 | 3.05 | −2.44 | 1.16 |
| rs2442581 | rs2442581 | 15.9 | 3.27 | −2.20 | 1.42 |
| rs7834209 | rs7834209 | 16.4 | 3.33 | −2.20 | 1.46 |
| rs2980438 | hCV11587790 | 17.8 | 3.19 | −2.34 | 1.23 |
| rs506960 | hCV725401 | 18.5 | 3.09 | −2.28 | 1.19 |
| D8S1130 | GATA25C10 | 19.3 | 2.93 | −2.18 | 1.13 |
| D8S1106 | GATA23D06 | 24.7 | 2.25 | −1.96 | .75 |
| rs609020 | hCV1380 | 25.9 | 2.42 | −1.87 | .92 |
| rs7006666 | hCV1160271 | 26.6 | 2.46 | −1.76 | 1.02 |
| rs2017470 | hCV1854379 | 26.7 | 2.49 | −1.75 | 1.06 |
| rs1355305 | hCV283932 | 27.1 | 2.52 | −1.69 | 1.11 |
| rs4383970 | hCV10024049 | 27.3 | 2.56 | −1.66 | 1.16 |
| rs352783 | hCV2038032 | 28.0 | 2.49 | −1.43 | 1.21 |
| rs1871573 | hCV12104846 | 28.6 | 2.35 | −1.42 | 1.08 |
| rs822300 | rs822300 | 29.3 | 2.37 | −1.35 | 1.12 |
| rs2239879 | hCV2441245 | 30.4 | 2.62 | −1.22 | 1.40 |
| rs2904748 | hCV16157316 | 31.1 | 2.85 | −.98 | 1.73 |
| rs2237851 | rs2237851 | 31.5 | 2.82 | −.87 | 1.77 |
| rs1041983 | hCV8684085 | 32.1 | 2.80 | −.72 | 1.85 |
| D8S1145 | GATA72C10 | 32.2 | 2.90 | −.69 | 1.94 |
| rs3736273 | hCV1199888 | 32.9 | 2.94 | −.54 | 2.06 |
| rs4486225 | hCV11848800 | 34.0 | 2.96 | −.33 | 2.20 |
| rs7005767 | hCV9639133 | 34.7 | 3.03 | −.23 | 2.33 |
| NA | hCV1167258 | 35.7 | 3.10 | .06 | 2.55 |
| rs896044 | hCV1313416 | 36.3 | 3.05 | .18 | 2.59 |
| rs11991767 | hCV1682164 | 37.5 | 3.01 | .40 | 2.66 |
| D8S560 | AFMa127ye5 | 38.8 | 2.97 | .76 | 2.84 |
| rs7490 | hCV8805141 | 39.4 | 2.96 | .93 | 2.93 |
| rs9886594 | rs9886594 | 40.0 | 3.02 | 1.10 | 3.09 |
| rs2055824 | hCV12106772 | 41.9 | 2.86 | 1.35 | 3.10 |
| rs1002791 | hCV75221 | 42.3 | 2.94 | 1.46 | 3.23 |
| rs2928679 | hCV1382529 | 42.7 | 2.83 | 1.62 | 3.24 |
| rs1436148 | hCV8470964 | 43.1 | 2.70 | 1.68 | 3.17 |
| rs7836430 | hCV2995961 | 43.9 | 2.82 | 1.65 | 3.25 |
| rs196886 | hCV796208 | 44.4 | 2.91 | 1.66 | 3.32 |
| rs11998492 | hCV11857651 | 44.8 | 2.89 | 1.62 | 3.28 |
| D8S1771 | AFMb320va5 | 44.8 | 2.88 | 1.62 | 3.28 |
| rs2467698 | hCV2895454 | 45.1 | 2.66 | 1.56 | 3.07 |
| rs4872389 | hCV1158898 | 45.8 | 2.56 | 1.44 | 2.92 |
| NA | hCV3094290 | 46.2 | 2.53 | 1.38 | 2.85 |
| rs442103 | hCV1126721 | 46.8 | 2.51 | 1.33 | 2.81 |
| rs1351499 | hCV1714124 | 47.5 | 2.70 | 1.04 | 2.78 |
| rs11783299 | hCV2152299 | 47.5 | 2.70 | 1.04 | 2.78 |
| rs2071575 | hCV2045384 | 47.6 | 2.69 | 1.03 | 2.78 |
| rs2294092 | hCV1374353 | 48.1 | 2.61 | .97 | 2.67 |
| rs4732838 | hCV1140451 | 48.8 | 2.18 | .90 | 2.28 |
| NA | hCV2049967 | 49.2 | 2.13 | .91 | 2.25 |
| rs6558073 | hCV3275985 | 49.6 | 2.02 | .94 | 2.17 |
| rs12056328 | hCV25645594 | 50.4 | 1.74 | 1.04 | 2.01 |
| rs12676327 | hCV444094 | 51.2 | 1.76 | 1.10 | 2.06 |
| rs12386822 | hCV1584515 | 52.1 | 1.78 | 1.09 | 2.08 |
| rs2979487 | hCV15880850 | 52.9 | 1.68 | 1.14 | 2.02 |
| NA | hCV1379657 | 53.2 | 1.63 | 1.25 | 2.04 |
| NA | hCV2779153 | 53.5 | 1.67 | 1.32 | 2.13 |
| rs763551 | hCV2259922 | 53.9 | 1.80 | 1.46 | 2.31 |
| rs1481762 | hCV8868705 | 54.2 | 1.89 | 1.42 | 2.35 |
| D8S1477 | GGAA20C10 | 54.3 | 1.90 | 1.41 | 2.36 |
| NA | hCV2870374 | 54.5 | 1.91 | 1.32 | 2.31 |
| rs4733142 | hCV8280633 | 54.8 | 1.96 | 1.28 | 2.33 |
| rs938641 | hCV445693 | 55.1 | 1.98 | 1.20 | 2.30 |
| rs7463110 | hCV1943749 | 55.4 | 1.94 | 1.10 | 2.21 |
| rs6468305 | rs6468305 | 55.6 | 1.80 | .98 | 2.02 |
| rs7826624 | hCV1374319 | 56.1 | 1.71 | .95 | 1.93 |
| rs670603 | hCV2446103 | 56.5 | 1.62 | 1.04 | 1.92 |
| NA | hCV2445923 | 56.8 | 1.57 | 1.03 | 1.88 |
| rs1111387 | hCV2445746 | 56.9 | 1.54 | 1.05 | 1.86 |
| rs2298320 | hCV2445274 | 57.4 | 1.26 | 1.04 | 1.63 |
| rs7812866 | hCV1718918 | 57.6 | 1.18 | 1.02 | 1.56 |
| rs1043782 | hCV8844184 | 57.8 | 1.13 | .99 | 1.50 |
| rs7816768 | hCV2159104 | 58.2 | 1.05 | .93 | 1.39 |
| rs7832578 | hCV95287 | 58.4 | 1.05 | .89 | 1.37 |
| rs3852339 | hCV8896420 | 58.8 | 1.03 | .92 | 1.37 |
| rs2589916 | hCV2045597 | 60.1 | .74 | .84 | 1.10 |
| rs868586 | hCV1593263 | 60.5 | .72 | .77 | 1.05 |
| rs7823929 | hCV448973 | 61.0 | .64 | .66 | .92 |
| rs7837006 | hCV2896129 | 61.1 | .63 | .67 | .91 |
| rs906290 | hCV3201067 | 61.2 | .57 | .64 | .85 |
| rs732612 | hCV2533651 | 61.3 | .47 | .50 | .69 |
| rs2304297 | hCV15974192 | 61.5 | .40 | .49 | .63 |
| NA | hCV2896764 | 61.5 | .39 | .48 | .61 |
| rs7003908 | hCV1841761 | 63.2 | .02 | .20 | .16 |
| rs1027966 | hCV8809457 | 63.3 | .02 | .20 | .15 |
| rs2279301 | hCV333405 | 63.3 | .01 | .19 | .15 |
| rs11990854 | hCV1563801 | 63.7 | −.05 | .20 | .09 |
| rs4873375 | rs4873375 | 64.0 | −.11 | .24 | .07 |
| rs7002235 | hCV1145366 | 64.0 | −.11 | .24 | .07 |
| rs4382493 | hCV174458 | 64.5 | −.14 | .26 | .07 |
| rs1367819 | hCV2049503 | 65.2 | −.28 | .00 | −.20 |
| rs718251 | hCV9319187 | 65.5 | −.26 | −.01 | −.19 |
| rs1365245 | hCV7478079 | 66.1 | −.09 | −.04 | −.07 |
| D8S1110 | GATA8G10 | 66.6 | −.12 | −.09 | −.11 |
| rs10504146 | rs10504146 | 66.8 | −.13 | −.10 | −.13 |
| rs545125 | hCV3140037 | 67.1 | −.13 | −.10 | −.12 |
| rs1477958 | hCV7480634 | 68.0 | −.11 | −.20 | −.17 |
| rs7824575 | hCV1951523 | 68.5 | −.18 | −.25 | −.25 |
| rs311390 | hCV3034570 | 68.5 | −.19 | −.19 | −.22 |
| NA | hCV2770809 | 68.6 | −.19 | −.10 | −.18 |
| rs7837220 | hCV1854535 | 69.2 | .04 | .15 | .14 |
| rs867531 | hCV8967543 | 69.3 | .08 | .14 | .17 |
| rs12678930 | hCV1320563 | 69.4 | .11 | .15 | .19 |
| rs1821814 | hCV11365409 | 69.7 | .16 | .11 | .21 |
| rs1834413 | hCV11922708 | 70.2 | .13 | −.03 | .10 |
| rs1317836 | rs1317836 | 70.7 | .07 | −.12 | .00 |
| rs4397388 | hCV117013 | 71.0 | .10 | −.17 | .00 |
| rs6471680 | hCV2826238 | 71.3 | −.11 | −.19 | −.18 |
| rs1023651 | hCV2914108 | 71.6 | −.17 | −.21 | −.25 |
| 10p: | |||||
| rs2448384 | hCV1904568 | 1.2 | 1.77 | .86 | 1.99 |
| rs7912017 | rs7912017 | 1.5 | 1.74 | .82 | 1.93 |
| rs10904587 | hCV27050 | 2.0 | 1.67 | .80 | 1.88 |
| rs4256905 | hCV358407 | 2.2 | 1.61 | .80 | 1.83 |
| rs1500965 | hCV7886666 | 2.4 | 1.56 | .80 | 1.78 |
| rs2387662 | hCV488722 | 2.5 | 1.58 | .81 | 1.81 |
| rs2813440 | hCV241818 | 2.6 | 1.62 | .86 | 1.86 |
| rs1029182 | rs1029182 | 4.4 | 2.26 | .85 | 2.36 |
| rs10751855 | hCV1759096 | 5.5 | 2.36 | .82 | 2.41 |
| rs826472 | hCV7431936 | 5.7 | 2.37 | .81 | 2.41 |
| rs729397 | hCV3162530 | 5.8 | 2.36 | .79 | 2.40 |
| rs2279768 | hCV11648249 | 9.5 | 1.99 | .49 | 1.91 |
| D10S1435 | GATA88F09 | 10.1 | 1.95 | .45 | 1.85 |
| rs9630129 | hCV1904916 | 11.0 | 2.20 | .22 | 1.92 |
| rs1325586 | hCV1977337 | 11.5 | 2.30 | .13 | 1.94 |
| rs10795055 | hCV3003439 | 12.2 | 2.45 | .10 | 2.05 |
| rs1886946 | hCV11373985 | 13.5 | 2.50 | .05 | 2.08 |
| rs1022768 | hCV8914257 | 14.2 | 2.50 | −.01 | 2.05 |
| rs1751277 | hCV1853650 | 14.6 | 2.51 | −.05 | 2.04 |
| rs4266965 | hCV7873798 | 15.6 | 2.49 | .00 | 2.07 |
| rs1155931 | hCV1803410 | 16.2 | 2.52 | −.01 | 2.10 |
| rs4881450 | hCV399566 | 16.9 | 2.42 | .05 | 2.06 |
| rs2386638 | hCV278985 | 17.5 | 2.31 | .07 | 2.00 |
| rs7072398 | hCV1882638 | 18.3 | 2.10 | .09 | 1.87 |
| rs3750671 | hCV9615599 | 18.6 | 2.01 | .06 | 1.79 |
| rs7902155 | rs7902155 | 18.8 | 1.90 | .06 | 1.69 |
| rs3815975 | rs3815975 | 19.4 | 1.72 | .03 | 1.51 |
| rs2255088 | hCV16011810 | 19.6 | 1.64 | .04 | 1.44 |
| D10S189 | AFM063xf4 | 19.8 | 1.65 | .04 | 1.45 |
| 10q: | |||||
| D10S1237 | GATA48G07 | 134.1 | 1.49 | .23 | 1.42 |
| rs11196798 | hCV1765605 | 134.2 | 1.52 | .14 | 1.40 |
| rs703349 | hCV3085761 | 134.8 | 1.58 | .29 | 1.58 |
| rs658530 | hCV10013467 | 134.8 | 1.58 | .29 | 1.58 |
| rs1264798 | rs1264798 | 134.8 | 1.58 | .29 | 1.58 |
| rs10159620 | hCV2647718 | 135.3 | 1.52 | .35 | 1.57 |
| rs1017822 | hCV3088272 | 135.6 | 1.47 | .38 | 1.54 |
| rs180552 | hCV3088334 | 135.9 | 1.61 | .48 | 1.70 |
| rs2420257 | hCV15801819 | 136.3 | 1.76 | .59 | 1.87 |
| rs11197672 | hCV2065828 | 136.6 | 1.72 | .61 | 1.83 |
| rs1867991 | hCV12127228 | 136.9 | 1.76 | .51 | 1.80 |
| rs740598 | hCV3254784 | 137.1 | 1.78 | .42 | 1.76 |
| rs2118207 | hCV15820611 | 137.4 | 1.81 | .42 | 1.78 |
| hCV55714 | hCV55714 | 137.6 | 1.87 | .44 | 1.84 |
| rs1935168 | hCV11744008 | 137.8 | 1.92 | .40 | 1.85 |
| rs71003 | hCV2981626 | 138.0 | 2.04 | .36 | 1.93 |
| rs1343418 | hCV11212949 | 139.2 | 2.07 | .42 | 1.99 |
| rs853600 | rs853600 | 140.1 | 1.94 | .47 | 1.90 |
| rs1857269 | hCV11757499 | 140.6 | 1.88 | .51 | 1.87 |
| rs4752182 | hCV8171237 | 140.9 | 1.85 | .49 | 1.83 |
| rs7905458 | hCV1484613 | 141.5 | 1.94 | .47 | 1.89 |
| rs884970 | hCV1802876 | 142.0 | 1.71 | .44 | 1.69 |
| rs10886515 | hCV2101894 | 142.2 | 1.68 | .56 | 1.73 |
| rs378150342 | hCV1310103 | 142.5 | 1.66 | .50 | 1.68 |
| rs1537219 | hCV2731394 | 143.2 | 1.61 | .59 | 1.70 |
| rs12769643 | hCV8174365 | 143.8 | 1.47 | .71 | 1.65 |
| rs7082695 | hCV8175290 | 144.4 | 1.36 | .62 | 1.51 |
| rs732640 | hCV2118990 | 144.9 | 1.36 | .53 | 1.46 |
| rs2288336 | hCV11309727 | 146.2 | 1.36 | .57 | 1.49 |
| rs12571870 | hCV1821946 | 147.0 | 1.34 | .38 | 1.36 |
| rs7920046 | hCV3006292 | 147.2 | 1.34 | .36 | 1.34 |
| rs6599619 | hCV1821764 | 148.0 | 1.20 | .31 | 1.19 |
| rs9804347 | hCV367398 | 148.2 | 1.17 | .31 | 1.17 |
| rs10751744 | hCV2102486 | 148.4 | 1.16 | .29 | 1.15 |
| rs766295 | hCV2102652 | 148.6 | 1.01 | .30 | 1.04 |
| rs11597880 | hCV1250945 | 151.3 | .60 | .88 | 1.06 |
| D10S1656 | AFMa184xd9 | 151.4 | .61 | .91 | 1.08 |
| rs12569498 | hCV251049 | 151.6 | .62 | 1.02 | 1.17 |
| rs10736889 | hCV1662136 | 152.0 | .53 | 1.16 | 1.20 |
| rs2015178 | rs2015178 | 152.6 | .47 | 1.20 | 1.20 |
| rs875748 | hCV8892668 | 153.8 | .35 | 1.31 | 1.14 |
| rs768761 | hCV1703535 | 154.3 | .39 | 1.35 | 1.19 |
| rs2281955 | hCV2017094 | 155.2 | .45 | 1.31 | 1.19 |
| rs10901596 | hCV1419866 | 156.7 | .44 | 1.31 | 1.17 |
| rs1436804 | rs1436804 | 157.3 | .36 | 1.35 | 1.13 |
| rs7909756 | rs7909756 | 157.8 | .37 | 1.35 | 1.13 |
| rs10741150 | hCV372978 | 158.5 | .47 | 1.34 | 1.21 |
| rs2483853 | hCV1779324 | 158.7 | .47 | 1.37 | 1.25 |
| rs943207 | hCV8197363 | 159.2 | .48 | 1.41 | 1.27 |
| D10S217 | AFM212XD6 | 159.9 | .54 | 1.31 | 1.23 |
| rs1410064 | hCV2009015 | 160.4 | .46 | 1.16 | 1.08 |
| rs10764754 | hCV1801130 | 160.7 | .45 | 1.16 | 1.07 |
| rs1001990 | hCV8901459 | 163.1 | .52 | .77 | .90 |
| rs7085203 | rs7085203 | 164.8 | .62 | .90 | 1.05 |
| rs1869251 | hCV3014410 | 165.4 | .68 | .90 | 1.11 |
| rs1861860 | rs1861860 | 166.4 | .71 | .91 | 1.13 |
| rs7898151 | hCV3158009 | 168.1 | .75 | 1.00 | 1.20 |
| rs573370 | hCV1769819 | 168.8 | .79 | .91 | 1.19 |
| D10S1248 | GGAA23C05 | 169.2 | .93 | .72 | 1.18 |
| rs447363 | rs447363 | 170.0 | 1.04 | .75 | 1.29 |
| rs448669 | hCV1166802 | 170.4 | 1.07 | .71 | 1.29 |
| rs964681 | rs964681 | 172.0 | 1.37 | .65 | 1.49 |
| rs11017736 | hCV363155 | 172.8 | 1.43 | .61 | 1.52 |
| rs754575 | hCV3232964 | 173.2 | 1.46 | .62 | 1.55 |
| hCV94022 | hCV94022 | 175.4 | 1.59 | .66 | 1.68 |
| rs870552 | hCV9496352 | 176.0 | 1.66 | .68 | 1.75 |
| rs11146353 | hCV12058853 | 176.6 | 1.67 | .66 | 1.75 |
| D10S212 | AFM198zb4 | 177.2 | 1.61 | .58 | 1.65 |
| 11: | |||||
| D11S1392 | GATA6B09 | 50.6 | .73 | −.66 | .20 |
| rs2288667 | hCV15879519 | 51.2 | .75 | −.54 | .29 |
| rs1570214 | hCV7619035 | 51.8 | .99 | −.51 | .50 |
| rs650693 | hCV995778 | 52.7 | 1.38 | −.41 | .87 |
| rs10836555 | hCV2898958 | 53.7 | 1.72 | −.16 | 1.29 |
| rs179801 | rs179801 | 55.4 | 1.50 | .34 | 1.39 |
| ss2946400 | hCV12037009 | 56.0 | 1.40 | .46 | 1.38 |
| rs1038254 | hCV9821215 | 56.3 | 1.35 | .47 | 1.34 |
| rs10837120 | hCV1177638 | 56.3 | 1.35 | .47 | 1.34 |
| rs2953308 | hCV1625511 | 57.0 | 1.32 | .46 | 1.32 |
| rs1371671 | hCV8894006 | 57.2 | 1.32 | .45 | 1.32 |
| rs1381578 | hCV1204510 | 57.5 | 1.33 | .47 | 1.34 |
| rs959632 | hCV2162237 | 57.8 | 1.43 | .51 | 1.45 |
| rs1484960 | rs1484960 | 58.2 | 1.42 | .51 | 1.46 |
| rs2042684 | hCV2897320 | 58.8 | 1.37 | .56 | 1.47 |
| D11S1993 | ATA1B07 | 59.2 | 1.34 | .58 | 1.46 |
| rs10838149 | hCV289210 | 59.3 | 1.34 | .56 | 1.45 |
| rs178503 | hCV12032785 | 59.8 | 1.34 | .59 | 1.46 |
| rs1344709 | hCV2836372 | 61.5 | 1.10 | .87 | 1.44 |
| rs2289448 | hCV9653411 | 63.0 | .87 | .92 | 1.29 |
| rs2945999 | hCV1717146 | 63.4 | .90 | .96 | 1.34 |
| D11S1344 | AFM298vc9 | 63.5 | .90 | .97 | 1.35 |
| rs12284298 | hCV2898441 | 63.5 | .93 | .99 | 1.39 |
| rs1055447 | hCV9636495 | 63.7 | .84 | 1.06 | 1.37 |
| rs11039402 | rs11039402 | 63.8 | .88 | 1.06 | 1.41 |
| rs753095 | hCV11668059 | 63.8 | .88 | 1.06 | 1.41 |
| rs692607 | hCV1182084 | 63.9 | .95 | 1.07 | 1.46 |
| rs11227802 | hCV1870167 | 64.0 | .87 | 1.07 | 1.40 |
| rs514385 | hCV1000052 | 64.2 | .86 | 1.03 | 1.36 |
| rs2441952 | rs2441952 | 64.2 | .81 | 1.03 | 1.31 |
| rs1938596 | hCV3185821 | 64.2 | .80 | 1.03 | 1.31 |
| rs1938781 | hCV3160372 | 64.5 | .96 | 1.03 | 1.43 |
| rs507035 | hCV2138493 | 64.9 | 1.07 | 1.01 | 1.51 |
| rs198436 | hCV2575631 | 67.0 | 1.42 | .96 | 1.76 |
| rs10897237 | hCV1243327 | 67.6 | 1.71 | 1.02 | 2.03 |
| rs2428549 | hCV3186395 | 68.0 | 1.72 | 1.03 | 2.04 |
| rs515213 | hCV826909 | 68.9 | 1.69 | 1.01 | 2.01 |
| rs11231531 | rs11231531 | 69.2 | 1.79 | 1.01 | 2.09 |
| rs7938819 | hCV9153521 | 69.7 | 1.94 | 1.00 | 2.21 |
| rs7940569 | hCV3012560 | 70.3 | 2.01 | .99 | 2.25 |
| rs1467110 | hCV8376844 | 70.6 | 2.02 | .99 | 2.26 |
| rs1784223 | hCV3093494 | 71.0 | 2.05 | 1.03 | 2.31 |
| rs593525 | rs593525 | 71.3 | 2.14 | 1.01 | 2.38 |
| rs2279861 | hCV11667604 | 71.6 | 2.23 | 1.04 | 2.47 |
| rs7108388 | hCV1346738 | 71.9 | 2.28 | 1.03 | 2.51 |
| rs1638588 | hCV1250035 | 72.1 | 2.32 | 1.05 | 2.55 |
| rs105147 | hCV2623925 | 72.3 | 2.34 | 1.21 | 2.66 |
| rs10896364 | hCV209229 | 73.2 | 2.61 | 1.21 | 2.87 |
| rs1551305 | hCV8762660 | 74.0 | 2.83 | 1.21 | 3.04 |
| rs4275647 | hCV12034770 | 74.6 | 2.80 | 1.32 | 3.08 |
| rs734102 | rs734102 | 75.7 | 2.66 | 1.24 | 2.92 |
| rs2276068 | hCV12039531 | 76.2 | 2.55 | 1.15 | 2.79 |
| rs1207142 | hCV8896286 | 76.9 | 2.62 | 1.09 | 2.81 |
| rs3924047 | hCV12040954 | 77.5 | 2.60 | 1.11 | 2.80 |
| hCV2139440 | hCV2139440 | 78.4 | 2.38 | 1.08 | 2.60 |
| rs1000620 | hCV8893770 | 78.8 | 2.43 | .99 | 2.58 |
| D11S2371 | GATA90D07 | 79.3 | 2.35 | .96 | 2.50 |
| rs11235876 | hCV153313 | 80.4 | 2.45 | .80 | 2.49 |
| rs1044782 | hCV7985452 | 80.7 | 2.48 | .83 | 2.53 |
| rs1315265 | hCV7625202 | 80.9 | 2.41 | .85 | 2.49 |
| rs554202 | hCV1045980 | 81.2 | 2.27 | .97 | 2.45 |
| rs663746 | hCV3245441 | 81.6 | 2.32 | 1.03 | 2.51 |
| rs7926553 | hCV3242281 | 83.0 | 2.31 | 1.03 | 2.50 |
| rs595654 | hCV1249332 | 83.3 | 2.39 | 1.11 | 2.61 |
| rs230656 | hCV3152042 | 83.7 | 2.23 | 1.09 | 2.47 |
| rs663520 | hCV8749748 | 83.8 | 2.21 | 1.10 | 2.46 |
| rs552604 | hCV1514816 | 85.4 | 1.87 | 1.47 | 2.40 |
| rs672600 | hCV573098 | 86.4 | 1.66 | 1.58 | 2.30 |
| D11S2002 | GATA30G01 | 87.3 | 1.78 | 1.76 | 2.49 |
| rs1037398 | hCV8755557 | 87.4 | 1.82 | 1.75 | 2.52 |
| rs1013273 | hCV8755635 | 88.4 | 2.01 | 1.61 | 2.61 |
| rs605335 | hCV949590 | 88.5 | 1.99 | 1.59 | 2.57 |
| rs7940296 | hCV3245775 | 89.0 | 1.72 | 1.49 | 2.30 |
| rs11233632 | hCV2077671 | 89.2 | 1.58 | 1.50 | 2.19 |
| rs10751099 | hCV11327826 | 89.5 | 1.49 | 1.51 | 2.12 |
| rs1399619 | hCV7624437 | 89.8 | 1.33 | 1.50 | 1.99 |
| rs290202 | hCV1379796 | 90.3 | 1.12 | 1.45 | 1.81 |
| ss1510291 | hCV8753222 | 90.4 | 1.08 | 1.48 | 1.79 |
| rs4144615 | hCV2946456 | 90.9 | .82 | 1.56 | 1.63 |
| rs2226844 | hCV1511121 | 91.8 | .62 | 1.68 | 1.55 |
| rs12270038 | hCV2965095 | 92.3 | .69 | 1.81 | 1.69 |
| rs11020488 | rs11020488 | 92.4 | .71 | 1.82 | 1.71 |
| rs10830237 | hCV1423593 | 92.5 | .80 | 1.85 | 1.80 |
| rs1369953 | hCV8371719 | 92.6 | .72 | 1.86 | 1.74 |
| rs1892887 | rs1892887 | 92.9 | .65 | 1.86 | 1.71 |
| rs10830528 | rs10830528 | 93.0 | .60 | 2.00 | 1.76 |
| rs1528767 | rs1528767 | 93.5 | .55 | 1.87 | 1.65 |
| rs2514288 | rs2514288 | 93.8 | .52 | 1.68 | 1.49 |
| rs1793089 | hCV8370494 | 93.9 | .55 | 1.62 | 1.47 |
| rs606462 | hCV3160944 | 95.4 | .76 | 1.25 | 1.40 |
| rs493390 | hCV2134445 | 95.5 | .75 | 1.19 | 1.36 |
| rs10501817 | hCV1558437 | 96.1 | .71 | 1.05 | 1.23 |
| rs568878 | hCV3243095 | 97.5 | .55 | 1.09 | 1.11 |
| rs483884 | hCV8366620 | 98.1 | .49 | .99 | .99 |
| rs1023849 | hCV1374954 | 99.7 | .49 | .73 | .83 |
| rs1840054 | hCV3013255 | 100.7 | .44 | .45 | .62 |
| rs2597565 | hCV1828315 | 100.9 | .47 | .40 | .61 |
| rs7925640 | rs7925640 | 101.6 | .40 | .37 | .54 |
| rs901540 | hCV1832533 | 102.0 | .32 | .23 | .38 |
| rs1494471 | rs1494471 | 102.5 | .33 | .25 | .39 |
| rs6590589 | hCV1369078 | 102.8 | .27 | .20 | .32 |
| rs1502277 | hCV1505893 | 103.1 | .22 | .10 | .22 |
| rs566351 | hCV3182870 | 103.5 | .15 | .15 | .19 |
| rs10791487 | hCV372177 | 104.0 | −.23 | .48 | .08 |
| rs10895277 | hCV12072893 | 104.1 | −.22 | .55 | .14 |
| rs1784410 | hCV7492851 | 104.3 | −.22 | .70 | .22 |
| rs634192 | hCV944666 | 104.7 | −.21 | .90 | .35 |
| rs716273 | hCV1335221 | 104.9 | −.12 | 1.06 | .52 |
| D11S2000 | GATA28D01 | 105.2 | −.21 | 1.23 | .55 |
| rs10750684 | hCV1951845 | 105.3 | −.49 | 1.25 | .33 |
| rs578169 | hCV3148094 | 105.7 | −.41 | 1.30 | .43 |
| rs2028816 | hCV3247421 | 106.4 | −.31 | 1.37 | .57 |
| rs6591147 | hCV1636106 | 106.6 | −.26 | 1.36 | .61 |
| rs2000930 | hCV3014100 | 106.9 | −.18 | 1.32 | .63 |
| rs669715 | hCV3250963 | 107.5 | −.32 | 1.17 | .43 |
| rs228592 | hCV2609424 | 107.9 | −.48 | 1.11 | .26 |
| rs10890898 | hCV1849328 | 108.2 | −.57 | 1.08 | .16 |
| rs7116461 | hCV9412214 | 108.6 | −.54 | 1.09 | .19 |
| rs11606636 | hCV3096929 | 109.0 | −.63 | 1.18 | .16 |
| rs1509317 | hCV8910331 | 109.5 | −.70 | 1.40 | .25 |
| D11S1391 | GATA4E01 | 109.7 | −.71 | 1.40 | .24 |
| rs167685 | hCV2073302 | 110.0 | −.73 | 1.36 | .19 |
| rs7926485 | hCV1366793 | 110.6 | −.87 | 1.25 | .02 |
| rs2043055 | hCV11481682 | 111.2 | −.85 | 1.25 | .04 |
| rs321146 | rs321146 | 112.5 | −.61 | 1.20 | .20 |
| rs1022084 | hCV1568571 | 113.1 | −.46 | 1.18 | .32 |
| rs387977 | hCV1057713 | 114.7 | −.27 | 1.10 | .44 |
| rs1792379 | hCV1717400 | 115.2 | −.22 | 1.11 | .48 |
| rs4445669 | hCV12072002 | 115.8 | −.20 | 1.09 | .49 |
| rs1961328 | hCV1254535 | 116.5 | −.20 | 1.02 | .45 |
| rs2846903 | hCV1683155 | 116.5 | −.20 | 1.01 | .45 |
| rs1263149 | hCV2679516 | 117.6 | −.17 | 1.04 | .48 |
| rs528508 | hCV7501076 | 118.7 | −.25 | 1.01 | .40 |
| D11S1998 | GATA23E06 | 120.0 | −.34 | .85 | .23 |
cM values refer to the STRP map of deCODE.
Acknowledgments
We thank the patients and families for their participation. Data and biomaterials from the NIMH Genetics Initiative for Schizophrenia MGS1 families were collected in nine projects. From 1999 to 2004, the principal investigators (PIs) and coinvestigators were: University of Chicago, Chicago (grant R01 MH59571): P.V.G. (collaboration coordinator and PI) and A.R.S.; Baylor College of Medicine, Houston (grant R01 MH59587): F.A. (PI); University of California at Irvine, Irvine (grant R01 MH60870): W.F.B. (PI); University of Iowa, Iowa City (grant R01 MH59566): D.W.B. (PI) and R.R.C.; Washington University, St. Louis (grant R01 MH60879): C.R.C. (PI); University of Colorado, Denver (grant R01 MH59565): R.F. (PI) and A.O.; University of Pennsylvania, Philadelphia (grant R01 MH61675): D.F.L. (PI), and subcontract to Louisiana State University, New Orleans: N.G.B. (subcontract PI); University of Queensland, Brisbane (grant R01 MH59588): B.J.M. (PI); and Mt. Sinai School of Medicine, New York (grant R01 MH59586): J.M.S. (PI). We also thank the individuals at each participating institution for their laboratory, database, and/or clinical contributions, especially Roberta Fishman, Layla Kassem, Eric B. Carpenter, Gregory J. Burrell, Christos Pantelis, Robert J. Barrett, and Douglas A. Fugman. Genotyping was performed at CIDR (for STRPs) and at Evanston Northwestern Healthcare Research Institute (for SNPs).
Appendix A
Table A1.
Example of Renumbering to Assure that Allelic Designations Are Comparable between Waves I and II for D6S2427[Note]
|
Allele Frequency |
OriginalAllelicDesignation |
RenumberedAllelicDesignation |
||||
| FragmentSize(bp) | Wave I | Wave II | Wave I | Wave II | Wave I | Wave II |
| 187 | .003 | .003 | 1 | 1 | 1 | 1 |
| 195 | .004 | … | 2 | … | 2 | … |
| 199 | .025 | .019 | 3 | 2 | 3 | 3 |
| 200 | … | .002 | … | 3 | … | 4 |
| 202 | .057 | … | 4 | … | 5 | … |
| 203 | … | .028 | … | 4 | … | 5 |
| 206 | .116 | … | 5 | … | 6 | … |
| 207 | .003 | .148 | 6 | 5 | 7 | 6 |
| 209 | … | .001 | … | 6 | … | 8 |
| 210 | .283 | .247 | 7 | 7 | 9 | 9 |
| 211 | .003 | .003 | 8 | 8 | 10 | 10 |
| 213 | … | .001 | … | 9 | … | 11 |
| 214 | .229 | .256 | 9 | 10 | 12 | 12 |
| 215 | .005 | .002 | 10 | 11 | 13 | 13 |
| 218 | .153 | .153 | 11 | 12 | 14 | 14 |
| 219 | … | .001 | … | 13 | … | 15 |
| 222 | .060 | .072 | 12 | 14 | 16 | 16 |
| 226 | .038 | .045 | 13 | 15 | 17 | 17 |
| 227 | .001 | … | 14 | … | 18 | … |
| 230 | .021 | .016 | 15 | 16 | 19 | 19 |
| 234 | .001 | .002 | 16 | 17 | 20 | 20 |
Note.— The presence of rare alleles in one wave but not the other means that the original allelic designations from CIDR cannot be used when the waves are combined. For instance, allele 10 in wave I has a frequency of 0.5%, whereas, in wave II, it has a frequency of 25.6%. In fact, the fragment of size 214 bp (relabled as allele 12 when the waves are combined) has a frequency of 24.7% in this sample. When a large proportion of families have ungenotyped parents, as is the case here, the results of a linkage analysis necessarily will depend on the estimates of the allele frequencies. Accordingly, when genotypes are produced in different waves, by different laboratories, or by use of different technologies, it is critically important to assure that the alleles align correctly.
Table A2.
Example of Allelic Renumbering for D15S128 Used to Combine the EA and AA Families[Note]
|
Allele Frequency |
||||
| FragmentSize(bp) | AlleleNumber | EA | AA | Renumbered Allele(AA Only) |
| 196 | 1 | .0005 | … | … |
| 198 | 2 | .0011 | … | … |
| 200 | 3 | .0643 | .0141 | 14 |
| 202 | 4 | .0050 | .1141 | 15 |
| 204 | 5 | .0050 | .1043 | 16 |
| 206 | 6 | .2494 | .3424 | 17 |
| 208 | 7 | .2629 | .0826 | 18 |
| 210 | 8 | .1661 | .1065 | 19 |
| 212 | 9 | .1275 | .0576 | 20 |
| 213/214 | 10 | .0833 | .0620 | 21 |
| 215/216 | 11 | .0263 | .0185 | 22 |
| 217 | 12 | .0062 | .0011 | 23 |
| 219 | 13 | .0022 | .0032 | 24 |
| 221 | 14 | … | .0924 | 25 |
| 225 | 15 | … | .0011 | 26 |
Note.— Shown are the allele frequencies for D15S128 in the EA and AA families. The original recoding of this STRP resulted in 15 alleles. Alleles 1–13 are the only alleles present in the EA sample. Accordingly, we renumbered the alleles of the genotypes in the AA families, as indicated. In the linkage analysis of the combined sample, the allele frequencies sum to 200%, resulting in the correct computation of conditional probabilities of the genotypes of missing parents.
Figure A1.
Illustration of the counting of sib pairs. Because there is no optimal method for counting independent affected half sib pairs when there are full and half sibs in the same family, all possible half sib pairs (AHAPs) have been counted in these cases. Consider, for instance, the two families shown here. Pedigree A is counted as two statistically independent AFSPs and three AHAPs. Pedigree B is counted as two AFSPs and four AHAPs.
Table A3.
Previous Chromosome 8p SZ Linkage Reports[Note]
|
Best Single-Point Result |
Best Multipoint Result |
|||||||||||||
| Study | Ethnicity | No. of ASPs | No. of Families | Affection | P | Marker | Cytogenetic | cM | Mb | P | Marker | Cytogenetic | cM | Mb |
| Initial major study: | ||||||||||||||
| Blouin et al. 1998a | EA | … | 54 | SZ and SA | … | … | … | … | … | .00019 | D8S1771 | 8p21.2 | 50.05 | 25.5 |
| Follow-up sample, Blouin et al. 1998b | EA | … | 51 | SZ and SA | … | … | … | … | … | .023 | D8S1752 | 8p21.2 | 46.26 | 22.7 |
| Supportive studies: | ||||||||||||||
| SLCG 1996 (for chromosomes 3, 6, and 8)c | EA | 633 | 384 | SZ and SA | .0014 | D8S261 | p22 | 37.04 | 17.8 | .005 | D8S133 | p21.3 | 41.55 | 22.0 |
| Kaufmann et al. 1998d | AA | 42 | 30 | SZ and SADT | .01333 | D8S1819 | p23.1 | 9.96 | 6.7 | … | … | … | … | … |
| Brzustowicz et al. 2000e | EA | … | 22 | SZ, SA, and SSD | >.05 | D8S136 | p21.3 | 43.96 | 22.5 | >.05 | D8S136 | p11.21 | 64.96 | 40.3 |
| Gurling et al. 2001f | EA | … | 13 | SZ, SA, and UFP | .0001 | D8S503 | p23.1 | 16.19 | 9.3 | .0005 | D8S1771 | p21.2 | 50.05 | 25.5 |
| Straub et al. 2002g | EA | … | 270 | Very broad (D1–D9) | … | … | … | … | … | .005 | D8S1731 | p22 | 31.73 | 15.2 |
| Stefansson et al. 2002h | EA | … | 33 | SZ, SA, and UFP | … | … | … | … | … | Suggestive | D8S278 | p12 | 60.87 | 32.6 |
| DeLisi et al. 2002i | EA | 130 | 95 | Broad | … | … | … | … | … | .016 | Unknown | p23.1 | 18.20 | 9.8 |
| Moises et al. 1995j | EA | … | 5 | SZ and SA | .04 | D8S298 | p21.3 | 43.96 | 21.8 | … | … | … | … | … |
| Lindholm et al. 2001k | EA | … | 1 | SZ | Suggestive | D8S550 | p23.1 | 21.33 | 10.9 | … | … | … | … | … |
| Park et al. 2004l | EA | … | 40 | Psychotic BP | Suggestive | D8S382 | p12 | 51.15 | 26.0 | Suggestive | D8S382 | p12 | 51.15 | 26.0 |
Note.— cM values refer to the Marshfield map; cytogenetic locations and Mb on chromsome 8 are from the UCSC July 2003 freeze. Predominant ethnicities are listed; see footnotes for each study for more detail. ASPs are noted when available; otherwise, the number of families is listed. Affection status is listed for the most significant model if there is more than one. Psychotic BP = bipolar I, SA manic type (Park et al. 2004); SADT = SA depressed type; SSD = schizophrenia spectrum disorders (nonaffective psychotic disorder, schizotypal personality disorder, or paranoid personality disorder); UFP = unspecified functional psychosis. Very broad (D1–D9) refers to SZ, SA–poor outcome, “simple SZ,” schizotypal personality disorder (PD), schizophreniform disorder, delusional disorder, atypical psychosis, SA–good outcome, psychotic affective disorders, paranoid PD, avoidant PD, schizoid PD, nonpsychotic affective disorders, anxiety disorders, alcoholism, and the rest of the PDs. Broad refers to SZ, SA, psychosis not otherwise specified, schizotypal PD, and paranoid PD (DeLisi et al. 2002). The lowest P values—sometimes limited to a notation of “suggestive,” per standard criteria (Lander and Kruglyak 1995)—are listed, when available, for single- and multipoint analyses, along with marker and location information.
The initial major study by Blouin et al. (1998) (also reported earlier by Pulver et al. [1995]), both genome scan and replication samples, was performed on mostly EA samples (the 54 scan families and the 51 replication families derived from a total of 105 families composed of 44 EA from Maryland, 31 additional EA from throughout the United States, along with 6 AA, 5 Ashkenazim, 1 Amish, 10 Italian, 3 Polish, and 5 Greek families). The genome-scan nonparametric linkage (NPL) Zall=3.64 (P=.00019) near D8S1771, and the 95% CI was noted to be 18 cM wide.
The same study by Blouin et al. (1998) also had a replication sample. The replication study NPL Zall=1.95 (P=.023) at D8S1752.
This was a large multisite replication study (SLCG 1996), analyzed with and without the Johns Hopkins families reported elsewhere (Pulver et al. 1995; Blouin et al. 1998), that was composed of predominantly EA families (samples from 13 EA sites and 1 South African Bantu-speaking sample). The replication portion—that is, the “new” families, as far as linkage reported for chromosome 8—comprised (depending on available genotypes) 633 ASPs (independent) and 384–463 families. The single-point analysis showed a recessive heterogeneity LOD (HLOD) of 2.22 (P=.0014) for the new sample at D8S261. The multipoint analyses showed a maximum LOD score (MLS) for ASPs of 1.58 (P=.005) for the new sample near D8S133.
In this study (Kaufmann et al. 1998), 42 ASPs (some half siblings) from 30 families showed the strongest single-point finding in the region with NPL Zmax=2.27 (P=.0133) at D8S1819.
In this study (Brzustowicz et al. 2000), 22 families (21 Celtic and 1 German) were reported without P values, but all linkages were noted to be nonsignificant on empirical testing. The best single-point score was at D8S136, with a homogeneity Zmax=2.09 and a heterogeneity Zmax=2.16 (0.80 linked), and the best multipoint (three-point) score was at ∼21 cM centromeric to D8S136, with a heterogeneity Zmax=2.80 (0.90 linked).
In this study (Gurling et al. 2001), 13 families (5 British and 8 Icelandic) were reported with a recessive HLOD=3.6 (P=.0001) at D8S503 (note that there was a broad region with positive markers) and a recessive five-point HLOD=3.2 (P=.0005) at D8S1771 (note that homogeneity LOD=3.0).
In this study (Straub et al. 2002), 270 EA (Irish) families showed a maximum multipoint NPL score of 2.56 (P=.005) near D8S1731.
In this study (Stefansson et al. 2002), 33 EA (Icelandic) families were reported without P values, but linkage was clearly stated to be at the “suggestive” level. Both multipoint analyses showed peaks at D8S278: allele-sharing LOD=2.53 and parametric affecteds-only LOD=3.48.
In this study (DeLisi et al. 2002), 130 ASPs (independent, some half siblings) from 95 EA families (from the central valley of Costa Rica surrounding the city of San Jose, which originated from Spanish settlers, with some characteristics of a population isolate) showed a nonparametric multipoint MLS=1.19 (P=.016), with the multipoint peak reported to be at 18.2 cM.
In this study (Moises et al. 1995), five large EA (Icelandic) families showed a weighted-rank pairwise correlation (WRPC) score of 120.05 and WRPC variance of 4,630 (P=.04) at D8S298.
In this study (Lindholm et al. 2001), one very large EA (northern Sweden) family was reported without P values, but the linkage finding was clearly stated to be at the “suggestive” level via simulations that pointed to an MLS >2.2 as suggestive and an MLS >3.7 as significant. At D8S550, an MLS of 2.90 at recombination fraction 0.15 was noted when allele frequencies from the local control population were used; when the allele frequencies from the pedigree itself were used, the MLS was 0.70.
In this study (Park et al. 2004), 40 mostly EA families (29 EA and 11 Middle-Eastern) were reported without P values, but the linkage finding was clearly stated to be at the “suggestive” level by identification of a two-point LOD >1.9 as suggestive (and a two-point LOD >3.3 as significant). At D8S382, LOD=2.09 and MLS=3.46.
Table A4.
Previous Chromosome 5 Pericentromeric SZ Linkage Reports[Note]
|
Best Single-Point Result |
Best Multipoint Result |
||||||||||||
| Study | Ethnicity | No. of Families | Affection | P | Marker | Cytogenetic | cM | Mb | P | Marker | Cytogenetic | cM | Mb |
| Initial major study: | |||||||||||||
| Bespalova et al. 2005a | EA | 1 | SZ and STPD | Sig. | D5S111 | 5p13.3 | 47.09 | 33.3 | Significant | D5S111 | 5p13.3 | 47.09 | 33.3 |
| Supportive studies: | |||||||||||||
| Cooper-Casey et al. 2005b | EA | 1 | SZ and SA | Suggestive | D5S426 | 5p13.2 | 51.99 | 34.8 | Suggestive | D5S426 | 5p13.2 | 51.99 | 34.8 |
| Garver et al. 2001c | AA and EA | 30 | SZ, SADT, and CA | … | … | … | … | … | .015 | D5S426 | 5p13.2 | 51.99 | 34.8 |
| Straub et al. 2002d | EA | 90 | Narrow (D1–D2) | … | D5S1470 | 5p13.3 | 45.34 | 32.5 | … | … | … | … | … |
| Paunio et al. 2001e | EA | 53 | Broad (LC4) | … | D5S2504 | 5q11.1 | 59f | 49.9 | … | … | … | … | … |
| Gurling et al. 2001g | EA | 13 | SZ, SA, UFP, STPD, and SDPD | .002 | D5S407 | 5q11.2 | 64.67 | 56.0 | .003 | D5S407 | 5q11.2 | 64.67 | 56.0 |
Note.— cM values refer to the Marshfield map; cytogenetic locations and Mb on chromsome 8 are from the UCSC July 2003 freeze. Predominant ethnicities are listed; see notes for each study for more detail. ASPs are noted when available; otherwise, the number of families is listed. Affection status is listed for the most significant model if there is more than one. CA = cluster A personality disorders (SDPD, STPD, and paranoid personality disorder); SADT = SA depressed type; SDPD = schizoid personality disorder; STPD = schizotypal personality disorder; UFP = unspecified functional psychosis. Narrow (D1–D2) refers to SZ, SA–poor outcome, and “simple SZ.” Broad (LC4) refers to SZ, SA, CA, schizophreniform disorder, delusional disorder, brief psychotic disorder, psychosis not otherwise specified, and severe major affective disorders. The lowest P values—sometimes limited to a notation of “suggestive” or “significant,” per standard criteria (Lander and Kruglyak 1995)—are listed, when available, for single- and multipoint analyses, along with marker and location information.
The fine-mapping follow-up (Bespalova et al. 2005) of the initial major study (Silverman et al. 1996) was performed on a single large Puerto Rican family, primarily descended from Spanish ancestors. Analyses showed a maximum two-point LOD=3.72 at D5S111 under dominant inheritance at a recombination fraction of 0.01 and a 90% penetrance (Silverman et al. 1996); at the same marker, the multipoint LOD=4.37 (Bespalova et al. 2005), although no P values were provided.
This study was performed (Cooper-Casey et al. 2005) on a single large family from the central valley of Costa Rica primarily descended from Spanish ancestors. Analyses showed a maximum two-point LOD=1.80 at D5S426 under dominant inheritance; the nonparamteric multipoint LOD=2.70 at the same marker, with the results characterized as suggestive evidence, although no P values were provided.
This study (Garver et al. 2001) of 30 families (23 AA and 7 EA) showed a multipoint nonparametric linkage score of 2.21 (P=.015) at D5S426.
This study (Straub et al. 2002) of 270 EA (Irish) families showed a maximum pairwise heterogeneity LOD (HLOD) of 2.18 at D5S1470 under dominant inheritance and the narrow (D1–D2) affection status in the family set A (90 families).
This study (Paunio et al. 2001) of 53 EA (Finnish) families from the country’s “internal isolate” showed Zmax=2.33 at D5S395 and Zmax=2.36 at D5S2504 under dominant inheritance and affection liability class 4 (LC4), which was the broadest affection status model (Paunio et al. 2001).
Estimated.
This study (Gurling et al. 2001) of 13 EA families (5 British and 8 Icelandic) reported a dominant HLOD=2.5 (P=.002) at D5S407 under the spectrum affection status and a dominant five-point HLOD=2.2 (P=.003) peaking at the same marker (note that LOD=2.3 in a single pedigree as well).
Web Resources
The URLs for data presented herein are as follows:
- Center for Inherited Disease Research (CIDR), http://www.cidr.jhmi.edu/
- Consensus Coding DNA Sequence (CCDS) database, http://www.ncbi.nlm.nih.gov/projects/CCDS/ (for March 2005 report)
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for build 34)
- HapMap, http://www.hapmap.org/
- NIMH Center for Collaborative Genetics Studies of Mental Disorders, http://zork.wustl.edu/nimh/
- NIMH Human Genetics Initiative for Schizophrenia, http://zork.wustl.edu/nimh/NIMH_initiative/NIMH_initiative_link.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SZ) [PubMed]
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for May 2004 assembly of the human genome)
- UniSTS, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unists (for comprehensive database of sequence-tagged sites)
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association (APA) (1994) Diagnostic and Statistical Manual of Mental Disorders. APA, Washington, DC [Google Scholar]
- Badner JA, Gershon ES (2002) Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry 7:405–411 10.1038/sj.mp.4001012 [DOI] [PubMed] [Google Scholar]
- Bespalova IN, Angelo GW, Durner M, Smith CJ, Siever LJ, Buxbaum JD, Silverman JM (2005) Fine mapping of the 5p13 locus linked to schizophrenia and schizotypal personality disorder in a Puerto Rican family. Psychiatr Genet 15:205–210 10.1097/00041444-200509000-00012 [DOI] [PubMed] [Google Scholar]
- Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, et al (1998) Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet 20:70–73 10.1038/1734 [DOI] [PubMed] [Google Scholar]
- Boehnke M (1991) Allele frequency estimation from data on relatives. Am J Hum Genet 48:22–25 [PMC free article] [PubMed] [Google Scholar]
- Boehnke M, Cox NJ (1997) Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet 61:423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Matsumoto N, Giglio S, Martin CL, Roseberry JA, Zuffardi O, Ledbetter DH, Weber JL (2003) Common long human inversion polymorphism on chromosome 8p. In: Goldstein DR (ed) Science and statistics: a festschrift for Terry Speed. Vol 40. IMS lecture notes—monograph series. Institute of Mathematical Statistics, Beachwood, OH, pp 237–245 [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Weber JL (1998) Estimation of pairwise relationships in the presence of genotyping errors. Am J Hum Genet 63:1563–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS (2000) Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science 288:678–682 10.1126/science.288.5466.678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno AG, Marshall EJ, Coid B, Macdonald AM, Ribchester TR, Davies NJ, Venturi P, Jones LA, Lewis SW, Sham PC, Gottesman II, Farmer AE, McGuffin P, Reveley AM, Murray RM (1999) Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Arch Gen Psychiatry 56:162–168 10.1001/archpsyc.56.2.162 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Cloninger CR (2002) The discovery of susceptibility genes for mental disorders. Proc Natl Acad Sci USA 99:13365–13367 10.1073/pnas.222532599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloninger CR, Kaufmann CA, Faraone SV, Malaspina D, Svrakic DM, Harkavy-Friedman J, Suarez BK, Matise TC, Shore D, Lee H, Hampe CL, Wynne D, Drain C, Markel PD, Zambuto CT, Schmitt K, Tsuang MT (1998) Genome-wide search for schizophrenia susceptibility loci: the NIMH Genetics Initiative and Millennium Consortium. Am J Med Genet 81:275–281 [DOI] [PubMed] [Google Scholar]
- Cohen J (1960) A coefficient of agreement for nominal scales. Educ Psychol Meas 20:37–46 [Google Scholar]
- Cooper-Casey K, Mesen-Fainardi A, Galke-Rollins B, Llach M, Laprade B, Rodriguez C, Riondet S, Bertheau A, Byerley W (2005) Suggestive linkage of schizophrenia to 5p13 in Costa Rica. Mol Psychiatry 10:651–656 10.1038/sj.mp.4001640 [DOI] [PubMed] [Google Scholar]
- Craddock N, O’Donovan MC, Owen MJ (2005) The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet 42:193–204 10.1136/jmg.2005.030718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Sham PC (1996) Population stratifications can cause false positive linkage results if founders are untyped. Ann Hum Genet 60:261–263 [DOI] [PubMed] [Google Scholar]
- DeLisi LE, Mesen A, Rodriguez C, Bertheau A, LaPrade B, Llach M, Riondet S, Razi K, Relja M, Byerley W, Sherrington R (2002) Genome-wide scan for linkage to schizophrenia in a Spanish-origin cohort from Costa Rica. Am J Med Genet 114:497–508 10.1002/ajmg.10538 [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc 39:1–38 [Google Scholar]
- Duan J, Martinez M, Sanders AR, Hou C, Krasner AJ, Schwartz DB, Gejman PV (2005) Neuregulin 1 (NRG1) and schizophrenia: analysis of a US family sample and the evidence in the balance. Psychol Med 35:1599–1610 10.1017/S0033291705005428 [DOI] [PubMed] [Google Scholar]
- Dunn G, Bertelsen S, Bierut L, Hinrichs A, Jin C, Kauwe J, Suarez BK. Microsatellites versus SNPs in linkage analysis for quantitative and qualitative measures. BMC Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver DL, Holcomb J, Mapua FM, Wilson R, Barnes B (2001) Schizophrenia spectrum disorders: an autosomal-wide scan in multiplex pedigrees. Schizophr Res 52:145–160 10.1016/S0920-9964(01)00157-8 [DOI] [PubMed] [Google Scholar]
- Gerber DJ, Hall D, Miyakawa T, Demars S, Gogos JA, Karayiorgou M, Tonegawa S (2003) Evidence for association of schizophrenia with genetic variation in the 8p21.3 gene, PPP3CC, encoding the calcineurin gamma subunit. Proc Natl Acad Sci USA 100:8993–8998 10.1073/pnas.1432927100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon ES, DeLisi LE, Hamovit J, Nurnberger JI Jr, Maxwell ME, Schreiber J, Dauphinais D, Dingman CW, Guroff JJ (1988) A controlled family study of chronic psychoses: schizophrenia and schizoaffective disorder. Arch Gen Psychiatry 45:328–336 [DOI] [PubMed] [Google Scholar]
- Giglio S, Broman KW, Matsumoto N, Calvari V, Gimelli G, Neumann T, Ohashi H, Voullaire L, Larizza D, Giorda R, Weber JL, Ledbetter DH, Zuffardi O (2001) Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet 68:874–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard KA, Hopkins PJ, Hall JM, Witte JS (2000) Linkage disequilibrium and allele-frequency distributions for 114 single-nucleotide polymorphisms in five populations. Am J Hum Genet 66:216–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Bertelsen A (1989) Confirming unexpressed genotypes for schizophrenia: risks in the offspring of Fischer’s Danish identical and fraternal discordant twins. Arch Gen Psychiatry 46:867–872 [DOI] [PubMed] [Google Scholar]
- Gottesman II, Shields J (1982) Schizophrenia: the epigenetic puzzle. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- Grigull J, Alexandrova R, Paterson AD (2001) Clustering of pedigrees using marker allele frequencies: impact on linkage analysis. Genet Epidemiol 21:S61–S66 [DOI] [PubMed] [Google Scholar]
- Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, Petursson H, Curtis D (2001) Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet 68:661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heston LL (1966) Psychiatric disorders in foster home reared children of schizophrenic mothers. Br J Psychiatry 112:819–825 [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307:1072–1079 10.1126/science.1105436 [DOI] [PubMed] [Google Scholar]
- Holmans P, Zubenko GS, Crowe RR, DePaulo JR Jr, Scheftner WA, Weissman MM, Zubenko WN, Boutelle S, Murphy-Eberenz K, MacKinnon D, McInnis MG, Marta DH, Adams P, Knowles JA, Gladis M, Thomas J, Chellis J, Miller E, Levinson DF (2004) Genomewide significant linkage to recurrent, early-onset major depressive disorder on chromosome 15q. Am J Hum Genet 74:1154–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Shete S, Amos CI (2004) Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet 75:1106–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns LC, Cannon M, Singleton N, Murray RM, Farrell M, Brugha T, Bebbington P, Jenkins R, Meltzer H (2004) Prevalence and correlates of self-reported psychotic symptoms in the British population. Br J Psychiatry 185:298–305 10.1192/bjp.185.4.298 [DOI] [PubMed] [Google Scholar]
- Kaufmann CA, Suarez B, Malaspina D, Pepple J, Svrakic D, Markel PD, Meyer J, Zambuto CT, Schmitt K, Matise TC, Harkavy Friedman JM, Hampe C, Lee H, Shore D, Wynne D, Faraone SV, Tsuang MT, Cloninger CR (1998) NIMH Genetics Initiative Millenium Schizophrenia Consortium: linkage analysis of African-American pedigrees. Am J Med Genet 81:282–289 [DOI] [PubMed] [Google Scholar]
- Kendler KS, McGuire M, Gruenberg AM, O’Hare A, Spellman M, Walsh D (1993a) The Roscommon Family Study. I. Methods, diagnosis of probands, and risk of schizophrenia in relatives. Arch Gen Psychiatry 50:527–540 [DOI] [PubMed] [Google Scholar]
- ——— (1993b) The Roscommon Family Study. III. Schizophrenia-related personality disorders in relatives. Arch Gen Psychiatry 50:781–788 [DOI] [PubMed] [Google Scholar]
- ——— (1993c) The Roscommon Family Study. IV. Affective illness, anxiety disorders, and alcoholism in relatives. Arch Gen Psychiatry 50:952–960 [DOI] [PubMed] [Google Scholar]
- Kendler KS, McGuire M, Gruenberg AM, Spellman M, O’Hare A, Walsh D (1993d) The Roscommon Family Study. II. The risk of nonschizophrenic nonaffective psychoses in relatives. Arch Gen Psychiatry 50:645–652 [DOI] [PubMed] [Google Scholar]
- Kety SS, Rosenthal D, Wender PH, Schulsinger F (1971) Mental illness in the biological and adoptive families of adopted schizophrenics. Am J Psychiatry 128:302–306 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- Lange K, Weeks D, Boehnke M (1988) Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol 5:471–472 10.1002/gepi.1370050611 [DOI] [PubMed] [Google Scholar]
- Leckman JF, Sholomskas D, Thompson WD, Belanger A, Weissman MM (1982) Best estimate of lifetime psychiatric diagnosis: a methodological study. Arch Gen Psychiatry 39:879–883 [DOI] [PubMed] [Google Scholar]
- Levinson DF, Holmans P. The effect of linkage disequilibrium on linkage analysis of incomplete pedigrees. BMC Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Mowry BJ, Escamilla MA, Faraone SV (2002) The Lifetime Dimensions of Psychosis Scale (LDPS): description and interrater reliability. Schizophr Bull 28:683–695 [DOI] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, et al (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am J Hum Genet 73:34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm E, Ekholm B, Shaw S, Jalonen P, Johansson G, Pettersson U, Sherrington R, Adolfsson R, Jazin E (2001) A schizophrenia-susceptibility locus at 6q25, in one of the world’s largest reported pedigrees. Am J Hum Genet 69:96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay EA, Morris MA, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, Shprintzen R, Antonarakis SE, Baldini A, Pulver AE (1995) Schizophrenia and chromosomal deletions within 22q11.2. Am J Hum Genet 56:1502–1503 [PMC free article] [PubMed] [Google Scholar]
- Maier W, Lichtermann D, Minges J, Hallmayer J, Heun R, Benkert O, Levinson DF (1993) Continuity and discontinuity of affective disorders and schizophrenia: results of a controlled family study. Arch Gen Psychiatry 50:871–883 [DOI] [PubMed] [Google Scholar]
- Matise TC, Sachidanandam R, Clark AG, Kruglyak L, Wijsman E, Kakol J, Buyske S, et al (2003) A 3.9-centimorgan-resolution human single-nucleotide polymorphism linkage map and screening set. Am J Hum Genet 73:271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell ME (1992) Family Interview for Genetic Studies (FIGS): a manual for FIGS. Clinical Neurogenetics Branch, Intramural Research Program, NIMH, Bethesda, MD [Google Scholar]
- Moises HW, Yang L, Kristbjarnarson H, Wiese C, Byerley W, Macciardi F, Arolt V, et al (1995) An international two-stage genome-wide search for schizophrenia susceptibility genes. Nat Genet 11:321–324 10.1038/ng1195-321 [DOI] [PubMed] [Google Scholar]
- Nurnberger JI Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T (1994) Diagnostic interview for genetic studies: rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry 51:849–859 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke DH, Gottesman II, Suarez BK, Rice J, Reich T (1982) Refutation of the general single-locus model for the etiology of schizophrenia. Am J Hum Genet 34:630–649 [PMC free article] [PubMed] [Google Scholar]
- Ott J (1985) A chi-square test to distinguish allelic association from other causes of phenotypic association between two loci. Genet Epidemiol 2:79–84 10.1002/gepi.1370020108 [DOI] [PubMed] [Google Scholar]
- Owen MJ, Williams NM, O’Donovan MC (2004) The molecular genetics of schizophrenia: new findings promise new insights. Mol Psychiatry 9:14–27 10.1038/sj.mp.4001444 [DOI] [PubMed] [Google Scholar]
- Park N, Juo SH, Cheng R, Liu J, Loth JE, Lilliston B, Nee J, Grunn A, Kanyas K, Lerer B, Endicott J, Gilliam TC, Baron M (2004) Linkage analysis of psychosis in bipolar pedigrees suggests novel putative loci for bipolar disorder and shared susceptibility with schizophrenia. Mol Psychiatry 9:1091–1099 10.1038/sj.mp.4001541 [DOI] [PubMed] [Google Scholar]
- Paunio T, Ekelund J, Varilo T, Parker A, Hovatta I, Turunen JA, Rinard K, Foti A, Terwilliger JD, Juvonen H, Suvisaari J, Arajarvi R, Suokas J, Partonen T, Lonnqvist J, Meyer J, Peltonen L (2001) Genome-wide scan in a nationwide study sample of schizophrenia families in Finland reveals susceptibility loci on chromosomes 2q and 5q. Hum Mol Genet 10:3037–3048 10.1093/hmg/10.26.3037 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver AE, Lasseter VK, Kasch L, Wolyniec P, Nestadt G, Blouin JL, Kimberland M, Babb R, Vourlis S, Chen H, Lalioti M, Morris MA, Karayiorgou M, Ott J, Meyers D, Antonarakis SE, Housman D, Kazazian HH (1995) Schizophrenia: a genome scan targets chromosomes 3p and 8p as potential sites of susceptibility genes. Am J Med Genet 60:252–260 10.1002/ajmg.1320600316 [DOI] [PubMed] [Google Scholar]
- Rosenthal D, Wender PH, Kety SS, Welner J, Schulsinger F (1971) The adopted-away offspring of schizophrenics. Am J Psychiatry 128:307–311 [DOI] [PubMed] [Google Scholar]
- Silverman JM, Greenberg DA, Altstiel LD, Siever LJ, Mohs RC, Smith CJ, Zhou G, Hollander TE, Yang XP, Kedache M, Li G, Zaccario ML, Davis KL (1996) Evidence of a locus for schizophrenia and related disorders on the short arm of chromosome 5 in a large pedigree. Am J Med Genet 67:162–171 [DOI] [PubMed] [Google Scholar]
- SLCG (1996) Additional support for schizophrenia linkage on chromosomes 6 and 8: a multicenter study. Schizophrenia Linkage Collaborative Group for Chromosomes 3, 6 and 8. Am J Med Genet 67:580–594 [DOI] [PubMed] [Google Scholar]
- Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A, et al (2005) A common inversion under selection in Europeans. Nat Genet 37:129–137 10.1038/ng1508 [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, et al (2002) Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet 71:877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stompe T, Willinger U, Fischer G, Meszaros K, Berger P, Strobl R, Berger K, Isenberg E, Todd RD, Cloninger CR, Reich T, Aschauer HN (1998) The unified biosocial model of personality in schizophrenia families and controls. Psychopathology 31:45–51 10.1159/000029022 [DOI] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, O’Neill FA, Walsh D, Kendler KS (2002) Genome-wide scans of three independent sets of 90 Irish multiplex schizophrenia families and follow-up of selected regions in all families provides evidence for multiple susceptibility genes. Mol Psychiatry 7:542–559 10.1038/sj.mp.4001051 [DOI] [PubMed] [Google Scholar]
- Strauss JS (1969) Hallucinations and delusions as points on continua function: rating scale evidence. Arch Gen Psychiatry 21:581–586 [DOI] [PubMed] [Google Scholar]
- Sugawara H, Harada N, Ida T, Ishida T, Ledbetter DH, Yoshiura K, Ohta T, Kishino T, Niikawa N, Matsumoto N (2003) Complex low-copy repeats associated with a common polymorphic inversion at human chromosome 8p23. Genomics 82:238–244 10.1016/S0888-7543(03)00108-3 [DOI] [PubMed] [Google Scholar]
- Taylor MA, Berenbaum SA, Jampala VC, Cloninger CR (1993) Are schizophrenia and affective disorder related? Preliminary data from a family study. Am J Psychiatry 150:278–285 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Speer M, Ott J (1993) Chromosome-based method for rapid computer simulation in human genetic linkage analysis. Genet Epidemiol 10:217–224 10.1002/gepi.1370100402 [DOI] [PubMed] [Google Scholar]
- van Os J, Hanssen M, Bijl RV, Ravelli A (2000) Strauss (1969) revisited: a psychosis continuum in the general population? Schizophr Res 45:11–20 10.1016/S0920-9964(99)00224-8 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]