Abstract
Congenital lactase deficiency (CLD) is a severe gastrointestinal disorder characterized by watery diarrhea in infants fed with breast milk or other lactose-containing formulas. We initially assigned the CLD locus by linkage and linkage disequilibrium on 2q21 in 19 Finnish families. Here we report the molecular background of CLD via characterization of five distinct mutations in the coding region of the lactase (LCT) gene. Twenty-seven patients out of 32 (84%) were homozygous for a nonsense mutation, c.4170T→A (Y1390X), designated “Finmajor.” Four rare mutations—two that result in a predicted frameshift and early truncation at S1666fsX1722 and S218fsX224 and two point mutations that result in substitutions Q268H and G1363S of the 1,927-aa polypeptide—confirmed the lactase mutations as causative for CLD. These findings facilitate genetic testing in clinical practice and enable genetic counseling for this severe disease. Further, our data demonstrate that, in contrast to common adult-type hypolactasia (lactose intolerance) caused by a variant of the regulatory element, the severe infancy form represents the outcome of mutations affecting the structure of the protein inactivating the enzyme.
Congenital lactase deficiency (CLD [MIM 223000]) is an autosomal, recessively inherited, severe gastrointestinal disorder of infants. Lactase activity in intestinal mucosa is decreased to 0–10 U/g of protein, leading to copious watery diarrhea shortly after breast-feeding or the introduction of lactose-containing formulas. However, duodenal morphology and the activities of maltase, sucrase, and isomaltase are normal. The severe osmotic diarrhea followed by dehydration, acidosis, and weight loss usually afflict sufferers during the first days of life. Despite the symptoms, CLD infants are vigorous and hungry. When they are on a lactose-free diet, the symptoms subside rapidly and the children have normal growth and psychomotor development (Launiala et al. 1966; Savilahti et al. 1983). CLD has an incidence of 1:60,000 and is one of the 36 rare monogenic disorders enriched in the Finnish population (Peltonen et al. 1999; Norio 2003a, 2003b). However, cases have also been reported elsewhere in the world (Holzel 1967).
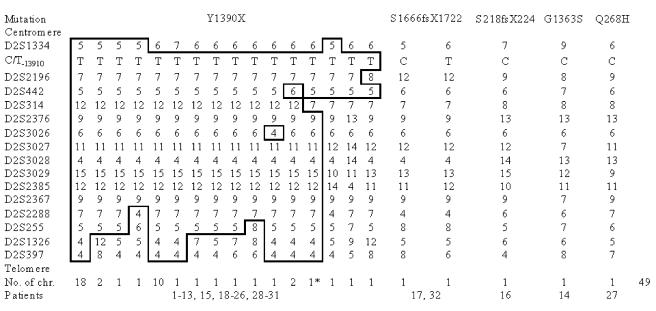
In our linkage report, we defined the critical DNA region for CLD as from D2S314 to D2S2385. These microsatellites shared haplotypes from both parents in 16 of 19 studied families and gave the highest linkage disequilibrium (LD) in disease alleles (Järvelä et al. 1998). However, sequence analyses of regional transcripts failed to reveal disease-causing mutations (data not shown). Importantly, the critical locus position excluded the LCT (MIM 603202) gene itself. Further, a published report of the sequence analysis of the LCT gene in a Finnish CLD patient failed to reveal disease-causing mutations in the coding region (Poggi and Sebastio 1991). However, since no mutations were found in transcripts on the linkage region, we wanted to confirm the position of the CLD locus with additional markers providing higher resolution. We genotyped 15 microsatellite markers covering 5.88 cM on 2q21–2q22 in 24 families with a total of 32 affected children. We were able to construct 21 different haplotypes in 48 disease chromosomes. One major haplotype, cen-5-T−13910-7-5-12-9-6-11-4-15-12-9-7-5-4-4-tel, was present in 18 disease chromosomes, representing the founder haplotype from which 15 of the remaining 20 different haplotypes have been derived (fig. 1). Five disease chromosomes, each of which carried a different haplotype, did not relate to the major haplotype. On the basis of information about shared haplotypes, we concluded that a critical region would contain the LCT gene, and we proceeded to perform sequence analysis of this gene. The transcript map of the critical DNA region for CLD is shown in figure 2.
Figure 1.
CLD haplotypes ordered according to each identified mutation. Patients 14, 16, 17, 27, and 32 are compound heterozygous for Y1390X and their respective rare mutations. Microsatellite mutation of D2S314 in one patient increased the total number of CLD chromosomes to 49. Microsatellite markers were searched for and ordered on the basis of genetic maps of the Marshfield Clinic Research Foundation, as well as the physical map of the UCSC Genome Browser (December 2001 assembly). The Baylor College of Medicine (BCM) Search Launcher repeat-masker algorithm was used to identify novel microsatellites from clones between D2S314 and D2S2385 (Smith et al. 1996). Finally, 15 microsatellites and C/T−13910, a SNP associated with adult-type hypolactasia, were analyzed, covering 5.88 cM of the CLD region. DNA was extracted by a standard protocol (Vandenplas et al. 1984) from the peripheral blood samples. PCR was performed and the genotypes were assigned as described elsewhere (Ylisaukko-oja et al. 2004). Haplotypes were constructed using the GENEHUNTER 2.1 program (Kruglyak et al. 1996). The CLD founder haplotype was generated with the assumption that there were a minimum number of recombinations.
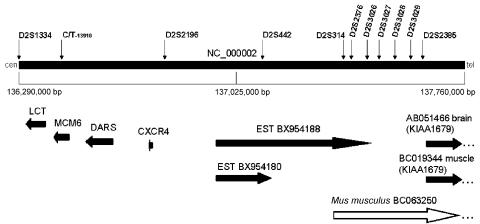
Figure 2.
Transcript map of the critical DNA region for CLD. The physical map was constructed using public (International Human Genome Sequencing Consortium) Human Genome Project annotations. Evidence for transcripts and for conserved amino acids was searched with BLAST against different databases, including the dbEST and Refseq protein databases. Complete sequence annotations were performed by the NIX program (UK Human Genome Mapping Project Resource Centre) and the UCSC Genome Browser. LCT, minichromosome maintenance deficient 6 (MCM6 [MIM 601806]), aspartyl-tRNA synthetase (DARS [MIM 603084]), chemokine receptor type 4 (CXCR4 [MIM 162643]), and KIAA1679 are the known genes in the region (horizontal arrows). The mouse ortholog (BC063250) of human KIAA1679 suggests that ESTs BX954188 and BX954180 may be related to transcripts of KIAA1679. Microsatellite markers are indicated by vertical arrows. The centromere is on the left.
We identified five distinct mutations in the LCT gene in 32 patients from 24 families. Three mutations were predicted to lead to a premature truncation of lactase, and two were missense mutations that resulted in amino acid substitutions (table 1 and figs. 3 and 4). All the chromosomes with the major disease haplotype carried a nonsense mutation, c.4170T→A (Finmajor), resulting in Y1390X, a premature stop codon in exon 9 predicting the truncation of 537 amino acids. Finmajor was detected in 27 of 32 patients who were homozygous for the conserved founder haplotype or its modification. A total of 38 parents (5 did not participate) and 14 healthy siblings were found to be carriers of the Finmajor mutation. Two patients had a deletion of four nucleotides, c.4998_5001delTGAG, in their paternal disease chromosome in exon 14, leading to a frameshift and a premature stop codon after 55 altered amino acids (S1666fsX1722). These patients were compound heterozygous for Finmajor and S1666fsX1722. On the basis of genealogical studies, both of these mutations are known to originate from central and northern Finland. The third mutation, c.653_654delCT, is a deletion of two nucleotides in exon 2, predicting a frameshift change at codon 218 and protein truncation at codon 224, S218fsX224. One patient carried this mutation in her paternal disease chromosome. The fourth mutation is a c.804G→C transversion at codon 268, leading to an amino acid substitution of histidine for glutamine, Q268H, in the last nucleotide of exon 3. One patient carried this mutation in her maternal disease chromosome. The fifth mutation is a c.4087G→A transition resulting in a missense substitution of serine for an uncharged glycine, G1363S, at codon 1363 in exon 9. One patient carried this mutation in his maternal disease chromosome. A healthy sibling of the family also carried this rare mutation. These patients were compound heterozygous for the Finmajor and their respective rare mutations. On the basis of the birthplaces of the great-grandparents of the patients, mutations three, four, and five would be assumed to originate from eastern Finland. The CLD mutations and the corresponding lactase activities measured from duodenal biopsy specimens are shown in table 1.
Table 1.
CLD Mutations in the LCT Gene and Observed Lactase Activities of Duodenal Biopsy Specimens
| DNA Mutation | Protein Mutation | Exon | Patient | Mean LactaseProtein Activity(U/g of Protein) | Genotype |
| c.4170T→A | Y1390X | 9 | 1–13, 15, 18–26, 28–31 | 2.1 (range 0–7)a | Homozygote |
| c.4998_5001delTGAG | S1666fsX1722 | 14 | 17, 32 | 5 (range 3–7) | Compound heterozygoteb |
| c.653_654delCT | S218fsX224 | 2 | 16 | 2 | Compound heterozygoteb |
| c.804G→C | Q268H | 3 | 27 | 5 | Compound heterozygoteb |
| c.4087G→A | G1363S | 9 | 14 | 3 | Compound heterozygoteb |
Lactase activities of patients 11, 18, 20, 26, and 29 were not available.
Compound heterozygote for Y1390X.
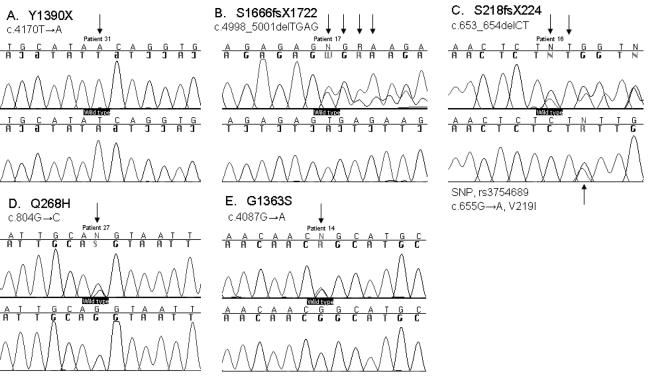
Figure 3.
CLD mutations in the LCT gene. A–E, DNA sequence chromatograms of five identified CLD mutations. One mutation, Y1390X, (A) is a homozygote, whereas the rest of the mutations are heterozygotes. The first row shows affected sequences and the second row wild-type sequences. In addition, c.655G→A, a SNP (rs3754689) leading to V219I, is shown in panel E (Boll et al. 1991). PCR was performed using the genomic DNA-amplifying promoter region, exons, flanking intron sequences, and 3′-UTRs of the LCT gene (primers are available on request). Sequencing was performed in both directions, and the sequenced products were electrophoresed on an ABI 3730 DNA analyzer (Applied Biosystems) in accordance with the manufacturer’s instructions and were analyzed using ABI Sequencing Analysis 3.3 (Applied Biosystems) and Sequencher 4.1 (Gene Godes).
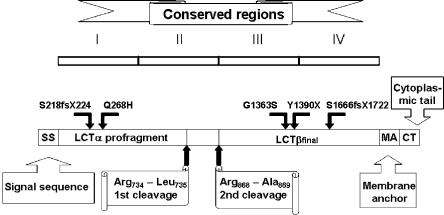
Figure 4.
Structure of lactase and the location of the identified CLD mutations. The genomic size of the LCT gene is ∼55 kb and is composed of 17 exons (Boll et al. 1991). The size of messenger RNA (mRNA) is 6,274 bases, and the primary translation product (prolactase) is 1,927 amino acids. The prolactase contains a cleavable signal sequence from Met1 to Gly19 that guides the polypeptide to the endoplasmic reticulum (von Heijne 1986; Mantei et al. 1988). The region from Ser20 to Thr1882 consists of four homologous domains (I–IV). The prolactase is processed by two proteolytic cleavages. The first, an intracellular cleavage, occurs between Arg734 and Leu735. The second cleavage in the intestinal lumen between Arg868 and Ala869 generates lactaseβfinal, the mature enzyme (Jacob et al. 1996; Wüthrich et al. 1996). The figure is modified from Naim (2001).
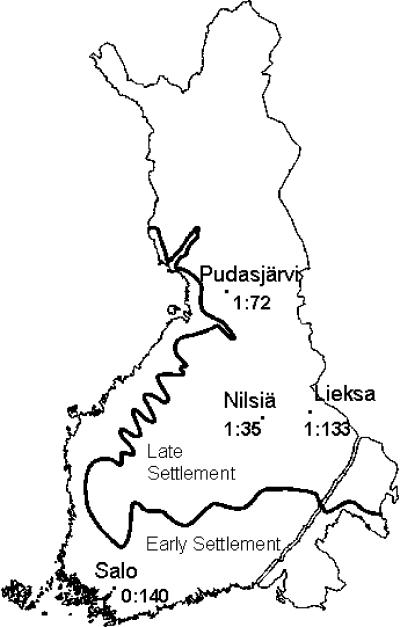
Ninety percent (43/48) of the disease chromosomes carried the Finmajor mutation. The carrier frequency of Finmajor was determined among 556 anonymous blood donors representing both the early and the later settlement regions of Finland. The highest carrier frequency of 1:35 (4/140) was seen in a little town, Nilsiä, in central Finland (fig. 5). This particular geographical region shows an enrichment of ancestors on genealogical studies (Järvelä et al. 1998). No carriers for other mutations (S218fsX224, S1666fsX1722, Q268H, and G1363S) were observed in any regional subpopulation screened.
Figure 5.
Carrier frequency of Y1390X in four subpopulations in the early and later settlement regions of Finland. The carrier frequencies of the CLD mutations were determined from 556 anonymous blood donors obtained from the Finnish Red Cross Blood Transfusion Service.
The nascent lactase polypeptide contains four (I-IV) conserved structural and functional regions, but after posttranslational processing only two (III-IV) remain in the mature lactase enzyme (Mantei et al. 1988). The polypeptide has two catalytically active sites: phlorizin hydrolase activity is located in region III and lactase activity in region IV (Zecca et al. 1998; Arribas et al. 2000). Finmajor, Y1390X, truncates lactase polypeptide in the beginning of region IV. We employed an allele-specific minisequencing method to characterize the potential impact of the mutation at the steady-state transcript level of the LCT allele carrying the nucleotide change resulting in Y1390X (Rasinperä et al. 2005). We used a duodenal biopsy sample of a patient who was heterozygous for the nonsense mutation resulting in Y1390X and heterozygous for C/T−13910, the SNP associated with adult-type hypolactasia (MIM 223100) (Enattah et al. 2002; Rasinperä et al. 2005). We were able to determine that the transcript level of the allele carrying the premature stop codon was at the same level as the transcript of the C−13910 allele, which typically represents ca 8% of the expressed LCT mRNA (Kuokkanen et al. 2003). We expect this to be caused by nonsense-mediated mRNA decay, a pathway which degrades mRNA induced by premature termination codons, eliminating the production of harmful truncated proteins (Maquat 2005). This mechanism has been seen to modulate human disease phenotypes caused by disease-associated premature nonsense or frameshift mutations (Baserga and Benz 1988; Frischmeyer and Dietz 1999; Inoue et al. 2004). Furthermore, three of our CLD patients with the Y1390X mutation were included in an earlier study in which protein patterns of brush-border fragments were preliminarily examined (Freiburghaus et al. 1976). CLD patients had a complete or nearly complete absence of brush-border lactase in studied duodenal biopsies (Freiburghaus et al. 1976), which is in good agreement with our result that the mutant Y1390X LCT allele is down-regulated by nonsense-mediated decay. The second mutation, S1666fsX1722, truncates lactase in the middle of region IV, and the third mutation, S218fsX224, truncates polypeptide early, at region I. In theory, these mutant alleles may also be subjected to nonsense-mediated decay. The fourth mutation, Q268H, hits region I, substituting the basic histidine for an uncharged glutamine. Histidine has an imidazole ring that can be uncharged or positively charged, depending on its microenvironment. Although Q268H does not affect the mature enzyme, it may affect the structure of the lactase α-profragment, which has been demonstrated to act during lactase-folding processes as an intramolecular chaperone towards the mature enzyme (Naim et al. 1994; Jacob et al. 2002). The importance of the Q268 residue is also implied by cross-species conservation from human through chimpanzee, rabbit, cow, rat, and mouse. The fifth mutation, G1363S, replaces the smallest amino acid, glycine, with serine, which has a hydroxyl group that makes it polar and reactive. The replacement of the small glycine by the larger serine may alter the three-dimensional structure of the polypeptide. Since the G1363S mutation is located in the mature lactase at the end of region III, near the catalytically active sites, it may have serious functional consequences. Also, G1363 is conserved across species. The schematic structure of lactase polypeptide and the location of the five CLD mutations are illustrated in fig. 4.
Opposite to the rare congenital lactase deficiency, adult-type hypolactasia (MIM 223100) is the most common enzyme deficiency worldwide. It is caused by developmental down-regulation of lactase activity during childhood or early adulthood (Sahi 1994). The decline of lactase activity is a normal physiological phenomenon; however, the majority of northern Europeans have the ability to maintain lactase activity and digest lactose throughout life (lactase persistence). We previously identified C/T−13910, a variant that is located 13.9 kb upstream of the LCT gene and is associated with the adult-type hypolactasia trait (Enattah et al. 2002). The down-regulation of lactase activity operates at the transcriptional level. LCT mRNA quantitated by minisequencing method showed that the T−13910 allele allows lactase gene expression and maintains lactase activity, whereas the C−13910 allele lets the down-regulation operate normally (Kuokkanen et al. 2003; Rasinperä et al. 2005). The lactase activity in Finns declines at 5–12 years of age (Rasinperä et al. 2004, 2005). It is of interest that the CLD Finmajor mutation, Y1390X in lactase, was systematically found here on the background of the lactase-persistent allele, T−13910, in Finns. Thus, some CLD Finmajor carriers with the CT−13910 genotype may suffer from lactose-induced symptoms after down-regulation of the C−13910 allele.
In conclusion, this study confirms that both human lactase deficiencies are related to DNA variants affecting the LCT gene. Interestingly, the mutations resulting in the severe congenital form have direct consequences for the polypeptide leading to nonsense-mediated mRNA decay in at least 90% of the disease alleles, whereas the critical nucleotide for the milder adult-type phenotype represents a distal enhancer regulating the transcript levels in intestinal cells (Kuokkanen et al. 2003; Olds and Sibley 2003; Troelsen et al. 2003; Rasinperä et al. 2005). Importantly, our study facilitates direct DNA-based diagnosis and carrier identification of CLD, which has so far been based on clinical symptoms supplemented by lactase activity assays from intestinal biopsy specimens.
Acknowledgments
We thank the families for participating in the study. Kaija-Leena Kolho is thanked for providing a CLD family for the study. Teemu Perheentupa, Elli Kempas, and Ulla Sundström are acknowledged for technical assistance. The Finnish Cultural Foundation, the Center of Excellence in Disease Genetics of the Academy of Finland, the Sigrid Jusélius Foundation of Helsinki, and Helsinki University Research Funding are gratefully acknowledged for their financial support.
Web Resources
URLs for data presented herein are as follows:
- Baylor College of Medicine (BCM) Search Launcher, http://searchlauncher.bcm.tmc.edu/
- Marshfield Clinic Research Foundation Genetic Map, http://research.marshfieldclinic.org/
- National Center for Biotechnology Information BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- Arribas JC, Herrero AG, Martin-Lomas M, Canada FJ, He S, Withers SG (2000) Differential mechanism-based labeling and unequivocal activity assignment of the two active sites of intestinal lactase/phlorizin hydrolase. Eur J Biochem 267:6996–7005 10.1046/j.1432-1327.2000.01784.x [DOI] [PubMed] [Google Scholar]
- Baserga SJ, Benz EJ Jr (1988) Nonsense mutations in the human beta-globin gene affect mRNA metabolism. Proc Natl Acad Sci USA 85:2056–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll W, Wagner P, Mantei N (1991) Structure of the chromosomal gene and cDNAs coding for lactase-phlorizin hydrolase in humans with adult-type hypolactasia or persistence of lactase. Am J Hum Genet 48:889–902 [PMC free article] [PubMed] [Google Scholar]
- Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Järvelä I (2002) Identification of a variant associated with adult-type hypolactasia. Nat Genet 30:233–237 10.1038/ng826 [DOI] [PubMed] [Google Scholar]
- Freiburghaus AU, Schmitz J, Schindler M, Rotthauwe HW, Kuitunen P, Launiala K, Hadorn B (1976) Protein patterns of brush-border fragments in congenital lactose malabsorption and in specific hypolactasia of the adult. N Engl J Med 294:1030–1032 [DOI] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC (1999) Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 8:1893–1900 10.1093/hmg/8.10.1893 [DOI] [PubMed] [Google Scholar]
- Holzel A (1967) Sugar malabsorption due to deficiencies of disaccharidase activities and of monosaccharide transport. Arch Dis Child 42:341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Khajavi M, Ohyama T, Hirabayashi S, Wilson J, Reggin JD, Mancias P, Butler IJ, Wilkinson MF, Wegner M, Lupski JR (2004) Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet 36:361–369 10.1038/ng1322 [DOI] [PubMed] [Google Scholar]
- Jacob R, Peters K, Naim HY (2002) The prosequence of human lactase-phlorizin hydrolase modulates the folding of the mature enzyme. J Biol Chem 277:8217–8225 10.1074/jbc.M111500200 [DOI] [PubMed] [Google Scholar]
- Jacob R, Radebach I, Wüthrich M, Grünberg J, Sterchi EE, Naim HY (1996) Maturation of human intestinal lactase-phlorizin hydrolase: generation of the brush border form of the enzyme involves at least two proteolytic cleavage steps. Eur J Biochem 236:789–795 10.1111/j.1432-1033.1996.t01-1-00789.x [DOI] [PubMed] [Google Scholar]
- Järvelä I, Enattah NS, Kokkonen J, Varilo T, Savilahti E, Peltonen L (1998) Assignment of the locus for congenital lactase deficiency to 2q21, in the vicinity of but separate from the lactase-phlorizin hydrolase gene. Am J Hum Genet 63:1078–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kuokkanen M, Enattah NS, Oksanen A, Savilahti E, Orpana A, Järvelä I (2003) Transcriptional regulation of the lactase-phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut 52:647–652 10.1136/gut.52.5.647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launiala K, Kuitunen P, Visakorpi JK (1966) Disaccharidases and histology of duodenal mucosa in congenital lactose malabsorption. Acta Paediatr Scand 55:257–263 [DOI] [PubMed] [Google Scholar]
- Mantei N, Villa M, Enzler T, Wacker H, Boll W, James P, Hunziker W, Semenza G (1988) Complete primary structure of human and rabbit lactase-phlorizin hydrolase: implications for biosynthesis, membrane anchoring and evolution of the enzyme. Embo J 7:2705–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE (2005) Nonsense-mediated mRNA decay in mammals. J Cell Sci 118:1773–1776 10.1242/jcs.01701 [DOI] [PubMed] [Google Scholar]
- Naim HY (2001) Molecular and cellular aspects and regulation of intestinal lactase-phlorizin hydrolase. Histol Histopathol 16:553–561 [DOI] [PubMed] [Google Scholar]
- Naim HY, Jacob R, Naim H, Sambrook JF, Gething MJ (1994) The pro region of human intestinal lactase-phlorizin hydrolase. J Biol Chem 269:26933–26943 [PubMed] [Google Scholar]
- Norio R (2003a) Finnish disease heritage I: characteristics, causes, background. Hum Genet 112:441–456 [DOI] [PubMed] [Google Scholar]
- ——— (2003b) Finnish disease heritage II: population prehistory and genetic roots of Finns. Hum Genet 112:457–469 [DOI] [PubMed] [Google Scholar]
- Olds LC, Sibley E (2003) Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet 12:2333–2340 10.1093/hmg/ddg244 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T (1999) Molecular genetics of the Finnish disease heritage. Hum Mol Genet 8:1913–1923 10.1093/hmg/8.10.1913 [DOI] [PubMed] [Google Scholar]
- Poggi V, Sebastio G (1991) Molecular analysis of the lactase gene in the congenital lactase deficiency. Am J Hum Genet Suppl 49:105 [Google Scholar]
- Rasinperä H, Kuokkanen M, Kolho KL, Lindahl H, Enattah NS, Savilahti E, Orpana A, Järvelä I (2005) Transcriptional downregulation of the lactase (LCT) gene during childhood. Gut 54:1660–1661 10.1136/gut.2005.077404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasinperä H, Savilahti E, Enattah NS, Kuokkanen M, Tötterman N, Lindahl H, Järvelä I, Kolho KL (2004) A genetic test which can be used to diagnose adult-type hypolactasia in children. Gut 53:1571–1576 10.1136/gut.2004.040048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahi T (1994) Hypolactasia and lactase persistence: historical review and the terminology. Scand J Gastroenterol Suppl 202:1–6 [DOI] [PubMed] [Google Scholar]
- Savilahti E, Launiala K, Kuitunen P (1983) Congenital lactase deficiency: a clinical study on 16 patients. Arch Dis Child 58:246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RF, Wiese BA, Wojzynski MK, Davison DB, Worley KC (1996) BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res 6:454–462 [DOI] [PubMed] [Google Scholar]
- Troelsen JT, Olsen J, Moller J, Sjostrom H (2003) An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology 125:1686–1694 10.1053/j.gastro.2003.09.031 [DOI] [PubMed] [Google Scholar]
- Vandenplas S, Wiid I, Grobler-Rabie A, Brebner K, Ricketts M, Wallis G, Bester A, Boyd C, Mathew C (1984) Blot hybridisation analysis of genomic DNA. J Med Genet 21:164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G (1986) A new method for predicting signal sequence cleavage sites. Nucleic Acids Res 14:4683–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich M, Grünberg J, Hahn D, Jacob R, Radebach I, Naim HY, Sterchi EE (1996) Proteolytic processing of human lactase-phlorizin hydrolase is a two-step event: identification of the cleavage sites. Arch Biochem Biophys 336:27–34 10.1006/abbi.1996.0528 [DOI] [PubMed] [Google Scholar]
- Ylisaukko-oja T, Nieminen-von Wendt T, Kempas E, Sarenius S, Varilo T, von Wendt L, Peltonen L, Järvelä I (2004) Genome-wide scan for loci of Asperger syndrome. Mol Psychiatry 9:161–168 10.1038/sj.mp.4001385 [DOI] [PubMed] [Google Scholar]
- Zecca L, Mesonero JE, Stutz A, Poiree JC, Giudicelli J, Cursio R, Gloor SM, Semenza G (1998) Intestinal lactase-phlorizin hydrolase (LPH): the two catalytic sites; the role of the pancreas in pro-LPH maturation. FEBS Lett 435:225–228 10.1016/S0014-5793(98)01076-X [DOI] [PubMed] [Google Scholar]