Abstract
Based on molecular information theory, 10 T7-like promoter models were built for the T7 group of phages and used to scan their host genomes and closely related genomes. 38 genomes were scanned and 12 clusters of tandem promoters were identified in nine enteropathogens. Comparative analysis of these tandem promoter-bearing regions reveals that they are similar to each other, forming prophage-like islands of 4–13 kb. Each island appears to contain two or three tandem T7-like promoters within a stretch of 150–620 bases, but there are no corresponding RNA polymerase (RNAP) genes. The promoters would transcribe two to five putative phage-related proteins, but none of these resemble known phage structural proteins. An integrase belonging to the Int family of site-specific recombinases is encoded upstream of the tandem promoters. A direct repeat of 17–24 bases was found on the ends of all 12 islands. Comparative analysis of the islands shows that these islands appear to have recombined with each other. These results suggest that the islands could encode a group of satellite phages. Activation and function of the islands may depend on transcription by a T7-like RNAP after infection by a T7-like phage or foreign DNA that encodes a T7-like RNAP.
INTRODUCTION
The T7 group contains phages that have a strategy of infection similar to the prototype phage T7 (1,2). This strategy is largely determined by a T7-like transcription system, which consists of a phage-encoded RNA polymerase (RNAP) and a set of highly conserved T7-like promoters scattered across the phage genome (2). The phage RNAP is a single-chain protein, which specifically recognizes its cognate promoters and carries out all the steps of transcription by itself. Distant relatives of T7 RNAP are found in mitochondria and chloroplasts of many eukaryotes (3–5), however, these RNAPs recognize weakly conserved promoters and the promoter pattern is distinct from the T7-like promoters (6). Compared with the multi-subunit transcription systems of bacteria, archaea and eukarya, the T7-like transcription system is relatively simple and highly efficient, so it has been widely used in the laboratory. However, so far genetic systems with a T7-like RNAP and highly conserved cognate promoters have only been found in the T7 group of phages; no other form of utilization has been found yet.
Many phages have been shown to be involved in host-pathogen interactions by providing virulence or fitness factors to the pathogens (7). Because of the simplicity and high efficiency of the T7-like transcription system, it could also be caught and utilized by other organisms during evolution. However, it has been suggested that the highly efficient T7-like transcription systems cause the host cells to be inviable, probably by using up the ribonucleoside triphosphates (8–12). So it seems unlikely that a T7-like RNAP and a set of conserved promoters recognized by the RNAP can coexist in the cell under natural conditions. This was further indicated by the discovery of several T7-like prophages (13,14) in bacterial genomes (15–17). These prophages encode a T7-like RNAP, however, none of these have a set of conserved T7-like promoters. The promoter model for the prophage Ppu40 has only 19.1 bits, significantly lower than promoter models of the T7 group phages, which are about 33.4 ± 1.7 bits (2). This suggests that Ppu40 promoters have partially decayed since divergence, which could allow this prophage to coexist with its host.
Decayed promoters may provide a way for bacterial hosts to utilize the components of T7 group phages. Strong repressed expression of T7 RNAP could be another mechanism, though no cases have been observed yet. Another way could be to separate the T7-like promoters from their recognizers, thus making the transcription at the promoters be temporally and spatially controlled. However, so far no T7-like promoters have been reported in any genomes other than the T7 group of phages.
Molecular information theory provides us with a universal measurement of sequence conservation (in bits) at DNA-binding sites, and information theory-based DNA-binding site models can be used to sensitively find new binding sites. To explore the possible distribution of T7-like promoters in microbial genomes, we used T7-like promoter models that had been built in a previous study (2) to scan phage hosts and closely related genomes; 38 genomes were scanned using these models and more than 40 strong T7-like promoters were found. Within these, 12 clusters of tandem promoters were found to be located within a novel group of genomic islands in nine pathogenic enterobacteria.
With more and more genome sequences accumulating in the databank, both interspecies and intraspecies genome comparisons became practical and provided insights about bacterial genome evolution. Many genomic islands, such as pathogenic islands (PAIs) and prophages, have been identified by comparative genome analysis (14,18,19). Our detailed comparative analyses of the 12 tandem promoter bearing islands showed that these are similar to each other, share most features of a prophage and encode an integrase and several putative phage-related proteins. This group of islands may encode satellite phages. The helper phages could be T7-like or any genetic elements that encode a T7-like RNAP. The helper phages could not only trigger transcription of the islands, thereby activating them, but could also provide structural proteins for the satellite phages. To our knowledge, this is the first time that T7-like promoters and tandem promoter bearing islands have been found in bacterial genomes in the absence of a cognate RNAP. The identification of these islands strongly suggests a new form of utilization of T7-like transcription systems.
MATERIALS AND METHODS
Programs
Most programs used in this study are available at http://www.ccrnp.ncifcrf.gov/~toms/ (20). Two new features were developed for the lister program, which was used to generate sequence walkers http://www.ccrnp.ncifcrf.gov/~toms/paper/walker/ (21). A sequence walker (e.g. Fig. 2) shows the contribution that individual bases in a sequence give to the total information content of a binding site http://www.ccrnp.ncifcrf.gov/~toms/paper/ri/ (22). Letters that project upwards contribute positively, while those that point downwards decrease the total. The scale is in bits, as indicated by a light green bar that runs from −2 to +2 bits. A purple rectangle behind a letter indicates that the information contribution of that base is below −2 bits. A black rectangle indicates that the base is not observed in the original data set used to construct the model.
Figure 2.
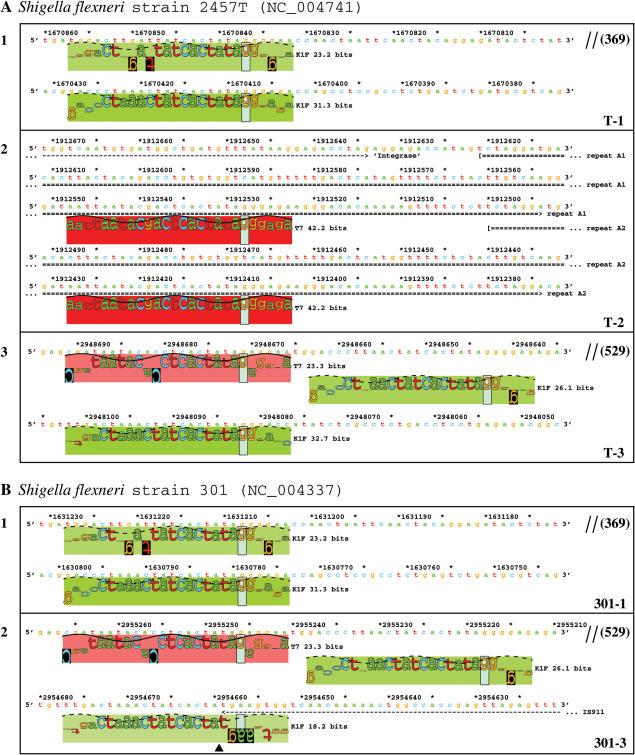
Tandem T7-like promoters in S.flexneri genomes. A total of 12 T7-like promoters that appear in tandem are identified in the genomes of S.flexneri strain 2457T (A) and strain 301 (B), and the sequence walkers (21) are shown. Different colored rectangles indicate different promoters (by hue) and their strengths (by saturation). Each cluster corresponds to one island and the island names are given in the lower right corner of each box. A double slash means that an intervening sequence is not shown; the size (in bp) is given in parenthesis. A black triangle in B-2 indicates the insertion point of IS911.
The first new feature is that colored rectangles, called ‘petals’ can be placed behind each sequence walker (see Fig. 2). The color is determined by hue, saturation and brightness with values ranging from 0 to 1. The hue of the rectangles corresponds to the kind of binding site and the saturation is determined from the strength of the site by dividing the individual information by the consensus information, which is the strongest individual information possible http://www.ccrnp.ncifcrf.gov/~toms/paper/ri/ (22). The petals are set to be fully bright. For example, a strong site would be red, a medium site would be pink and weak sites fade to white against the white background. We noticed that in many T7 promoters the sites were close to the consensus so letters such as t, which is displayed as red, would not be visible against a red background. The second feature is to allow letters of the walkers to be outlined so that they can be distinguished against the petals of strong sites.
Other programs used include Blast2 (23), BlastP (24), PSI-Blast (25–27), GAP (28), T_Coffee (29), GeneMark.hmm (30) and PROTDIST, NEIGHBOR, ProML and DRAWTREE (from the PHYLIP 3.65 package http://evolution.gs.washington.edu/phylip.html) (31,32).
Building promoter models and scanning genomes
In a previous study (2), nine T7-like promoter models were built for phages T7, φA1122, T3, φYeO3-12, gh-1, K11, SP6, K1-5 and VP4; a combined 76-site model was also built. In this study, a T7-like promoter model was built for the recently sequenced coliphage K1F (NC_007456) (Supplementary Figure S1) (33). The program scan was used for genome scanning, and genhis, genpic and xyplo were used to plot the individual information distribution from each scan http://www.ccrnp.ncifcrf.gov/~toms/paper/ri/ (22). The program lister was used to generate sequence walkers http://www.ccrnp.ncifcrf.gov/~toms/paper/walker/ (21) for the promoters found. Initially, each of the 10 individual promoter models was used to scan its host genome or some closely related genomes, and then we expanded our scanning to the available complete enterobacteria genome sequences in GenBank. The 76-site model was used to scan certain genome regions of interest.
Comparative analysis of the promoter-bearing regions
The promoter-containing regions and their encoding proteins were compared using the program Blast2 (23). The programs BlastP (24) and PSI-Blast (25–27) were used for similarity searches with the proteins encoded within the islands. Several online databases, including one about genomic islands (Islander) (34), and one about comparative genomics (coliBASE) (35), were also used.
Phylogenetic analysis
Phylogenetic analysis was performed for several sets of homologous proteins that are encoded by some or all of the islands. The protein sequences were aligned using the T_Coffee program (29). To reduce noise in the alignments (Supplementary Figures S2–S4), only conserved blocks that have no gaps and contain 10 or more positions were retained for further analysis (36,37). The program PROTDIST was used to calculate the distances among the aligned proteins and the resulting distance matrix was used to infer an unrooted tree using the neighbor-joining method (38) by the program NEIGHBOR. A bootstrap analysis of 1000 replicates was performed to test the statistical significance of the trees. Phylogenetic analysis was also carried out using the maximum likelihood method implemented in the program ProML (31,32).
RESULTS
Genome scanning with the T7-like promoter models
A total of 10 T7-like promoter models were built for T7 group phages [(2) and Supplementary Figure S1]. These models can be used to identify new T7-like promoters, and when they were scanned across the corresponding phage genomes, a significant gap between the promoters (>23 bits) and the background (<15 bits) was found in the individual information (Ri) distribution (2). When scanning bacterial genomes, the background will certainly be larger. For any given model and a given genome size of g bp, theoretically the background can be calculated as Rb ≤ log22g bits. So for a genome of 5 Mb, the background (Rb) should be lower than 23.3 bits. To test this, random sequences from 1 to 10 Mb were generated and scanned with the 10 models. As the sequence size increases from 1 to 10 Mb, the highest information obtained from these scans increases from 19 to 23 bits. Since all available bacterial genomes in GenBank are <10 Mb, this suggests that 23 bits can be used as a threshold for bacterial genome scanning with the 10 models. Considering that the lowest information for a T7-like promoter by its own model is 23 bits, 23 bits appears to be a natural cutoff, which suggests that the T7 group phages have maintained their promoters so well conserved that the T7-like RNAPs can distinguish their cognate promoters from any host genome background during infection.
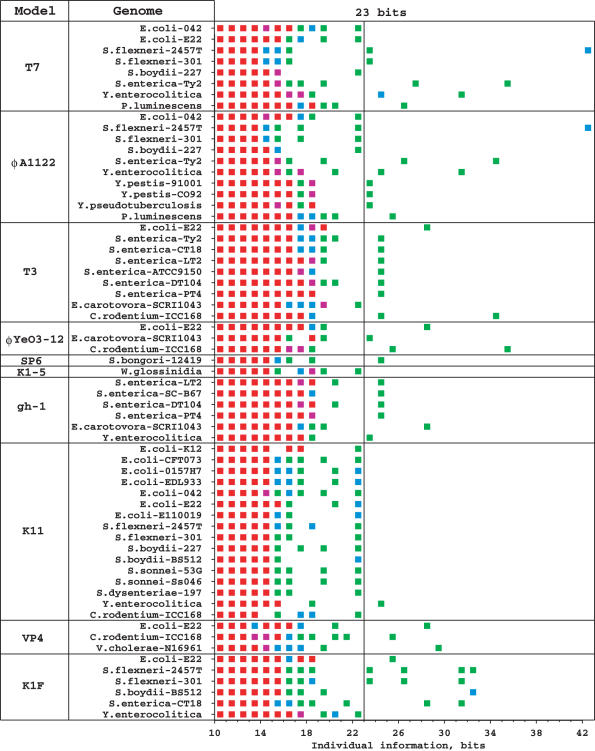
Initially, each of the 10 models was used to scan its own host genome or some closely related genomes, and then we expanded our scanning to all available enterobacterial genomes in GenBank. A total of 38 hosts and closely related genomes (Supplementary Table S1) were scanned, and 43 T7-like promoters above 23 bits were found (Fig. 1). Within these, 12 clusters of two or three tandem promoters were found in the genomes of Shigella flexneri strain 2457T (39), Shigella flexneri strain 301 (40), Salmonella enterica strain Ty2 (41), Salmonella enterica strain CT18 (42), Erwinia carotovora strain SCRI1043 (43), Yersinia enterocolitica strain 8081, Escherichia coli strain E22, Shigella boydii strain BS512 and Citrobacter rodentium strain ICC168. In each of these clusters, promoters have the same orientation and generally exist within a region of 150 to 620 bases. Sequence walkers http://www.ccrnp.ncifcrf.gov/~toms/paper/walker/ (21) for all tandem promoters are shown in Figures 2, 5 and 6.
Figure 1.
Bacterial genome scanning with the 10 T7-like promoter models. A total of 38 enterobacterial and closely related genomes (Supplementary Table S1) were scanned on both strands with each of the 10 models. The individual information distributions were plotted for each scan by using colored squares (Green, 1 site; blue, 2 sites; purple, 3 sites; red, 4 or more sites). Only genomes that have at least one site above 22 bits by any of the 10 models are listed. The cutoff of 23 bits is marked with a vertical line. In this figure 56 sites appear above 23 bits, however, 13 were picked up by more than one model, so actually there are 43 unique sites.
Figure 5.
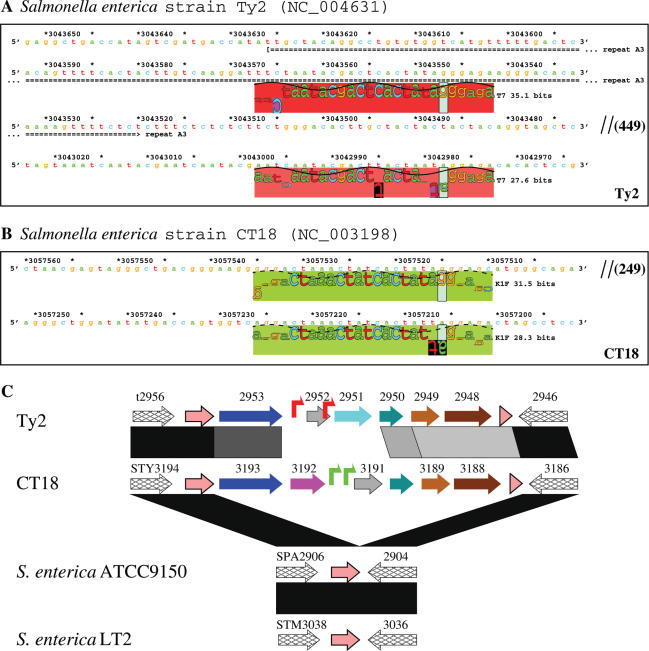
Tandem T7-like promoters in Salmonella genomes and genome organization of the islands Ty2 and CT18. (A) and (B) Sequence walkers (21) of tandem T7-like promoters in the islands Ty2 and CT18. The repeat A3 is a shortened copy of repeats A1 and A2 (Figure 2A-2). (C) Genome organization and comparison of the islands Ty2 and CT18. Symbol key is given in Figure 4.
Figure 6.
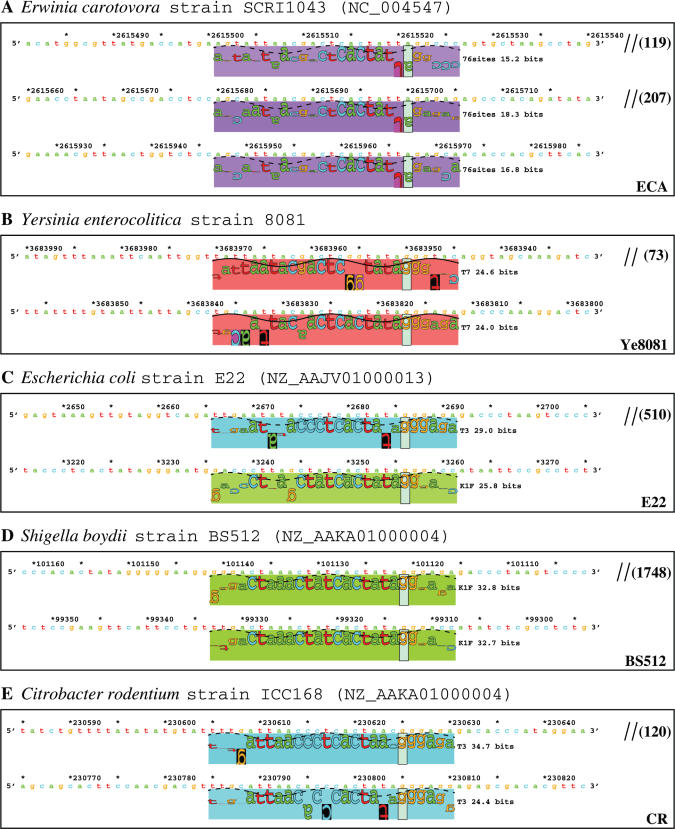
Tandem T7-like promoters in the genomes of E.carotovora (A), Y.enterocolitica (B), E.coli (C), S.boydii (D) and C.rodentium (E). Sequence walkers are shown for each cluster of tandem promoters. Different colored rectangles indicate different promoters (by hue) and their strengths (by saturation).
So far, more than 200 non-enterobacterial genomes have also been scanned with the 10 promoter models, and ∼80 T7-like promoters that appear alone were found (data not shown). In this study, we will only focus on the tandem promoters that were found in the nine pathogenic enterobacteria, the other promoters will not be discussed further.
T7 islands in enterobacterial genomes
The 12 tandem promoter bearing regions are similar to each other in genome organization and encoded proteins. All 12 regions were annotated in GenBank as containing a putative integrase gene, and the encoded proteins significantly match to several conserved integrase-related domains in the known integrases XerD, XerC and P4 integrase (see Supplementary Table S2), suggesting that the T7 island proteins belong to the Int family of site-specific recombinases (44).
These regions also encode several putative phage-related proteins. For eight regions, a tRNA-Gly gene is located upstream of the integrase gene. Since tRNA genes in bacterial genomes are frequently associated with foreign insertion islands (e.g. PAIs, prophages) (45–48), we believe that these promoter bearing regions may be foreign insertion islands. This was further supported by the identification of direct repeats at the ends of all 12 regions (Table 1). The eight islands that are adjacent to a tRNA-Gly gene share an almost identical direct repeat, which is actually the 3′ end of the tRNA gene. The direct repeats for the other four islands are different (Table 1). These islands were named by their respective strain name, and for convenience we call this group of islands T7 islands.
Table 1.
Features of the T7-like promoter containing islands
| Genome island | Genome (Accession no.) | Length (location) | Integration site | Direct repeata | Promoter |
|---|---|---|---|---|---|
| T-1 | S.flexneri 2457T (NC_004741) | 7762 (1674124-1666363) | ynfE | gtagttccagatg(g/a)act | K1F |
| T-2 | S.flexneri 2457T (NC_004741) | 3762 (1914374-239927)b | tRNA-Gly | tcgattcccgctgcccgctcca | T7 |
| T-3 | S.flexneri 2457T (NC_004741) | 6380 (2951093-2944714) | tRNA-Gly | tcccttcgcccgctcca | T7/K1F |
| 301-1 | S.flexneri 301 (NC_004337) | 7760 (1634495-1626736) | ynfE | gtagttccagatg(g/a)act | K1F |
| 301-3 | S.flexneri 301 (NC_004337) | 7636 (2957668-2950033) | tRNA-Gly | tcccttcgcccgctcca | T7/K1F |
| Ty2 | S.enterica Ty2 (NC_004631) | 5900 (3045460-3039561) | tRNA-Gly | tcgattcccttcgcccgctcca | T7 |
| CT18 | S.enterica CT18 (NC_003198) | 6307 (3059960-3053654) | tRNA-Gly | tcccttcgcccgctcca | K1F |
| ECA | E.carotovora SCRI1043 (NC_004547) | 6676 (2612648-2619323) | ECA2305/ispZ | gtaattttctgcaaattat | unknown |
| Ye8081 | Y.enterocolitica 8081 (Sanger)c | 3946 (3685769-3681824) | tRNA-Gly | ttcgattcccttcacccgctcca | T7 |
| E22 | E.coli E22 (NZ_AAJV01000013) | 13121 (159-13279) | GTPase | ctgacacaaggctgacacaaa | T3/K1F |
| BS512 | S.boydii BS512 (NZ_AAKA01000004) | 7893 (103574-95682) | tRNA-Gly | tcccttcgcccgctcca | K1F |
| CR | C.rodentium ICC168 (rod123h06.q1k) | 5371 (228046-233416)d | tRNA-Gly | gttcgattcccttcgcccgctcca | T3 |
aDirect repeats (DR) are located at the ends of each island. One kind of repeat is the 3′ end of the tRNA gene. For the most part the others do not resemble each other and are possibly duplications created during island integration. In this study the direct repeats were used to mark the endpoints of the corresponding islands.
bThis island was broken by insertion sequences (IS) and rearranged. To reconstruct this island, several pieces, 1 914 374 to 1 912 009, 1 910 756 to 1 910 232, and 240797 to 239927, were joined together, giving a length of 3762 bases. Some parts have been lost and cannot be found in the bacterial genome.
cThis sequence (with annotation) was obtained from the Sanger Institute (http://www.sanger.ac.uk/Projects/Microbes/); it has not been deposited in GenBank.
dThis is an unfinished genome obtained from the Sanger Institute. The island CR was found in one of the sequence contigs, so the coordinates given here are based on the contig rod123h06.q1k.
Each of the 12 T7 islands appears to contain two or three tandem T7-like promoters, which are located downstream of the integrase gene and would transcribe two to five putative phage-related genes. Using Blast for comparison, the integrases and five other sets of homologous proteins (for convenience we named these hypothetical proteins Hyp1 to Hyp5) encoded within these islands were found to be more conserved to each other than to other proteins in GenBank, so they can be used in conjunction with other features, such as the tandem T7-like promoters and the direct repeats, to identify new islands of this group. Though more than 100 T7-like promoters were found in both enterobacterial and non-enterobacterial genomes (data not shown), so far the tandem promoter-containing islands have been only found in certain strains of pathogenic enterobacteria, indicating that these islands might be involved in pathogenicity.
T7 islands in S.flexneri genomes
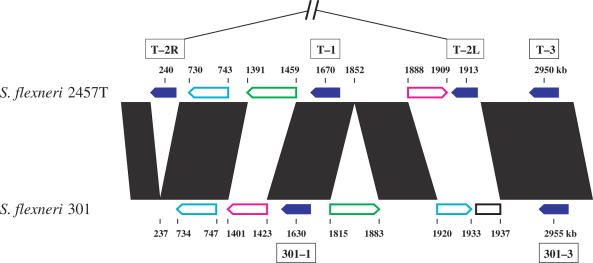
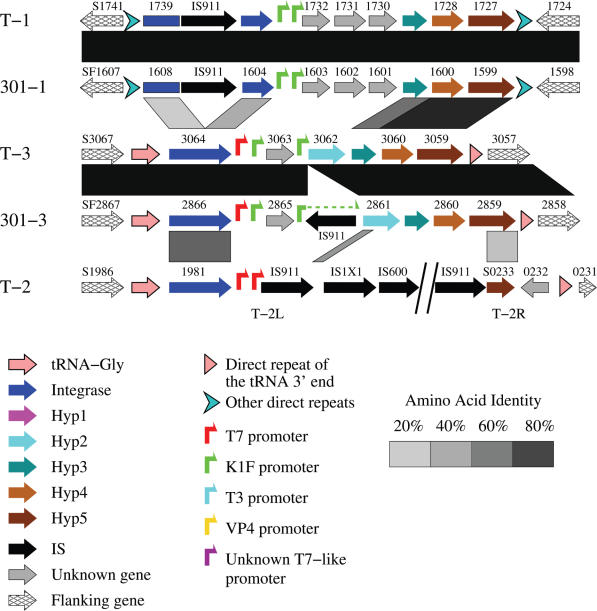
By scanning with the 10 T7-like promoter models, 12 T7-like promoters that appear in tandem were identified in the genomes of S.flexneri strain 2457T (39) and strain 301 (40) (Figure 2). These promoters appear in five clusters, with two or three in a cluster. Each cluster corresponds to one island, so a total of five tandem promoter containing islands were located in S.flexneri genomes, with three in strain 2457T (T-1, T-2 and T-3) and two in strain 301 (301-1 and 301-3) (Table 1). Chromosome locations for these islands are shown in Figure 3. Island T-1 corresponds to 301-1 and T-3 corresponds to 301-3. There is no equivalent to T-2 in strain 301.
Figure 3.
Chromosome locations of the five T7 islands in Shigella genomes. The five islands, found between genome positions 200 and 3000 kb, are represented by solid blue arrows. Notice that the island T-2 was broken into two pieces and rearranged, one piece (T-2L) is located at around 1913 kb, another piece (T-2R) is at around 240 kb. The open colored arrows (cyan, green and magenta) indicate several major rearrangements (larger than 10 kb) between the two Shigella chromosomes. The region in the genome of strain 301 indicated by an open black rectangle contains an IS sequence and prophage genes but no T7 island. Black parallelograms indicate highly similar regions of the two Shigella genomes. Locations of the islands and the rearrangements are given in kb, but the figure is not to scale.
In the genome of S.flexneri strain 2457T, two tandem K1F-like promoters, with a spacing of 430 bases, were located in island T-1, which covers a region from 1 674 124 to 1 666 363 (Figure 2A-1 and Table 1). The island T-2 contains two T7 promoters (Figure 2A-2 and Table 1); it will be discussed in detail below. Two sites of 26.1 and 32.7 bits were also picked up at 2 948 643 and 2 948 082 by the K1F promoter model (Figure 2A-3). These two sites are adjacent to a T7-like promoter of 23.3 bits, which was picked up at 2 948 671 by the T7 model. These three promoters are located within the island T-3, which covers a region from 2 951 093 to 2 944 714 (Table 1).
By comparing strains we determined that the island T-2 has been broken into at least two pieces and rearranged, one piece (T-2L) is located at ∼1913 kb and another piece (T-2R) is at ∼240 kb (Table 1, Figures 3 and 4). Two tandem T7 promoters, with a spacing of 120 bases, are located within T-2L at 1 912 408 and 1 912 528 (Figures 2A-2 and 4), respectively. These two promoters are identical to the strongest T7 promoter, φ10 (49), from position −21 to +6, relative to the transcription start. Each of them contains an individual information of 42.2 bits. Both of the major pieces of the island T-2 are associated with a copy of IS911, indicating that this insertion sequence might have been involved in the rearrangement.
Figure 4.
Genome organization and comparison of T7 islands in S.flexneri 2457T and S.flexneri 301. Note that one of the K1F-like promoters in 301-3 has been split by an insertion sequence, IS911. The integrase genes in the islands T-1 and 301-1 have also been split into N-terminal and C-terminal domains (see Supplementary Figure S2). Shaded parallelograms indicate regions of similarity (darker shades indicate higher levels of similarity). The first gene on the left is named by its GenBank locus_tag (e.g. S1741), while, to conserve space, the remaining genes are only numbered. The Hyp3 genes in the islands T-1, 301-1, T-3 and 301-3 were predicted in this study by using the ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and Blastx (against the Hyp3 protein of island BS512, see Figure 7). The symbol keys also apply to Figures 5C, 7, 8, 9 and Supplementary Figure S5.
The islands 301-1 and 301-3 are located in the genome of S.flexneri strain 301, and correspond to the islands T-1 and T-3 (Figures 3 and 4), respectively. 301-1 is almost identical to T-1, indicating that island integration happened before the differentiation of these two strains. 301-3 is also almost identical to T-3, except that a copy of IS911 is inserted within the island. The IS911 inserted into the third promoter within the island 301-3 at 2 954 659 (Figures 2B-2 and 4), resulting in a lower information (18.2 bits) for this split promoter. The insertion of IS911 may inactivate the third promoter and block the transcription of downstream genes. However, by precisely excising the IS911, the promoter could be recovered. The other four uninterrupted promoters in the islands 301-1 and 301-3 (Figure 2B-1 and B-2) are identical to the corresponding promoters in the islands T-1 and T-3 (Figure 2A-1 and A-3). Though these two islands are almost identical to their counterparts, no corresponding island for the two major parts of T-2 can be found in the genome of strain 301 (Figure 3), suggesting that the integration of T-2 happened after the differentiation of these two Shigella strains. Apparently several insertion events (IS911, IS1X1 and IS600, Figure 4) happened after the integration of the island T-2 into the 2457T genome.
Genome comparisons have shown that S.flexneri and E.coli are closely related and belong to the same species (39,40,50,51). By comparing the tandem promoter containing regions in 2457T and 301 with corresponding regions of E.coli K12 genome (52), the insertion islands in S.flexneri genomes were further confirmed. The islands T-1 and 301-1 are both inserted into ynfE within the ynfEFGHI operon (53). The islands T-3 and 301-3 are inserted into a tRNA-Gly gene at 2 997 kb of E.coli K12 (NC_000913).
T7 islands in Salmonella genomes
In the genome of S.enterica serovar Typhi strain Ty2 (41), two tandem T7-like promoters of 27.6 and 35.1 bits were found at 3 042 978 and 3 043 548, respectively (Figure 5A). These two promoters are located within the island Ty2, which covers a region from 3 045 460 to 3 039 561 (Table 1). For another strain (strain CT18) (42) of the same serovar, two K1F-like promoters of 28.3 and 31.5 bits were picked up at 3 057 206 and 3 057 515 (Figure 5B). These two promoters are located within the island CT18, which covers a region from 3 059 960 to 3 053 654 (Table 1).
The islands Ty2 and CT18 are located at similar regions of these two genomes. These two regions share a weak sequence similarity, while the respective flanking regions are almost identical, so these regions were noted by Deng et al. (41) as hypervariable regions. We compared these two regions in detail and found that they share a similar genome organization and encode several homologous proteins (Figure 5C), suggesting that both regions are foreign insertion islands of the same group.
To investigate the corresponding regions in several other available Salmonella genomic sequences, we compared these two islands and their flanking regions with the other genomes. The results clearly show that none of the other Salmonella genomes contain a similar island. By aligning the islands Ty2 and CT18 and their flanking regions with the corresponding genome regions of S.enterica serovar Typhimurium strain LT2 (54) and S.enterica serovar Paratyphi A strain ATCC9150 (55), we find that the islands Ty2 and CT18 are exact insertions at tRNA-Gly, which are absent in the genomes of S.enterica LT2 and S.enterica ATCC9150 (Figure 5C).
T7 islands in other genomes
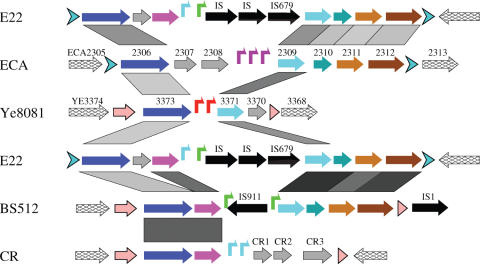
In the genome of an enterobacterial plant pathogen, E.carotovora strain SCRI1043 (43), we found a region (∼2618 kb) encoding several proteins that are highly similar to the proteins encoded in the island Ty2, suggesting that there exists a T7 island. However, none of the 10 T7-like promoter models found tandem sites in this region. Instead, only one site of 23.7 bits (by the YeO3-12 model) was found at 708 783, and another site of 28.3 bits (by the gh-1 model) was found at 1 606 534 (Figure 1). We then used the combined 76-site model to scan this region. Three sites of 15.2, 18.3 and 16.8 bits were found at 2 615 521, 2 615 700 and 2 615 967, respectively (Figure 6A). These three sites are weakly T7-like, but they are highly similar to each other. These sites could be a new class of T7-like promoters, which may be recognized by an unknown T7-like RNAP. We named this tandem promoter containing region as island ECA (Figure 7 and Table 1).
Figure 7.
Comparison of the islands ECA, Ye8081, E22, BS512 and CR. No sequence data are available for the upstream side of the island E22. Symbol key is given in Figure 4. The locus_tag names are given for the genes in the islands ECA and Ye8081, while because of space limitation, the names of the genes in the islands E22 and BS512 are not given in the figure. The gene names for the island E22 are (from left to right, excluding IS elements and tRNA genes): EcolE2_01002184, EcolE2_01002185, EcolE2_01002186, EcolE2_01002192, EcolE2_01002193, EcolE2_01002194, EcolE2_01002195 and EcolE2_01002197; the names for the island BS512 are: SboyB_01001168, SboyB_01001167, SboyB_01001166, SboyB_01001164, SboyB_01001163, SboyB_01001162 and SboyB_01001161. The island CR was found in an unfinished genome, C.rodentium strain ICC168 (Table 1 and Supplementary Table S1), and the genes were predicted in this study by using the programs GeneMark.hmm and ORF Finder. E22 is repeated for this comparison.
In the genome of Y.enterocolitica strain 8081, two tandem T7-like promoters of 24.6 and 24.0 bits are located at 3 683 950 and 3 683 817, respectively (Figure 6B). These two promoters are located within the island Ye8081, which covers a region from 3 685 769 to 3 681 824 (Table 1). Genome organization of the island is shown in Figure 7.
Similarity searches with the proteins encoded within the T7 islands indicate three more T7 islands in three unfinished genome sequences, E.coli strain E22, S.boydii strain BS512 and C.rodentium strain ICC168. Scanning these three genomes with the 10 T7-like promoter models revealed significant tandem T7-like promoters within corresponding regions (Figure 6C–E), thus confirming that these are T7 islands (Figure 7 and Table 1).
Comparative analysis of the T7 islands
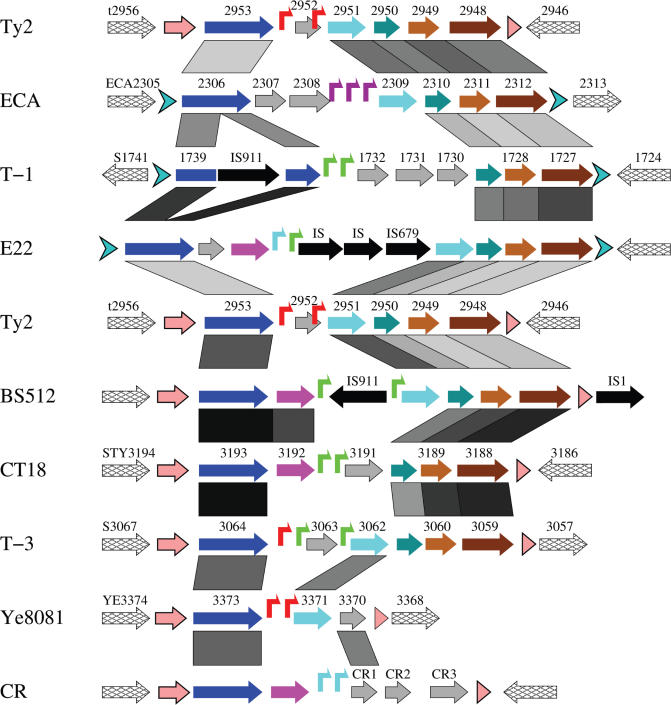
More extensive comparisons of the 12 T7 islands revealed that these islands are similar to each other in genomic organization and encoded proteins (Figures 4, 5C, 7 and 8), suggesting that they are prophage-like islands of the same group. Eight of the islands are located adjacent to a tRNA gene, and share an almost identical direct repeat (Table 1). These eight islands encode integrases that are highly similar to each other, while the other four islands encode relatively distantly related integrases (Figure 9A, marked in grey). This suggests that the direct repeats could be the target sites recognized by the corresponding integrases for integration or excision of the islands, and that these foreign genetic elements integrate into sites other than tRNA genes (47).
Figure 8.
Comparison of the T7 islands. Genome organization and comparison of nine of the 12 T7 islands. The islands 301-1 and 301-3 are not included because they are almost identical to T-1 and T-3, respectively (Figure 4). The broken island T-2 is also not included. Ty2 is repeated for this comparison. Symbol key is given in Figure 4.
Figure 9.
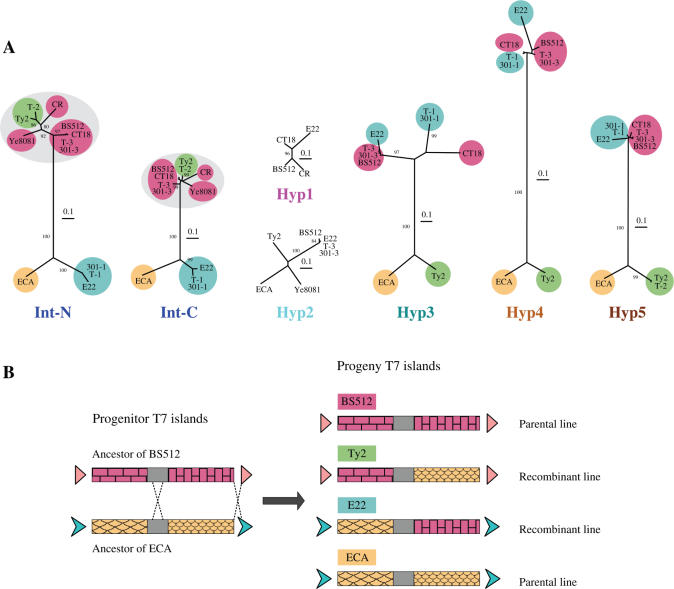
Phylogenetic analysis of six protein homologs shared by at least four of the 12 T7 islands. (A) Neighbor-joining trees of the six homologous island proteins are presented from left to right in the same order as the genetic map. The integrases were split into N-terminal and C-terminal domains (Int-N and Int-C), as indicated in Supplementary Figure S2, and the trees were generated for both domains. The eight islands that are adjacent to a tRNA gene encode closely related integrases (both Int-N and Int-C, circled in grey), while the other four integrases (ECA, 301-1, T-1 and E22) are distantly related. Reassortment along the genetic map of distances between sets of islands, shown by colored ellipses, indicates a major recombination between ancestors of ECA (orange, only on the bottom of the trees) and BS512 (red, only on the top) to give recombinants of E22 (blue, switches from bottom to top) and Ty2 (green, switches from top to bottom). All trees were normalized to the same scale, which represents a distance of 0.1 (in the Jones-Taylor-Thornton (JTT) amino acid replacement model). The bootstrap values given are percentages of 1000 replications, and only values greater than 80% are shown. (B) One possible double recombination between two distantly related T7 islands.
Using Blast2 (23) for protein comparisons, six homologous genes were found to be shared by four or more of the 12 islands, including an integrase and five hypothetical proteins (named Hyp1 to Hyp5). Similarity searches show that these proteins are, for the most part, more conserved to each other than to other proteins in the database (Supplementary Table S2). Searching the Conserved Domain Database (CDD) revealed that the integrases encoded in the 12 T7 islands contain the C-terminal catalytic domain of the Int family subgroup 2 (INT_SG2) (Supplementary Table S2) (56,57), so these integrases belong to INT_SG2. The Hyp2 proteins were identified as putative phage antirepressors, and a protein (ECA2308) in the island ECA was identified as a prophage antirepressor (Supplementary Table S2), suggesting that these T7 islands could encode satellite phages (58–61).
For each kind of protein we derived a phylogenetic tree based on sequence alignment, using the neighbor-joining method (Figure 9A) (38). The integrases are more conserved in their C-terminal domain than the N-terminal domain (57), so the alignment of the integrases was split into two parts (Supplementary Figure S2), and the trees were generated for both domains (Int-N and Int-C) (Figure 9A). All trees were normalized to the same scale and compared. The results (Figure 9A) show that the trees for Int-N and Int-C are highly similar to each other, except that the Int-N domains are more diversified than the Int-C domains. The trees for Hyp3, Hyp4 and Hyp5 are similar to each other, so these three genes appear to constitute a conserved block in each of the islands. Both the Int proteins and the Hyp3 to Hyp5 proteins fall into two distantly related clusters, while the Hyp1 and Hyp2 proteins all appear to be closely related, suggesting that these two genes may have different origins from the others. Compared with the trees of Int proteins, two branches, (T-2, Ty2) and (T-1, 301-1 and E22), are switched in the trees of Hyp3, Hyp4 and Hyp5, strongly suggesting that at least one recombination happened between ancestors of the two distantly related clusters, and that the recombination site was between the Int and Hyp3 genes. Phylogenetic analysis was also performed by using the maximum likelihood method, and the results are highly similar to Figure 9A (data not shown).
We do not know which islands represent parental or recombinant lines so, for example, suppose that islands ECA and BS512 were originally parental lines, and consider E22 and Ty2 as the recombinant lines. In Figure 9A, BS512 (circled in red) is always on the top halves of the trees, while ECA (orange) is always on the bottom; E22 (blue) is on the bottom for Int, but on the top for Hyp3 to Hyp5. In contrast, Ty2 (green) is on the top for Int, but on the bottom for Hyp3 to Hyp5. So in Figure 8 (middle), the left half of the comparison between Ty2 and BS512 shows a strong resemblance (∼75%) in the Int and Hyp2 proteins but low resemblance (21 to 43%) for Hyp3 to Hyp5. However, if we compare Ty2 with ECA, as shown in Figure 8 (top), the Int proteins have low resemblance (27%), while the Hyp3 to Hyp5 proteins have high resemblance (58 to 69%). Therefore, Ty2 appears to have originated from the left half of an ancestral BS512, combined with the right half of an ancestral ECA, as shown in Figure 9B.
Remarkably, a reciprocal recombinant exists in our sample of T7 islands. These are T-1 and E22. As can be seen in the middle of Figure 8, T-1 resembles E22 closely (59 to 90%). However, only the upstream half of E22 resembles ECA (Figure 7 top, 50 to 52% upstream versus 26 to 33% downstream), while the downstream half resembles BS512 (Figure 7 middle, 30% upstream versus 66 to 87% downstream). Therefore, E22 appears to have originated from the left half of an ancestral ECA, combined with the right half of an ancestral BS512, as shown in Figure 9B. However, E22 is not necessarily the exact reciprocal recombinant of Ty2, as they may have been generated from independent crossover events. More data will be required to map the recombination points.
It is clear from Figure 9 that a recombination has occurred at a site between Int and Hyp3. However, because the direct repeats are preserved on the ends of all T7 islands, there must also have been a second recombination between Hyp5 and the direct repeat at the 3′ end (Figure 9B). The two recombinations appear to have caused an exchange of the Hyp3 to Hyp5 genes between two distantly related lines of T7 islands. It is important to note that the islands that we have labeled parental and recombinant may actually be reversed; we do not know which are the parental lines, only that a double recombination occurred.
DISCUSSION
Genome scanning and identification of the T7 islands
Nine T7-like promoter models were built in a previous study (2), and a K1F promoter model was built in this study (Supplementary Figure S1). These 10 models were used to scan 38 enterobacteria and closely related genomes. More than 40 T7-like promoters were found above 23 bits, and within these, 12 clusters of tandem promoters were located in a novel group of genomic islands within nine enterobacterial pathogens.
All T7-like promoters in the 10 models are higher than 23 bits (by their own model) (2), and the highest information site obtained from random sequence (up to 10 Mb) scanning is 23 bits, suggesting that the phage RNAPs completely distinguish their promoters from the host genome background. Therefore 23 bits is a natural threshold for bacterial genome scanning with the 10 T7-like promoter models. This demonstrates that the promoters found in this study are significant.
All of the T7 islands encode an integrase, have several homologous phage-related proteins, contain a direct repeat at both ends and bear two or three tandem T7-like promoters (Figure 8). Furthermore, eight of the 12 islands are inserted into the TψC loop of a tRNA-Gly gene (34,47), and the corresponding integrase gene is oriented in the same direction as the tRNA gene, suggesting that the T7 islands are similar to P4-like prophages, which also integrate into the TψC loop at the 3′ end of tRNA genes (47,48). Although there are closer matches, the T7 island integrases resemble the P4-related integrases (E-value = 2 × 10−10, Supplementary Table S2). These features suggest that the T7 islands are prophage-like.
Distribution of the T7 islands
So far, the T7 islands have only been found in certain pathogenic strains. Only one of the seven E.coli genomes analyzed in this study (Supplementary Table S1) contains a T7 island (E22). Although an integrase (ZP_00718924) from an unfinished genome of E.coli strain E110019 was found to be highly similar (>70% identity) to the integrases of several T7 islands (Supplementary Table S2), no homologs of other island proteins were found in this genome. Furthermore, scanning this unfinished genome with the 10 T7-like promoter models found no significant sites (Figure 1). The integrase gene is located downstream of a tRNA-Gly gene at the 5′ end of one sequence contig (NZ_AAJW01000031) of E.coli strain E110019, and it points to the 5′ direction, so there may be other parts of a T7 island in that direction, in the unsequenced region.
For the seven Shigella genomes (Supplementary Table S1), two strains of S.flexneri serotype 2a were found to contain the five T7 islands shown in Figures 2–4. Since no genome sequences are available for other serotypes of S.flexneri, it is not known whether the other serotypes contain T7 islands. Another Shigella genome, S.boydii serotype 18 strain BS512, was found to contain a T7 island (BS512) while a closely related strain, S.boydii serotype 4 strain 227, and all other Shigella genomes analyzed in this study do not have a T7 island.
The islands Ty2 and CT18 are located in the genomes of S.enterica strain Ty2 and strain CT18, respectively. These two strains belong to the serovar Typhi of S.enterica, which has been shown to contain human specific pathogens (62). The other six Salmonella genomes (Supplementary Table S2) analyzed in this study do not contain a T7 island. Several more unpublished Salmonella genomes were searched with the proteins encoded in T7 islands, but no significant matches were found (data not shown). These results suggest that the T7 islands might be specific to the serovar Typhi of S.enterica.
The T7 islands appear to be specific to certain bacterial strains within certain species, however, so far the strain preference of the T7 islands remains largely unclear. To address this will require investigating many more bacterial strains.
As we were finishing this paper, we detected a putative T7 island (O395) in one piece (NZ_AAKG01000002) of an unfinished genome, Vibrio cholerae strain O395 (NZ_AAKG00000000). This island encodes an integrase and homologs of Hyp4 and Hyp5 (Supplementary Table S2).It also contains four VP4 promoters, with one upstream and three downstream of the integrase gene (Supplementary Figure S5). This island is adjacent to a super-integron (63,64). There is also a distant fragment containing a Hyp2 homologue. By comparing O395 with V.cholerae strain N16961 (65) one more putative T7 island (N16961) was found (Supplementary Figure S5B). However, both islands do not have direct repeats on their ends, and the island N16961 contains only one promoter, so these two islands are putative T7 islands. There might be a group of T7 islands in Vibrio genomes that are distantly related to the 12 T7 islands found in enterobacteria.
Function of the T7 islands
As discussed above, the T7 islands are similar to prophages in their genomic organization and encoded proteins. However, none of the islands encode structural proteins that resemble phage capsids, which implies that these prophage-like islands may be satellite phages. The islands do not carry any RNAP genes, so the presence of tandem T7-like promoters in each of the 12 islands strongly suggests that the corresponding helper phages may be T7-like phages, or some other unknown genetic elements that encode a T7-like RNAP, and that the function and activation of the islands may depend on the helper phages. A practical consideration is that introducing T7 RNAP-based expression constructs into a bacterial strain may have serious unanticipated consequences.
All T7 islands were found in pathogenic strains, indicating that these islands could be involved in pathogenicity. Of the nine pathogens that bear a T7 island, one is a plant pathogen, one is a mouse pathogen, and all others are human pathogens. Because the databases contain many pathogenic strains, these observations may reflect a bias in the databases rather than a correlation with pathogenicity.
The identification of this group of prophage-like islands raises many interesting questions: are these islands inducible and if so would infection by a T7-like phage induce them? What happens after induction of the islands? One interesting possibility is that these islands contain suicide genes. If a colony of pathogenic bacteria is infected with T7 or some foreign DNA which encodes a T7-like RNA polymerase, this region would be transcribed just after infection, killing the cells and thereby protecting the remainder of the colony. Alternatively, or perhaps in addition, the island may be mobilized and packaged as a satellite phage. Further investigation of the T7 islands may shed new light on phage-pathogen-host interactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
We thank Danielle Needle and Ilya Lyakhov for useful discussions, and anonymous reviewers for useful suggestions. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Funding to pay the Open Access publication charges for this article was provided by NIH/NCI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hausmann R. The T7 Group. In: Calendar R., editor. The Bacteriophages. Vol. 1. NY: Plenum Press; 1988. pp. 259–289. [Google Scholar]
- 2.Chen Z., Schneider T.D. Information theory based T7-like promoter models: classification of bacteriophages and differential evolution of promoters and their polymerases. Nucleic Acids Res. 2005;33:6172–6187. doi: 10.1093/nar/gki915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cermakian N., Ikeda T.M., Cedergren R., Gray M.W. Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res. 1996;24:648–654. doi: 10.1093/nar/24.4.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cermakian N., Ikeda T.M., Miramontes P., Lang B.F., Gray M.W., Cedergren R. On the evolution of the single-subunit RNA polymerases. J. Mol. Evol. 1997;45:671–681. doi: 10.1007/pl00006271. [DOI] [PubMed] [Google Scholar]
- 5.Rousvoal S., Oudot M., Fontaine J., Kloareg B., Goer S.L. Witnessing the evolution of transcription in mitochondria: the mitochondrial genome of the primitive brown alga Pylaiella littoralis (L.) Kjellm. Encodes a T7-like RNA polymerase. J. Mol. Biol. 1998;277:1047–1057. doi: 10.1006/jmbi.1998.1679. [DOI] [PubMed] [Google Scholar]
- 6.Kuhn K., Weihe A., Borner T. Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 2005;33:337–346. doi: 10.1093/nar/gki179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brussow H., Canchaya C., Hardt W.D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McAllister W.T., Morris C., Rosenberg A.H., Studier F.W. Utilization of bacteriophage T7 late promoters in recombinant plasmids during infection. J. Mol. Biol. 1981;153:527–544. doi: 10.1016/0022-2836(81)90406-x. [DOI] [PubMed] [Google Scholar]
- 9.Tabor S., Richardson C.C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl Acad. Sci. USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Studier F.W., Moffatt B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 11.Schneider T.D., Stormo G.D. Excess information at bacteriophage T7 genomic promoters detected by a random cloning technique. Nucleic Acids Res. 1989;17:659–674. doi: 10.1093/nar/17.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon Y.S., Kim J., Kang C. Viability of E.coli cells containing phage RNA polymerase and promoter: interference of plasmid replication by transcription. Genet. Anal. 1998;14:133–139. doi: 10.1016/s1050-3862(98)00007-2. [DOI] [PubMed] [Google Scholar]
- 13.Hendrix R.W. Bacteriophage genomics. Curr. Opin. Microbiol. 2003;6:506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 15.Nelson K.E., Weinel C., Paulsen I.T., Dodson R.J., Hilbert H., Martins dos Santos V.A., Fouts D.E., Gill S.R., Pop M., Holmes M., et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 16.Wood D.W., Setubal J.C., Kaul R., Monks D.E., Kitajima J.P., Okura V.K., Zhou Y., Chen L., Wood G.E., Almeida N.F., Jr, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 17.da Silva A.C., Ferro J.A., Reinach F.C., Farah C.S., Furlan L.R., Quaggio R.B., Monteiro-Vitorello C.B., Van Sluys M.A., Almeida N.F., Alves L.M., et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- 18.Canchaya C., Proux C., Fournous G., Bruttin A., Brussow H. Prophage genomics. Microbiol. Mol. Biol. Rev. 2003;67:238–276. doi: 10.1128/MMBR.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt H., Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 2004;17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider T.D. Reading of DNA sequence logos: prediction of major groove binding by information theory. Meth. Enzym. 1996;274:445–455. doi: 10.1016/s0076-6879(96)74036-3. [DOI] [PubMed] [Google Scholar]
- 21.Schneider T.D. Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences [Erratum (1998) Nucleic Acids Res., 26, 1135.] Nucleic Acids Res. 1997;25:4408–4415. doi: 10.1093/nar/25.21.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider T.D. Information content of individual genetic sequences. J. Theor. Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. [DOI] [PubMed] [Google Scholar]
- 23.Tatusova T.A., Madden T.L. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 24.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S.F., Koonin E.V. Iterated profile searches with PSI-BLAST – a tool for discovery in protein databases. Trends. Biochem. Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 27.Schaffer A.A., Aravind L., Madden T.L., Shavirin S., Spouge J.L., Wolf Y.I., Koonin E.V., Altschul S.F. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Womble D.D. GCG: The Wisconsin Package of sequence analysis programs. Methods. Mol. Biol. 2000;132:3–22. doi: 10.1385/1-59259-192-2:3. [DOI] [PubMed] [Google Scholar]
- 29.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 30.Lukashin A.V., Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felsenstein J. PHYLIP – Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 32.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 33.Scholl D., Merril C. The genome of bacteriophage K1F, a T7-like phage that has acquired the ability to replicate on K1 strains of Escherichia coli. J. Bacteriol. 2005;187:8499–8503. doi: 10.1128/JB.187.24.8499-8503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantri Y., Williams K.P. Islander: a database of integrative islands in prokaryotic genomes, the associated integrases and their DNA site specificities. Nucleic Acids Res. 2004;32:D55–D58. doi: 10.1093/nar/gkh059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhuri R.R., Khan A.M., Pallen M.J. coliBASE: an online database for Escherichia coli, Shigella and Salmonella comparative genomics. Nucleic Acids Res. 2004;32:D296–D299. doi: 10.1093/nar/gkh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf Y.I., Rogozin I.B., Koonin E.V. Coelomata and not Ecdysozoa: evidence from genome-wide phylogenetic analysis. Genome Res. 2004;14:29–36. doi: 10.1101/gr.1347404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novichkov P.S., Omelchenko M.V., Gelfand M.S., Mironov A.A., Wolf Y.I., Koonin E.V. Genome-wide molecular clock and horizontal gene transfer in bacterial evolution. J. Bacteriol. 2004;186:6575–6585. doi: 10.1128/JB.186.19.6575-6585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Wei J., Goldberg M.B., Burland V., Venkatesan M.M., Deng W., Fournier G., Mayhew G.F., Plunkett G., III, Rose D.J., Darling A., et al. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 2003;71:2775–2786. doi: 10.1128/IAI.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Q., Yuan Z., Xu J., Wang Y., Shen Y., Lu W., Wang J., Liu H., Yang J., Yang F., et al. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 2002;30:4432–4441. doi: 10.1093/nar/gkf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng W., Liou S.R., Plunkett G., IIIrd, Mayhew G.F., Rose D.J., Burland V., Kodoyianni V., Schwartz D.C., Blattner F.R. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parkhill J., Wren B.W., Thomson N.R., Titball R.W., Holden M.T., Prentice M.B., Sebaihia M., James K.D., Churcher C., Mungall K.L., et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 43.Bell K.S., Sebaihia M., Pritchard L., Holden M.T., Hyman L.J., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., et al. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl Acad. Sci. USA. 2004;101:11105–11110. doi: 10.1073/pnas.0402424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nunes-Duby S.E., Kwon H.J., Tirumalai R.S., Ellenberger T., Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiter W.D., Palm P., Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989;17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hacker J., Kaper J.B. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 47.Williams K.P. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 2002;30:866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell A. Prophage insertion sites. Res. Microbiol. 2003;154:277–282. doi: 10.1016/S0923-2508(03)00071-8. [DOI] [PubMed] [Google Scholar]
- 49.Ikeda R.A. The efficiency of promoter clearance distinguishes T7 class II and class III promoters. J. Biol. Chem. 1992;267:11322–11328. [PubMed] [Google Scholar]
- 50.Pupo G.M., Lan R., Reeves P.R. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl Acad. Sci. USA. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang F., Yang J., Zhang X., Chen L., Jiang Y., Yan Y., Tang X., Wang J., Xiong Z., Dong J., et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blattner F.R., Plunkett G., III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 53.Lubitz S.P., Weiner J.H. The Escherichia coli ynfEFGHI operon encodes polypeptides which are paralogues of dimethyl sulfoxide reductase (DmsABC) Arch. Biochem. Biophys. 2003;418:205–216. doi: 10.1016/j.abb.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 54.McClelland M., Sanderson K.E., Spieth J., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Hou S., et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 55.McClelland M., Sanderson K.E., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Harkins C.R., et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nature Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- 56.Marchler-Bauer A., Bryant S.H. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marchler-Bauer A., Anderson J.B., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu T., Renberg S.K., Haggard-Ljungquist E. The E protein of satellite phage P4 acts as an anti-repressor by binding to the C protein of helper phage P2. Mol. Microbiol. 1998;30:1041–1050. doi: 10.1046/j.1365-2958.1998.01132.x. [DOI] [PubMed] [Google Scholar]
- 59.Eriksson S.K., Liu T., Haggard-Ljungquist E. Interacting interfaces of the P4 antirepressor E and the P2 immunity repressor C. Mol. Microbiol. 2000;36:1148–1155. doi: 10.1046/j.1365-2958.2000.01937.x. [DOI] [PubMed] [Google Scholar]
- 60.Davis B.M., Kimsey H.H., Kane A.V., Waldor M.K. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J. 2002;21:4240–4249. doi: 10.1093/emboj/cdf427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davis B.M., Waldor M.K. Filamentous phages linked to virulence of Vibrio cholerae. Curr. Opin. Microbiol. 2003;6:35–42. doi: 10.1016/s1369-5274(02)00005-x. [DOI] [PubMed] [Google Scholar]
- 62.Pascopella L., Raupach B., Ghori N., Monack D., Falkow S., Small P.L. Host restriction phenotypes of Salmonella typhi and Salmonella gallinarum. Infect. Immun. 1995;63:4329–4335. doi: 10.1128/iai.63.11.4329-4335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rowe-Magnus D.A., Guerout A.M., Mazel D. Super-integrons. Res. Microbiol. 1999;150:641–651. doi: 10.1016/s0923-2508(99)00127-8. [DOI] [PubMed] [Google Scholar]
- 64.Clark C.A., Purins L., Kaewrakon P., Focareta T., Manning P.A. The Vibrio cholerae O1 chromosomal integron. Microbiology. 2000;146:2605–2612. doi: 10.1099/00221287-146-10-2605. [DOI] [PubMed] [Google Scholar]
- 65.Heidelberg J.F., Eisen J.A., Nelson W.C., Clayton R.A., Gwinn M.L., Dodson R.J., Haft D.H., Hickey E.K., Peterson J.D., Umayam L., et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.