Abstract
Seventeen-marker simple tandem repeat genetic analysis of Irish Y chromosomes reveals a previously unnoted modal haplotype that peaks in frequency in the northwestern part of the island. It shows a significant association with surnames purported to have descended from the most important and enduring dynasty of early medieval Ireland, the Uí Néill. This suggests that such phylogenetic predominance is a biological record of past hegemony and supports the veracity of semimythological early genealogies. The fact that about one in five males sampled in northwestern Ireland is likely a patrilineal descendent of a single early medieval ancestor is a powerful illustration of the potential link between prolificacy and power and of how Y-chromosome phylogeography can be influenced by social selection.
Irish Y-chromosome haplotype analysis has highlighted the striking predominance of a SNP-delineated haplogroup, R1b3, that displays a cline of increasing frequency from the Middle East to northwestern Europe, peaking with near fixation in western parts of Ireland (Hill et al. 2000; Rosser et al. 2000). Within this SNP-defined haplogroup, higher-resolution studies have identified a 6-microsatellite haplotype common on the European Atlantic facade, consequently termed “the Atlantic modal haplotype” (AMH) (Wilson et al. 2001).
Several factors suggest that further Irish Y-chromosome analysis is warranted. First, its position at the European extreme in the cline of R1b3 frequency points to an unusual population history, perhaps with genetic signatures of past demographic events that are relatively undisturbed by migration. Second, as in many European societies, Y chromosomes and Irish surnames share (in the main) patrilineal transmission. These names are among the oldest cultural lineage markers in the world and emerged from an earlier tribal nomenclature that also emphasized patrilineal relationships. Hence, Y-chromosome comparisons between surnames may be informative with respect to older genealogical links that stretch back into Irish prehistory and mythology. Finally, the indigenous medieval Irish or “Gaelic” social order differed from that of much of Europe, survived until the 16th century, and was, arguably, highly patriarchal and pastoralist. These sociocultural features may have left a distinctive biological legacy.
We initially examined 796 Y chromosomes from all areas of Ireland. DNA samples were collected, along with written informed consent, via buccal swab from volunteers at a range of locations in Ireland (Hill et al. 2000; McEvoy et al. 2004). Each Y chromosome was typed hierarchically for 11 Y-linked binary polymorphisms that define common European haplogroups (Y Chromosome Consortium 2002; Jobling and Tyler-Smith 2003). The SNPs M269, SRY-2627, SRY-1532, M35, M9, 92R7, and TAT were genotyped using a PCR/RFLP method. M170, M26, and M89 were typed using Tetra-primer ARMS-PCR (Ye et al. 2001). Insertion/deletion polymorphism 12f2 can be assayed by PCR alone. The most common haplogroup in Ireland, R1b3, roughly equivalent to haplogroup 1 in an earlier nomenclature (Hill et al. 2000), is defined by a T→C transition at the M269 SNP (Cruciani et al. 2002). To allow higher-resolution studies of Irish Y chromosome lineages, samples were also typed for 17 microsatellites—DYS19, DYS388, DYS390, DYS391, DYS392, DYS393, DYS434, DYS435, DYS436, DYS437, DYS438, DYS439, DYS389I, DYS389B (calculated by subtracting the DYS389I repeat score from that of DYS389II), DYS460, DYS461, and DYS462—in three PCR multiplex reactions, essentially as described elsewhere (Bosch et al. 2002). Of the samples, 221 had been typed elsewhere for DYS19, DYS390, DYS391, DYS392, DYS393, and DYS389I (Hill et al. 2000). The Y-chromosome haplotypes of these were extended to include all of the above markers. Full genotype data from this study is available in online-only tab-delimited text files 1 and 2 (which can be downloaded and opened into an Excel spreadsheet) and at B.M.’s Web site.
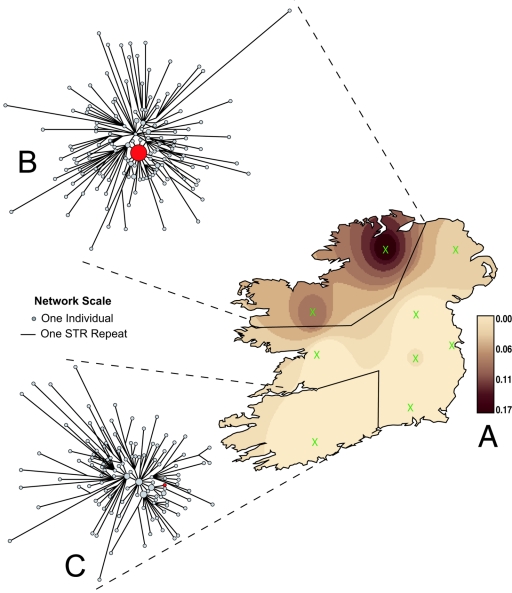
The vast majority (85.4%) of the Y chromosomes belonged to the R1b3 haplogroup. The greater resolution afforded by the 17-marker system disperses the AMH lineage into 94 different haplotypes, none of which predominates. However, it simultaneously uncovers a distinct alternative Irish modal haplotype (IMH), also within haplogroup R1b3, that, together with its one-mutational-step (IMH+1) neighbors, accounts for 8.2% of the island sample (IMH 17-marker haplotype with use of the loci as ordered above is 14-12-25-11-14-13-9-11-12-15-12-12-13-16-11-10-11). The IMH distribution is uneven, showing a distinct frequency peak in northwestern Ireland, where it accounts for 16.9% of Y chromosomes (21.5% when one-step derivatives are included) (fig. 1A). The inequitable distribution is further illustrated by comparison of two regional median-joining (MJ) networks of R1b3 Y chromosomes. These graphically display the phylogeny or interrelationship of different Y-chromosome haplotypes. In the northwest (fig. 1B), IMH+1 Y chromosomes form a predominant and phylogenetically coherent starlike cluster that is consistent with a single origin. This feature is absent from a corresponding network of Y chromosomes from southwestern Ireland (fig. 1C).
Figure 1.
Phylogeography of the IMH Y-chromosome lineage. A, Contour map displaying the frequency distribution of the IMH. It shows a clear focus on northwestern areas of the island, where it reaches a regional maximum of 16.9% (21.5% including one-repeat unit derivatives). MJ networks of 17-STR marker R1b3 Y-chromosome haplotypes, sampled in northwestern (n=166) (B) and southwestern (n=125) (C) Ireland. Each circle represents a different Y-chromosome haplotype, with area proportional to frequency. Line length between haplotypes shows mutational divergence. The IMH (filled red) and one-step neighbors predominate in the northwest, whereas this lineage is virtually absent in the southwest. The synthetic surface map was constructed using the ArcView package (version 3.2) (Environmental Systems Research Institute). Observed frequencies in nine Irish sample groupings of similar size (geographic coordinates marked with a green X) were used, according to the inverse-weighting method, to interpolate the frequency at other coordinates, generating 10 equal-interval surface contours.
The distribution of the IMH lineage was also examined in a broader British Isles context, through use of Y-chromosome data for 1,525 individuals across 22 sampling points reported by Capelli et al. (2003). However, since the former were typed for only six microsatellites (DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393), comparison was truncated to include only these loci. The 6-STR IMH is virtually absent from much of Britain but reaches frequencies of up to 7.3% (16.7% including likely one-step derivatives) in western and central Scottish locations. Scotland is known to have had substantial historical and prehistorical links with the northern part of Ireland; for example, the Celtic language still spoken in parts of western Scotland is the closest relative of Irish Gaelic.
The global occurrence of a 7-STR locus IMH (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, and DYS393) was next examined in 28,650 Y chromosomes from 249 geographically defined populations (but ignoring sample populations of <10 individuals) deposited in the Y Chromosome Haplotype Reference Database. The localization of the haplotype was evident with only 38 matches detected (a worldwide occurrence of just 0.13%), with an Irish population sample showing the highest frequency (4.8%). Interestingly, it was also observed in multiple North American population samples. For example, ∼2% of European American New Yorkers carry the truncated IMH. Large-scale emigration to North America from Ireland is well recorded. Indeed, given historically high rates of Irish emigration to North America and other parts of the world, it seems likely that the number of descendents worldwide runs to perhaps 2–3 million males.
An analogously predominant Y-chromosome lineage is found in Central Asian populations at a frequency of ∼8% (Zerjal et al. 2003). The level of diversity assessed using 15 microsatellite markers supported an origin ∼1,000 year ago. This, together with details of its geography, suggested that the lineage’s rapid rise to predominance occurred through an association with the male-line descendents of Genghis Khan and the dynasties he founded in the region. Several observations led us to propose that the high frequency of the IMH in these data may be the result of a similar linkage to an ancient, enduring dynasty.
Gaelic society placed great emphasis on family relationships organized around a strongly patrilineal system (derbhfine) in which land and title could be handed down to successors chosen from within a kin group of male-lineage relatives. This wider inheritance cohort resulted in a decreased likelihood of dissociation of lineage from power (O’Croinin 1995). Also, whereas medieval Ireland was Christian, earlier marriage customs persisted and allowed divorce and concubinage. One feature of these customs was that illegitimate sons were claimed and had rights protected by law (Jaski 2000). As in other polygynous societies, the siring of offspring was related to power and prestige (Betzig 1995). For example, Lord Turlough O’Donnell (d. 1423) had 18 sons with 10 different women and counted 59 grandsons in the male line (Connolly 2002).
Turlough and other O’Donnells were members of the most powerful and remarkably durable royal lineage in medieval Gaelic Ireland, the Uí Néill, literally translated as “descendents of Niall.” Gaelic genealogies were important records used to validate claims to prestige and power and linked most ruling families in the northern part of Ireland, prior to the Elizabethan conquest, to the Uí Néill, who claimed high-kingship of Ireland from the 7th to the 11th century ad (Pender 1951). The ultimate origin of this dynasty is attributed to the conquering sons of the eponymous and possibly mythological 5th-century warlord, Niall of the Nine Hostages. The historical region under Uí Néill power coincides with the peak in the frequency of the IMH.
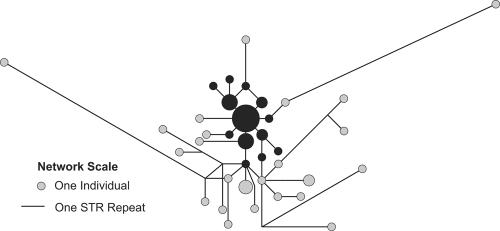
In line with the early Irish emphasis on ancestry, it is not surprising that Ireland has one of the largest surviving bodies of early genealogical records. These records range from the solidly historical to the dubiously mythical. Furthermore, since the main purpose of the records was to validate claims to power and property, they were often altered or forged to accord with prevailing political circumstances. Nonetheless, they do present the opportunity to directly test the circumstantial geographic association of the IMH lineage and the Uí Néill dynasty. Modern Irish surnames began in the 10th century ad but often arose from the pre-existing dynastic/population groupings. The Y chromosomes of 59 men possessing names with a purported common origin within the Uí Néill genealogies were collected and genotyped for 17 Y-chromosome STRs. The phylogenetic relationships between these Y-chromosome haplotypes, shown in figure 2, were reconstructed using MJ networks (Bandelt et al. 1999) with the software NETWORK, version 4.1 (Fluxus Engineering). To simplify networks, a reduced median (Bandelt et al. 1995) algorithm was initially applied to the data, followed by analysis with the MJ method. A large coherent cluster centered on the IMH is predominant. The time to the most-recent common ancestor (TMRCA) of this lineage was estimated with the ρ statistic (Morral et al. 1994) in NETWORK, with use of a mutation rate of 1 per 2,131 years for a 17-marker haplotype (Zhivotovsky et al. 2004). At 1,730 (SD 670) years ago, it is at least consistent with an early-medieval time frame. A similar TMRCA analysis of the IMH lineage in the general northwestern population sample (shown in fig. 1B) is also roughly consistent with this time frame (1,010 years ago [SD 390). Furthermore, the presence of the IMH across several surnames, which are up to 1,000 years old, certainly suggests an origin predating their adoption. However, it may be that the rise in frequency did not begin with this TMRCA but rather was associated with a group of descendents sometime later. The residual diversity in the Uí Néill sample is probably the cumulative consequence of nonpaternity events and the induction into the clan structure of unrelated males. If any of the Y chromosomes associated with these events were similar to the IMH, because of local homogeneity and recurrent mutation, then they will not be distinguishable here.
Figure 2.
MJ network of Y chromosomes from men with Uí Néill–derived surnames. The sample population (n=59) features the IMH as its modal haplotype, which is ancestral to a phylogenetically coherent cluster (blackened circles) that accounts for over half (52.5%) of the sample group. The cluster was taken to extend to all haplotypes that could be continuously traced to the ancestral IMH Y chromosome via a pathway of increasing frequency, a topology roughly in line with expectations under a Poisson-distributed mutational process.
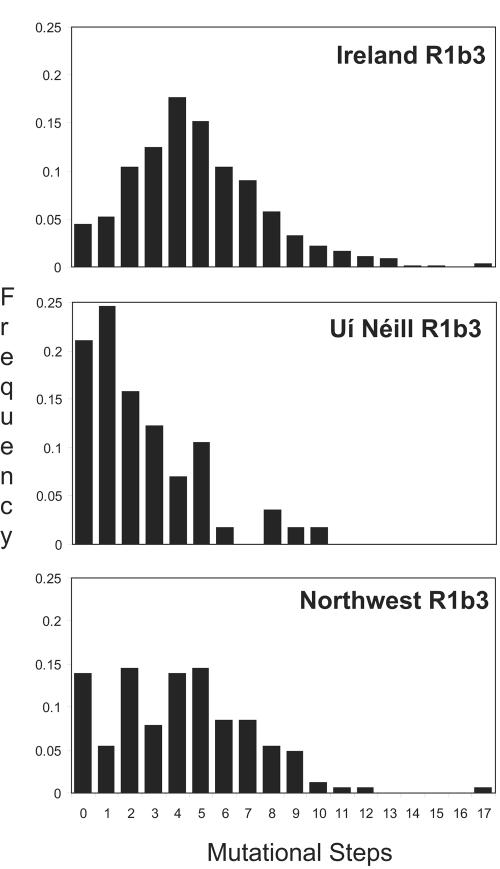
Figure 3 shows the distribution of mutational divergence from the IMH in haplogroup R1b3 samples from Ireland as a whole, the Uí Néill–surname population, and the northwestern region where they originated. The smooth distribution in Ireland generally is an obvious contrast to the low-divergence bias (zero and one-step mutational distance) seen in the Uí Néill sample. The disrupted distribution in the general northwestern population illustrates the marked impact of this single recent ancestor on the Y-chromosome structure of the entire region.
Figure 3.
Frequency distributions of STR mutational steps from the IMH of all haplotypes in three different groupings. In each case, a minority of non-R1b3 haplotypes was excluded as known outliers. Notably, the sample of subjects with surnames linked by genealogical tradition to the Uí Néill dynasty shows a significantly reduced pattern of divergence from this central haplotype when compared with the background normal distribution seen in the Irish population generally. The powerful legacy of the IMH ancestor on the population structure of the northwestern region is borne out by the disrupted distribution observed in this area. The Uí Néill surname samples were not genotyped for binary markers, since it became apparent during that general population survey that R1b3 chromosomes could be readily detected through STR profiles alone. The Uí Néill sample population was composed of the following surnames (sample number): (O’)Gallagher (12), (O’)Boyle (9), (O’)Doherty (5), O’Donnell (4), O’Connor (3), Cannon (3), Bradley (2), O’Reilly (2), Flynn (2), (Mc)Kee (2), Campbell (1), Devlin (1), Donnelly (1), Egan (1), Gormley (1), Hynes (1), McCaul (1), McGovern (1), McLoughlin (1), McManus (1), McMenamin (1), Molloy (1), O’Kane (1), O’Rourke (1), and Quinn (1).
The significance of IMH prominence in the Uí Néill cohort was next tested using a Mann-Whitney test, with the number of differences from the IMH for each haplotype (excluding outlying non-R1b3 chromosomes) used as a metric. The Uí Néill sample (n=57) showed a significantly higher affinity with the IMH (P<.001) than with a general R1b3 northwestern Ireland geographic population (n=166). Importantly, a genealogically based control (n=54) composed of R1b3 Y chromosomes from men with surnames originating in the northwest but not within our cohort of Uí Néill derivatives was also found to be significantly different (P=.006). Thus, it seems unlikely that the IMH rose to a high frequency because of a general feature of northwestern Irish demographic history. The rate of increase in the frequency of the Uí Néill lineage is ∼1.21 per generation, under the assumption of an evenly spread rise from ad ∼500 to the present (some 50 generations, under a 30-year paternal generation interval [Tremblay and Vézina 2000]). This rate is somewhat lower than that associated with the patrilineal legacy of Genghis Khan (Zerjal et al. 2003) but still represents a greater increase than that conferred on loci by many known episodes of natural selection, including the sickle cell trait (Li 1975) and lactose persistence (Bersaglieri et al. 2004).
Genealogical association together with the predominance and pattern of variation of the IMH strongly suggest a rise in frequency due to strong social selection associated with the hegemony of the Uí Néill dynasty and their descendents. Figures such as Niall of the Nine Hostages reside at the cusp of mythology and history, but our results do seem to confirm the existence of a single early-medieval progenitor to the most powerful and enduring Irish dynasty. They also lend support to the veracity and remarkable knowledge preservation of the genealogical and oral traditions of Gaelic Ireland and give a powerful and specific illustration of the link between prolificacy and power in one European society within the past 2 millennia.
Supplementary Material
Acknowledgments
We are indebted to those who volunteered DNA samples and to Emmeline Hill and other colleagues who facilitated their collection. We thank A. R. Freeman for assistance with figure preparation. This work was supported by a Wellcome Trust studentship in Bioarchaeology (to L.T.M.).
Web Resources
The URLs for data presented herein are as follows:
- B.M.’s Web site, http://www.gen.tcd.ie/molpopgen/data.htm (for full Y-chromosome genotypes)
- Fluxus Engineering, http://www.fluxus-engineering.com/sharenet.htm (for NETWORK)
- Y Chromosome Haplotype Reference Database, http://www.ystr.org/index.html
References
- Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN (2004) Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet 74:1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig L (1995) Medieval monogamy. J Fam Hist 20:181–216 [Google Scholar]
- Bosch E, Lee AC, Calafell F, Arroyo E, Henneman P, de Knijff P, Jobling MA (2002) High resolution Y chromosome typing: 19 STRs amplified in three multiplex reactions. Forensic Sci Int 125:42–51 10.1016/S0379-0738(01)00627-2 [DOI] [PubMed] [Google Scholar]
- Capelli C, Redhead N, Abernethy JK, Gratrix F, Wilson JF, Moen T, Hervig T, Richards M, Stumpf MP, Underhill PA, Bradshaw P, Shaha A, Thomas MG, Bradman N, Goldstein DB (2003) A Y chromosome census of the British Isles. Curr Biol 13:979–984 10.1016/S0960-9822(03)00373-7 [DOI] [PubMed] [Google Scholar]
- Connolly SJ (2002) The Oxford companion to Irish history. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, Wallace DC, Oefner PJ, Torroni A, Cavalli-Sforza LL, Scozzari R, Underhill PA (2002) A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 70:1197–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill EW, Jobling MA, Bradley DG (2000) Y-chromosome variation and Irish origins. Nature 404:351–352 10.1038/35006158 [DOI] [PubMed] [Google Scholar]
- Jaski B (2000) Early Irish kingship and succession. Four Courts Press, Dublin [Google Scholar]
- Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612 10.1038/nrg1124 [DOI] [PubMed] [Google Scholar]
- Li WH (1975) The first arrival time and mean age of a deleterious mutant gene in a finite population. Am J Hum Genet 27:274–286 [PMC free article] [PubMed] [Google Scholar]
- McEvoy B, Richards M, Forster P, Bradley DG (2004) The Longue Durée of genetic ancestry: multiple genetic marker systems and Celtic origins on the Atlantic facade of Europe. Am J Hum Genet 75:693–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morral N, Bertranpetit J, Estivill X, Nunes V, Casals T, Gimenez J, Reis A, et al (1994) The origin of the major cystic fibrosis mutation (δF508) in European populations. Nat Genet 7:169–175 10.1038/ng0694-169 [DOI] [PubMed] [Google Scholar]
- O’Croinin D (1995) Early medieval Ireland 400–1200. Longman, London [Google Scholar]
- Pender S (ed) (1951) The O’Clery book of genealogie. Analecta Hibernica, no 18. Dublin Stationary Office, Dublin [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, et al (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M, Vézina H (2000) New estimates of intergenerational time intervals for the calculation of age and origins of mutations. Am J Hum Genet 66:651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JF, Weiss DA, Richards M, Thomas MG, Bradman N, Goldstein DB (2001) Genetic evidence for different male and female roles during cultural transitions in the British Isles. Proc Natl Acad Sci USA 98:5078–5083 10.1073/pnas.071036898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 10.1101/gr.217602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Dhillon S, Ke X, Collins AR, Day IN (2001) An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 29:E88 10.1093/nar/29.17.e88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Xue Y, Bertorelle G, Wells RS, Bao W, Zhu S, Qamar R, Ayub Q, Mohyuddin A, Fu S, Li P, Yuldasheva N, Ruzibakiev R, Xu J, Shu Q, Du R, Yang H, Hurles ME, Robinson E, Gerelsaikhan T, Dashnyam B, Mehdi SQ, Tyler-Smith C (2003) The genetic legacy of the Mongols. Am J Hum Genet 72:717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioğlu C, Kayser M, Morar B, Kivisild T, Scozzari R, Cruciani F, Destro-Bisol G, Spedini G, Chambers GK, Herrera RJ, Yong KK, Gresham D, Tournev I, Feldman MW, Kalaydjieva L (2004) The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 74:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.