Abstract
Gallbladder disease (GBD) is one of the major digestive diseases. Its risk factors include age, sex, obesity, type 2 diabetes, and metabolic syndrome (MS). The prevalence of GBD is high in minority populations, such as Native and Mexican Americans. Ethnic differences, familial aggregation of GBD, and the identification of susceptibility loci for gallstone disease by use of animal models suggest genetic influences on GBD. However, the major susceptibility loci for GBD in human populations have not been identified. Using ultrasound-based information on GBD occurrence and a 10-cM gene map, we performed multipoint variance-components analysis to localize susceptibility loci for GBD. Phenotypic and genotypic data from 715 individuals in 39 low-income Mexican American families participating in the San Antonio Family Diabetes/Gallbladder Study were used. Two GBD phenotypes were defined for the analyses: (1) clinical or symptomatic GBD, the cases of cholecystectomies due to stones confirmed by ultrasound, and (2) total GBD, the clinical GBD cases plus the stone carriers newly diagnosed by ultrasound. With use of the National Cholesterol Education Program/Adult Treatment Panel III criteria, five MS risk factors were defined: increased waist circumference, hypertriglyceredemia, low high-density lipoprotein cholesterol, hypertension, and high fasting glucose. The MS risk-factor score (range 0–5) for a given individual was used as a single, composite covariate in the genetic analyses. After accounting for the effects of age, sex, and MS risk-factor score, we found stronger linkage signals for the symptomatic GBD phenotype. The highest LOD scores (3.7 and 3.5) occurred on chromosome 1p between markers D1S1597 and D1S407 (1p36.21) and near marker D1S255 (1p34.3), respectively. Other genetic locations (chromosomes 2p, 3q, 4p, 8p, 9p, 10p, and 16q) across the genome exhibited some evidence of linkage (LOD ⩾1.2) to symptomatic GBD. Some of these chromosomal regions corresponded with the genetic locations of Lith loci, which influence gallstone formation in mouse models. In conclusion, we found significant evidence of major genetic determinants of symptomatic GBD on chromosome 1p in Mexican Americans.
Gallbladder disease (GBD) is a common, economically burdensome digestive disease in the United States (Sandler et al. 2002). An estimated 20 million Americans are affected with GBD, and >700,000 cholecystectomies are performed every year (Hall and Lawrence 1998; Everhart et al. 1999; Lawrence and Hall 1999; Diehl 2000). GBD prevalence is high in some minority populations in the United States, including Native and Mexican Americans (Weiss et al. 1984a; Diehl and Stern 1989; Everhart et al. 2002; Méndez-Sánchez et al. 2004). Gallstones composed of cholesterol (cholelithiasis) are the common manifestations of GBD in Western countries, including the United States (Diehl et al. 1994; Nakeeb et al. 2002; Paigen and Carey 2002). Most people with gallstones, however, remain asymptomatic, or silent, through their lifetimes; only ∼10%–50% of individuals eventually develop symptoms (Paigen and Carey 2002).
The significant risk factors associated with GBD are age, female sex, obesity (especially central obesity), lipids, diet, parity, type 2 diabetes (T2DM), medications, and Mexican American ethnicity (Diehl 1991; Hanis et al. 1993; Misciagna et al. 1996; Everhart et al. 1999; Duggirala et al. 1999b; Paigen and Carey 2002; Lee 2004; Méndez-Sánchez et al. 2004). Rapid weight loss, smoking, and sedentary lifestyle were also identified as risk factors for GBD (Everhart 1993; Sahi et al. 1998; Leitzmann et al. 1999). The association between diabetes and GBD is controversial; some have suggested that hyperinsulinemia rather than diabetes may play a major role in the etiology of GBD (Haffner et al. 1993; Everhart 1995; Diehl 2000; Ruhl and Everhart 2000). Also, it has been shown that duration of diabetes and blood sugar control are associated with impaired gallbladder function (Haffner et al. 1993; Yang et al. 2002). There is increasing evidence that GBD is strongly related to metabolic syndrome (MS) and/or its major components, such as hyperinsulinemia, dyslipidemia, and abdominal adiposity (Boland et al. 2002; Grundy 2004; Tsai et al. 2004). The contribution of bacteria to the occurrence of gallstones has become an interesting area of research as well (Swidsinski and Lee 2001; Silva et al. 2003; Maurer et al. 2005).
The pathogenesis of cholesterol gallstones is unclear. Factors such as hypersecretion of hepatic cholesterol, supersaturation of bile with cholesterol, cholesterol crystal nucleation time, and hypomotility of the gallbladder appear to influence the formation of gallstones (Pomeranz and Shaffer 1985; Carey 1993; Portincasa et al. 1995; Méndez-Sánchez et al. 1996; Paigen and Carey 2002; Portincasa et al. 2003). Thus, the pathobiological mechanisms that underlie the phenotypic expression of GBD appear to be rather complex, and one or more defects could occur in genes that play critical roles in the diverse pathways leading to cholesterol gallstone formation.
It is generally thought that GBD is a complex, multifactorial disease influenced by genetic and environmental factors and their interactions. The available information based on family data, albeit limited, suggests that genetic factors play a key role in the development of GBD (Kesaniemi et al 1989; Sarin et al. 1995; Duggirala et al. 1999b; Nakeeb et al. 2002; Kosters et al. 2003). Using data from a large Swedish study of 43,141 twin pairs, Katsika et al. (2005) determined that genetic influences are major contributors to the variation in symptomatic gallstone disease. According to this study, genetic factors accounted for 25%, shared environmental factors for 13%, and unique environmental factors for 62% of the phenotypic variance among twins. In addition, varying prevalence on the basis of ethnicity has been considered to be indirect evidence of the genetic determination of GBD (Weiss et al. 1984a; Diehl and Stern 1989; Everhart et al. 2002; Paigen and Carey 2002; Méndez-Sánchez et al. 2004). In fact, Weiss et al. (1984b) proposed that there might be a genetic susceptibility association among complex diseases such as GBD, diabetes, and obesity, which cluster to form a “New World Syndrome” in populations with Native American ancestry. Aside from these observations in human populations, several mouse models identified various Lith (i.e., lithogenic) loci influencing gallstone formation (Khanuja et al. 1995; Paigen et al. 2000; Lammert et al. 2001; Hillebrandt et al. 2003; Kosters et al. 2003; Lyons et al. 2003, 2005).
Major susceptibility loci for GBD in human populations have not yet been identified. Therefore, we conducted a genetic epidemiologic investigation of GBD, using data from complex Mexican American families, as part of the San Antonio Family Diabetes/Gallbladder Study (SAFDGS). Using a 10-cM map and ultrasound-based information on GBD occurrence, we employed a variance-components linkage technique, using a liability model to map susceptibility genes for GBD in the Mexican American population.
Subjects and Methods
SAFDGS
Demographic and other phenotypic information was collected from 741 individuals drawn from 39 large Mexican American families that were enrolled in the San Antonio Family Gallbladder Study (SAFGS), a follow-up and extension of the San Antonio Family Diabetes Study (SAFDS). These studies are collectively referred to as the SAFDGS. The recruitment for the SAFGS was conducted between 1998 and 2001. Of these 741 individuals, 476 had been examined previously at baseline and/or follow-up in the SAFDS and were members of the 31 original SAFDS families (Duggirala et al. 1999a, 2001). An additional 265 individuals were recruited into the SAFGS; of these, 152 participants were newly recruited members of the original 31 SAFDS families, and 113 were members of 8 newly recruited SAFGS families. Recruitment of the new SAFGS families followed the same guidelines as were used originally in the SAFDS recruitment (Duggirala et al. 1999a). Probands were recruited from a random sample of low-income Mexican American individuals who had been identified in the earlier San Antonio Heart Study as having T2DM. All of the probands’ first-, second-, and third-degree relatives aged ⩾18 years were invited to participate in the study. Of the total 646 individuals who had taken part in the earlier SAFDS examinations, 54 died before SAFGS recruitment began. Of the 592 SAFDS survivors, 476 (∼80%) individuals participated in the present project. The Institutional Review Board of the University of Texas Health Science Center at San Antonio approved all procedures, and all subjects gave written informed consent.
Phenotype Data
For each individual, a detailed medical history of previous gallbladder problems, including cholecystectomy, was obtained. Ultrasound examinations were conducted at the Frederic C. Bartter General Clinical Research Center (GCRC), South Texas Veterans Healthcare System, Audie L. Murphy Division, in San Antonio. Ultrasound is widely regarded as the test of choice for screening for gallstones because of its high sensitivity and specificity in detecting gallstones (Rosenthal et al. 1994). Each participant was asked to fast for a minimum of 12 h before the ultrasound scan. Gallbladder ultrasonograms were obtained using the GCRC’s ATL 3000 ultrasound imaging unit (3.5 or 5.0 MHz transducer frequencies). Each ultrasonogram was performed by one of three technicians trained in screening gallbladder ultrasound, under supervision by an experienced radiologist. In each examination, the protocol included videotaped documentation of the gallbladder viewed in longitudinal and/or transverse views. Each view was obtained in supine and lateral positions, with both subcostal and intercostal approaches. To verify the technicians’ work as part of ongoing quality control, ∼20% of the sonograms were chosen for review and verification by the radiologist.
A participant was classified as having gallstones when one of the following three diagnostic criteria had been fulfilled: (1) gallbladder lumen with mobile nodular or dependent layering echoes that exhibited posterior acoustic shadowing, (2) gallbladder with hyperechoic shadowing material filling the gallbladder lumen with an appearance of the WES triad (i.e., the gallbladder wall, the echo of the stone, and the acoustic shadow—a specific ultrasonographic sign of gallstones used to make a reliable diagnosis of cholelithiasis [MacDonald et al. 1981; Rybicki 2000]), or (3) a history of cholecystectomy with no gallbladder lumen but with a scar consistent with a history of cholecystectomy. When the gallbladder lumen was found to have no echoes, the subject was considered unaffected. Because the reasons for cholecystectomy of 14 individuals were found to be ambiguous, we obtained medical records of these individuals for review by two physicians, to determine the indications for surgery. Of the 14 cases reviewed, GBD status was determined for 9 individuals. Individuals with cholecystectomies in the absence of gallstones were considered unaffected.
Of the 741 examined individuals, the GBD status of 8 individuals was indeterminable either because of a lack of documentation about whether prior cholecystectomy had been due to stones or because of the uncertainty of the current diagnosis of stones. Hence, their phenotypes were considered to be unknown. The pedigree data used for this study, however, contained 715 individuals with GBD data available, because 18 unrelated individuals (mainly spouses) were excluded from the analyses. Two GBD phenotypes were defined for the analyses: (1) clinical GBD, the cases in which participants self-reported cholecystectomies due to symptomatic stones and the cholecystectomy was subsequently confirmed by ultrasound at the time of the study examination, and (2) total GBD, the clinically diagnosed cases plus asymptomatic persons found to have gallstones on ultrasound.
For the SAFGS, a variety of metabolic, hemodynamic, anthropometric, and demographic variables were collected, by use of standard procedures, at the GCRC Laboratory. Blood samples were obtained after 12-h fasts, for the assessment of various metabolic traits, including fasting glucose concentrations, and they were collected again 2 h after a standardized oral glucose load, for the assessment of plasma glucose. T2DM was diagnosed in accordance with the 1999 criteria of the World Health Organization (World Health Organization 1999). Participants who did not meet these criteria but who reported that they were under treatment with either oral antidiabetic agents or insulin and who gave a history of diabetes were also considered to have T2DM.
Given the complex relationships between the components of MS (e.g., hyperglycemia, dyslipidemia, and obesity) and GBD, we used the MS risk-factor score as a single, composite covariate in all our GBD genetic analyses. MS was defined in accordance with the National Cholesterol Education Program/Adult Treatment Panel III (NCEP/ATPIII) recommendations (Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults 2001). The NCEP/ATPIII definition requires the presence of at least three of the following five risk factors: increased waist circumference (>102 cm in men and >88 cm in women), hypertriglyceredemia (⩾150 mg/dl), low high-density lipoprotein (HDL) cholesterol (<40 mg/dl in men and <50 mg/dl in women), hypertension (⩾130/85 mm Hg or people who were on hypertensive medication with normal blood pressure values), and high fasting glucose (⩾110 mg/dl or a diagnosis of T2DM, as defined above). For the genetic analyses, however, the total number of MS risk factors (range 0–5) for a given individual was considered as a covariate. Given the requirement of common MS risk-factor information for each of the individuals, ∼4.5% of the total 715 individuals had missing information for MS risk factors. The MS risk-factor score thus defined is significantly heritable (h2=39±7%, P<.0001) in our data (S. Puppala and R. Duggirala, unpublished data). Incorporation of such covariates is expected to increase power to localize disease-specific susceptibility gene(s) by removing some of the background noise due to the phenotypic correlations among the related traits (e.g., see Arya et al. 2001).
Genotype Data, Genetic Map, and Estimation of Identity-by-Descent (IBD) Matrices
A 10-cM genome scan was performed by the Center for Inherited Disease Research (CIDR) at Johns Hopkins University on ∼900 SAFDGS participants. DNA was prepared from lymphocytes for genotyping. The CIDR performed the genome scan, using automated fluorescent microsatellite analysis; its marker set was composed primarily of trinucleotide and tetranucleotide repeats across the genome. In the CIDR map, there were no gaps >18 cM, and the average marker heterozygosity was 0.76. The CIDR genetic map is similar to the genetic map provided by the Center for Genetics at Marshfield Medical Research Foundation. For the present study, we used CIDR genotypic data on 382 highly polymorphic autosomal markers. We used the genotypic information to check for genotyping errors and to verify pedigree relationships among our study participants. The CIDR routinely checked for genotype errors and possible pedigree relationship errors. However, as an added precaution, the microsatellite marker data were used to further correct potential errors. The program PREST (McPeek and Sun 2000) was used to resolve pedigree discrepancies. The data were checked for Mendelian inconsistencies by use of the PEDSYS (Dyke 1996) programs INFER and GENTEST, to eliminate typing errors. If the discrepancies continued to exist, the program SimWalk2 (Sobel and Lange 1996; Sobel et al. 2002), which used Markov Chain–Monte Carlo and simulated annealing algorithms to assign probabilities of mistyping to each genotype, was used to make decisions about the appropriate genotypes to blank (exclude). SimWalk2 detects and blanks genotypes that generate unlikely double recombinants that inflate map distances. To resolve potential double-recombinant problems, all genotypes with an error probability of ⩾0.25 were blanked. Overall, the blanking rate for errors was <0.5% of the total number of genotypes.
Maximum-likelihood techniques that account for pedigree structure were used to estimate allele frequencies. Frequency estimates obtained using samples containing related individuals can be significantly biased unless pedigree structure is taken into account (Boehnke 1991). For each genetic marker locus, the estimates of the allele frequencies and their SEs were obtained using SOLAR (Almasy and Blangero 1998). We constructed sex-averaged genetic maps, using the programs MultiMap and CRI-MAP (Lander and Green 1987; Matise et al. 1994). Locus-specific IBDs were calculated using the program SOLAR (Almasy and Blangero 1998), and multipoint IBD matrices were estimated using Markov Chain–Monte Carlo methods implemented in the program Loki (Heath 1997).
After our initial clinical GBD linkage analysis, we performed additional marker genotyping at seven chromosomal regions (chromosomes 1, 2, 4, 9, 10, 11p, and 11q) of interest. In total, 31 additional markers were typed, and ∼4 markers were typed on average for a given genetic location. In brief, DNA was extracted from white blood cells by use of proteinase K digestion/phenol extraction and alcohol precipitation in a semiautomated fashion on an ABI 341 RNA/DNA extractor. Genotyping used PCR of locus-specific microsatellite markers. The 31-marker genotypic data were checked for mistyping errors by use of the procedures described earlier, discrepancies were checked in the laboratory for mistyping, and marker genotypes for discrepant individuals were either corrected or blanked before an analysis. The blanking rate for errors was ∼1% of the total number of genotypes. Thus, our present linkage analyses were based on a data set that contained information from 413 microsatellite markers.
Variance-Components Linkage Analysis
The genetics of GBD were evaluated with a variance-components approach using the genetic information contained in the pedigrees (Hopper and Mathews 1982; Amos 1994; Almasy and Blangero 1998). This approach is based on specifying variances or covariances between relatives as a function of their genetic relationships. An extension of the variance-components approach to a threshold model (Duggirala et al. 1997; Burke et al. 2000) was used to analyze the dichotomous trait, GBD. According to this approach, it is assumed that an individual belongs to a specific disease category if an underlying, genetically determined risk or liability exceeds a certain threshold, T, on a normally distributed liability curve. The liability is assumed to have an underlying multivariate normal distribution with equal unit variances of liability both in the general population and in relatives of affected individuals. The correlation in liability between pairs of individuals is estimated using the affected status of unrelated individuals and various categories of relatives. Because the calculation of the likelihood for this multifactorial model requires high dimensional integration, we evaluated it approximately, using the Mendell-Elston algorithm (Mendell and Elston 1974). The variance components—such as heritability attributed to the susceptibility locus and heritability attributed to the residual additive genetic effects—and covariate effects for discrete traits were estimated in likelihood terms, and hypothesis tests were performed using likelihood ratio tests (Self and Lang 1987; Duggirala et al. 1999a). To obtain LOD scores, the ln likelihood values were converted into values of log10. The variance-components procedure for discrete traits was implemented in the computer program SOLAR. Because the SAFDGS families were ascertained through diabetic probands, as a conservative approach, all analyses were performed using SOLAR to correct for the ascertainment by conditioning the likelihood for the family data on the phenotype (i.e., GBD) of the proband (Boehnke and Lange 1984).
Results
The prevalence of clinical GBD and total GBD was 15% and 28%, respectively. As can be seen from table 1, ∼46% of the individuals affected with clinical GBD also had T2DM, and ∼42% of all subjects with total GDB were found to have T2DM. The GBD phenotypes were found to cluster more with MS. Of the individuals affected with clinical GBD, ∼64% also had MS, and ∼59% of total GDB-affected subjects were found to have MS (table 1). Prevalence rates of both T2DM and MS in unaffected individuals were low relative to those found in affected individuals. The occurrence of both clinical GBD and total GBD was higher in women than in men, and the affected individuals were ∼10 years older on average than the unaffected individuals. Also, the affected individuals were obese, as measured by BMI or waist circumference, compared with the unaffected individuals (table 1).
Table 1.
Clinical Characteristics of the Study Participants by GBD Status
| Participants and Variables | Affected | Unaffected |
| Clinical GBDa: | ||
| Female (%) | 81 | 57 |
| Male (%) | 19 | 43 |
| Mean age (years ± SD) | 54 ± 16 | 43 ± 16 |
| T2DM (%) | 46 | 24 |
| MSb (%) | 64 | 39 |
| Mean waist circumference (mm ± SD) | 1,060 ± 181 | 985 ± 163 |
| Mean BMI (kg/m2 ± SD) | 33 ± 8 | 31 ± 7 |
| Total GBDc: | ||
| Female (%) | 75 | 55 |
| Male (%) | 25 | 45 |
| Mean age (years ± SD) | 52 ± 17 | 42 ± 15 |
| T2DM (%) | 42 | 21 |
| MSb (%) | 59 | 37 |
| Mean waist circumference (mm ± SD) | 1,040 ± 175 | 979 ± 162 |
| Mean BMI (kg/m2 ± SD) | 32 ± 7 | 30 ± 7 |
For affected persons, n=104; for unaffected persons, n=610.
As defined by NCEP/ATPIII criteria.
For affected persons, n=202; for unaffected persons, n=513.
Heritabilities
Before linkage analyses were conducted, the discrete phenotypes—clinical and total GBD—were subjected to a variance-components technique using a threshold model to quantify the respective proportions of variance that were attributable to additive genetic factors (h2) (table 2). This analytical procedure used data from both affected and unaffected individuals. Although GBD data were available for 715 individuals (table 1), the requirement of common covariate information for each of the individuals resulted in slightly reduced sample sizes (table 2). The GBD phenotypes were subjected to genetic analyses using data from the total sample (i.e., diabetic and nondiabetic individuals) and the subsample of nondiabetic individuals. For convenience, these data sets are called set 1 (total sample) and set 2 (nondiabetics only).
Table 2.
Heritabilities (h2) of Clinical and Total GBD Phenotypes by Data Set
| Data Set and Phenotype | N | h2 ± SE | P | VarianceExplainedby Covariatesa(%) |
| Set 1b: | ||||
| Clinical GBD | 682 | 64 ± 15 | <.0001 | 12 |
| Total GBD | 683 | 26 ± 10 | .0008 | 13 |
| Set 2c: | ||||
| Clinical GBD | 489 | 77 ± 24 | .0007 | 7 |
| Total GBD | 491 | 53 ± 18 | .0004 | 9 |
Age, sex, and MS risk-factor score (0–5) were included as covariates, and the estimates of variance explained by covariates were based on Kullback-Leibler R2 values. The five MS risk factors, as defined by NCEP/ATPIII criteria, are increased waist circumference, hypertriglyceredemia, low HDL cholesterol, hypertension, and high fasting glucose.
Total sample, including diabetic and nondiabetic individuals.
Nondiabetic individuals only.
In set 1 data, after adjusting for the significant effects of age (P<.0001), sex (P<.0001), and MS risk-factor score (P=.0240), we detected high heritability for clinical GBD (h2=64%; P<.0001). However, the heritability for total GBD (h2=26%; P=.0008) was estimated to be low, after adjustment for the covariate effects of age (P<.0001), sex (P<.0001), and MS risk-factor score (P=.0002) (table 2). The covariates explained 12% and 13% of total phenotypic variation in clinical and total GBD, respectively. In set 2 data, the heritability was detected to be high for clinical GBD (h2=77%; P=.0007), after accounting for the covariate effects of age (P=.0077), sex (P=.0022), and MS risk-factor score (P=.0870); it was moderate for total GBD (h2=53%; P=.0004), after correction for the effects of age (P=.0005), sex (P<.0001), and MS risk-factor score (P=.0047). The covariates explained 7% and 9% of total phenotypic variation in clinical and total GBD phenotypes, respectively. Overall, the high heritabilities for clinical GBD in both sets suggest that the clinical or symptomatic GBD may be more informative for genetic analyses, perhaps because of the severe nature of the clinical GBD phenotype. In fact, our subsequent linkage analyses yielded stronger linkage signals with the clinical GBD phenotype than with the total GBD phenotype (tables 3 and 4). Hence, mainly the linkage results relating to clinical GBD in set 1 are discussed in the present article.
Table 3.
Chromosomal Regions Potentially Linked (LOD Scores ⩾1.2) to Clinical and Total GBD Phenotypes in the Total Sample of Diabetic and Nondiabetic Individuals (Set 1)[Note]
| Maximum LOD Scoreb |
||||
| Marker Region | Distance from p-ter(cM)a | Chromosomal Location | Clinical GBD | Total GBD |
| D1S1597–D1S407 | 34 | 1p36.21 | 3.7 | … |
| D1S255 | 65 | 1p34.3 | 3.5 | … |
| D2S1360 | 38 | 2p24.2 | 1.8 | … |
| D3S2427–D3S1262 | 188–201 | 3q26.31–q27.3 | 1.2 | … |
| D4S403 | 26 | 4p15.33 | 1.6 | … |
| D8S1130 | 22 | 8p23.1 | 1.4 | … |
| D9S2169 | 14 | 9p24.1 | 2.0 | … |
| D10S550 | 49 | 10p12.2 | 2.3 | 1.2 |
| D15S643 | 52 | 15q22.2 | … | 1.3 |
| D16S3096 | 99 | 16q23.1 | 1.8 | … |
Note.— Results are based on multipoint-linkage analyses.
Marshfield data (Kosambi cM), for the purpose of comparison.
Age, sex, and MS risk factors were considered as covariates.
Table 4.
Chromosomal Regions Potentially Linked (LOD Scores ⩾1.2) to Clinical and Total GBD Phenotypes in Nondiabetic Individuals (Set 2)[Note]
| Maximum LOD Scoreb |
||||
| Marker Region | Distance from p-ter(cM)a | Chromosomal Location | Clinical GBD | Total GBD |
| D1S1679–D1S1677 | 171–176 | 1q23.3 | 3.4 | 2.1 |
| D2S2976–D2S1780 | 4 | 2p25.3 | 1.6 | 1.6 |
| D2S1360 | 38 | 2p24.2 | 2.7 | … |
| D3S2409–D3S1600 | 71–86 | 3p21.31–p14.2 | 1.7 | … |
| D3S2406 | 103 | 3p13 | 1.4 | 1.8 |
| D3S2459–D3S3045 | 119–124 | 3q12.3–q13.12 | 2.1 | … |
| D3S1744–D3S1763 | 161–177 | 3q24–q26.1 | 1.3 | … |
| D4S1551 | 39 | 4p15.2 | 1.7 | … |
| D4S2623 | 114 | 4q25 | 1.1 | 1.7 |
| D6S1035–D6S1277 | 165–173 | 6q26 | 1.4 | 1.3 |
| D7S1804 | 137 | 7q32.3 | … | 1.6 |
| D9S2169 | 14 | 9p24.1 | 2.6 | … |
| D9S922 | 80 | 9q21.31 | 1.4 | … |
| D9S1786 | 104 | 9q22.32 | 1.9 | … |
| D10S2325 | 33 | 10p13 | 1.5 | … |
| D10S550 | 49 | 10p12.2 | … | 1.7 |
| D11S2000 | 101 | 11q22.3 | 2.0 | 2.6 |
| D11S4464 | 123 | 11q24.1 | … | 2.7 |
| D18S542 or AFM036ya1 | 41 | 18p11.21 | 2.2 | 1.6 |
Note.— Results are based on multipoint-linkage analyses.
Marshfield data (Kosambi cM), for the purpose of comparison.
Age, sex, and MS risk factors were considered as covariates.
Multipoint-Linkage Findings
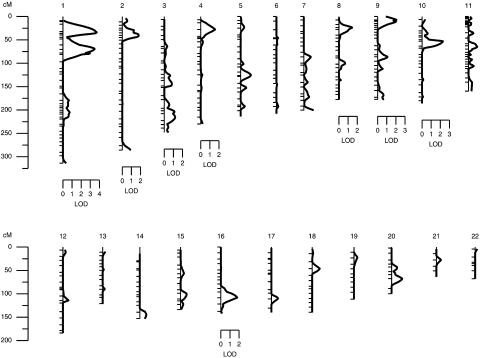
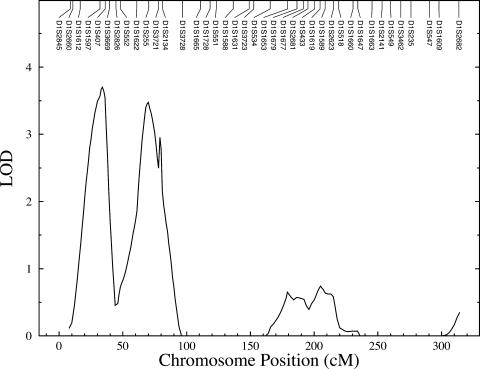
After the estimation of heritabilities, we performed multipoint linkage analyses of GBD phenotypes. In set 1, after correction for age, sex, and MS risk-factor score, potential evidence of linkage (i.e., LOD scores ⩾1.2) of clinical GBD was found at nine genetic locations representing eight chromosomes (fig. 1 and table 3). Potential linkages are considered as those genetic locations across the genome with nominal P values of ⩽.01 (i.e., LOD scores ⩾1.175). The strongest evidence of linkage (LOD = 3.7; P=.00002) of clinical GBD occurred at a genetic location between markers D1S1597 and D1S407 on chromosome 1p36.21 (set 1) (table 3 and figs. 1 and 2). The 1-LOD–unit support interval covers an ∼20-cM (or ∼9-Mb) chromosomal region between the markers D1S1612 and D1S3669. We also found strong evidence of linkage (LOD = 3.5; P=.00003) near marker D1S255 on chromosome 1p34.3 (set 1) (table 3 and figs. 1 and 2) for clinical GBD. The 1-LOD–unit support interval surrounding the linkage peak spans the ∼16-cM (or ∼11-Mb) chromosomal region between the markers D1S1622 and D1S3721. These two linkage peaks on chromosome 1p are separated by the ∼33-cM (or ∼23-Mb) chromosomal region.
Figure 1.
Summary of the clinical GBD linkage findings in Mexican Americans based on multipoint linkage analyses (LOD scores ⩾1.2) of the total data (set 1).
Figure 2.
Linkage findings of clinical GBD on chromosome 1p in Mexican Americans, by use of data from the total sample (set 1)
Suggestive evidence of linkage (LOD near or >1.9) of clinical GBD was found on chromosomes 10p near marker D10S550 (LOD = 2.3), 9p near marker D9S2169 (LOD = 2.0), 16q near marker D16S3096 (LOD = 1.8), and 2p near marker D2S1360 (LOD = 1.8) (set 1) (table 3 and fig. 1). However, for total GBD, only two genetic locations were found to exhibit potential evidence of linkage (LOD ⩾1.2) (table 3). Given that the heritabilities for GBD phenotypes were found to be moderate to high in set 2, multipoint linkage analyses were performed using the set 2 data (table 4). The strongest evidence of linkage (LOD = 3.4) of clinical GBD occurred at a genetic location between markers D1S1679 and D1S1677 on chromosome 1q23.3, whereas the highest LOD score of 2.7 for total GBD occurred at a genetic location near marker D11S4464 on chromosome 11q (table 4).
Discussion
In this study, to our knowledge the first in a human population to use a genome-scan and linkage approach, we have found strong evidence of a major locus near markers D1S1597 and D1S407 on chromosome 1p36.21 that influences variation in symptomatic or clinical GBD in the Mexican American population, after accounting for the significant covariate influences of age, sex, and MS risk factors. The evidence of linkage of clinical GBD to the chromosome 1p36.21 region is significant at the level of a genomewide scan (Lander and Kruglyak 1995). Because our data and that of others (e.g., Méndez-Sánchez et al. 2005) have revealed close association between GBD and MS (i.e., defined following NCEP/ATPIII criteria), the GBD phenotypes are adjusted for the effects of MS risk factors in our analyses. Another genetic location near marker D1S255 (chromosome 1p34.3) also exhibited strong evidence of linkage to clinical GBD, and it is also significant at the level of a genomewide scan. In consideration of the issues relating to localization, the two linkage peaks may correspond to the same susceptibility locus (Hauser and Boehnke 1997; Roberts et al. 1999; Hsueh et al. 2001a). However, the fact that they are ∼33 cM (or ∼23 Mb) apart from each other suggests that such a scenario is, effectively, very unlikely. Importantly, the 1-LOD–unit support intervals surrounding the two linkage peaks on chromosome 1p in our study approximately represent the cytogenetic locations 1p36.23-p36.13 and 1p35.3-p34.2. Such a scenario of no overlapping between the 1-LOD support intervals is suggestive of the occurrence of two loci on chromosome 1p.
In the absence of previous GBD genome-scan/linkage data in humans for comparison, we reviewed the literature for linkage studies of phenotypes correlated with GBD that implicated chromosome 1p to harbor susceptibility genes for such comorbid conditions. As shown in table 5, several studies have implicated a broad, overlapping region on chromosome 1p (i.e., 1p36.32-p32) as containing susceptibility loci for disease conditions that have relevance to GBD, especially the phenotypes related to the lipid/lipoprotein metabolism. In fact, using our data, we found weak evidence of linkage of total cholesterol and low-density lipoprotein (LDL) cholesterol near the two locations linked to GBD on chromosome 1p (table 5). Such linkage profiles of correlated phenotypes on chromosome 1p suggest the possibility of more than one susceptibility locus that could correspond to the findings reported in table 5.
Table 5.
Summary of Linkage Findings of the Phenotypes Related to GBD on Chromosome 1p[Note]
| Phenotype, Marker(s),and Populationa | Distance from p-ter(cM)b | Chromosomal Location | LOD | Reference |
| BMI: | ||||
| D1S468: | ||||
| Utah | 4 | 1p36.32 | 2.5c | Stone et al. 2002 |
| Whites | 4 | 1p36.32 | 1.4 | Liu et al. 2004 |
| LDL: | ||||
| D1S214–D1S228: | ||||
| Whites | 14–30 | 1p36.31-p36.21 | 2.4 | Elbein and Hasstedt 2002 |
| BMI: | ||||
| D1S508: | ||||
| Utah | 16 | 1p36.23 | 2.2c | Stone et al. 2002 |
| TC: | ||||
| D1S1612–D1S1597: | ||||
| Mexican Americans | 16–30 | 1p36.23-p36.21 | 1.3 | Present study |
| LDL: | ||||
| D1S1612–D1S1597: | ||||
| Mexican Americans | 16–30 | 1p36.23-p36.21 | 1.2 | Present study |
| Body size/adiposity: | ||||
| D1S1597: | ||||
| Mexican Americans | 30 | 1p36.21 | 2.5 | Cai et al. 2004 |
| GBD: | ||||
| D1S1597–D1S407: | ||||
| Mexican Americans | 30–34 | 1p36.21 | 3.7 | Present study |
| HT: | ||||
| TNFRSF1B D1S2834: | ||||
| Australian sib pairs | 31 | 1p36.22-p36.21 | 3.1 | Glenn et al. 2000 |
| FH: | ||||
| D1S2826–D1S513: | ||||
| Syrian family | 42–60 | 1p36.13-p36.12 | 3.1 | Al-Kateb et al. 2002 |
| TC: | ||||
| D1S552–D1S2843: | ||||
| Twins/parents, Berlin | 45–47 | 1p36.13-p36.12 | 1.8 | Al-Kateb et al. 2002 |
| LDL: | ||||
| D1S552–D1S2843: | ||||
| Twins/parents, Berlin | 45–47 | 1p36.13-p36.12 | 1.9 | Al-Kateb et al. 2002 |
| FH: | ||||
| D1S2725–D1S2787: | ||||
| Turkish and Asian Indians | 49–56 | 1p36.12-p35.3 | 5.3 | Eden et al. 2001 |
| TC: | ||||
| D1S1622: | ||||
| Mexican Americans | 57 | 1p35.3 | 1.0 | Present study |
| LDL: | ||||
| D1S1622–D1S255: | ||||
| Mexican Americans | 57–65 | 1p35.3-p34.3 | .5 | Present study |
| LDL-HDL ratio: | ||||
| D1S233–D1S193: | ||||
| Whites | 61–73 | 1p35.2-p34.2 | 2.1 | Elbein and Hasstedt 2002 |
| GBD: | ||||
| D1S255: | ||||
| Mexican Americans | 65 | 1p34.3 | 3.5 | Present study |
| FH: | ||||
| D1S2892–D1S2722: | ||||
| French families | 70–73 | 1p34.2 | 3.1 | Varret et al. 1999 |
| BMI: | ||||
| D1S3721: | ||||
| Old Order Amish | 73 | 1p34.2 | …d | Platte et al. 2003 |
| FH: | ||||
| D1S2134–D1S1661: | ||||
| Utah | 76–78 | 1p33-32 | 6.8 | Hunt et al. 2000 |
Note.— Findings from the present study are shown in bold italics. Some information was adapted from Bossé 2004.
HT = hypertension; FH = familial hypercholesterolemia; TC = total cholesterol.
Marshfield distance data (Kosambi cM), used for the purpose of comparison.
HLOD = heterogeneity LOD.
Evidence of linkage reported as P=.009.
The two distinct symptomatic GBD-linked regions on chromosome 1p, together with their flanking chromosomal regions, encompass a number of positional candidate genes, including TNFR2 (tumor necrosis factor receptor 2, 1p36.33-p36.2 [MIM 191191]), also called TNFRSF1B (tumor necrosis factor receptor subfamily, member 1B), SHP (small heterodimer partner, 1p36.1 [MIM 604630]), also called SHP1 or NROB2 (nuclear receptor subfamily 0, group B, member 2), and ARH (autosomal recessive hypercholesterolemia, 1p36-p35 [MIM 603813 and MIM 605747]). As shown in table 5, several studies reported that genetic locations near the TNFR2 gene are linked to obesity-related phenotypes (Stone et al. 2002; Liu et al. 2004). In another Mexican American family study, the marker region D1S1597 was found to be linked with the body size–adiposity factor (Cai et al. 2004). There is evidence that the genetic variation in and near TNFR2 could relate to familial combined hyperlipidemia-, hypertension-, and obesity-related phenotypes (Geurts et al. 2000; Glenn et al. 2000; Puga et al. 2005). Also, genetic variation in this gene was found to be associated with obesity phenotypes and insulin resistance (Fernandez-Real et al. 2000). The autosomal recessive hypercholesterolemia (ARH) has been found to be influenced by different loci, including the one mapped to chromosome 1p36.1-p35 (Eden et al. 2001; Al-Kateb et al. 2002). Garcia et al. (2001) cloned the ARH gene, which is located on chromosome 1p35 and encodes a putative LDL-receptor adapter protein.
The SHP gene (1p36.1) is located very close to our genetic region of interest on chromosome 1p36, which has striking functional relevance to GBD. There is close correspondence between this region and the chromosomal region in the mouse that harbors cholesterol gallstone–susceptibility locus Lith 8 (Wittenberg et al. 2003) (table 6). A positional candidate gene at the Lith 8 locus is SHP. SHP is an atypical nuclear receptor, a non-DNA binding protein, which plays a critical role in cholesterol/bile acid homeostasis (Kerr et al. 2002; Bhalla et al. 2004; Frank et al. 2005). SHP has been reported to repress the transcriptional activity of various nuclear receptors, such as retinoid X receptor (RXR), liver receptor homolog-1 (LRH-1), hepatocyte nuclear factor 4α (HNF-4α), and peroxisome proliferator-activated receptors (Brendel et al. 2002; Bhalla et al. 2004). Since SHP is a farnesoid X receptor (FXR) target gene and FXR is a key regulator of bile acid homeostasis, FXR and SHP play critical roles in feedback mechanisms of bile acid production (Davis et al. 2002; Schoonjans and Auwerx 2002; Wittenburg et al. 2003; Moschetta et al. 2004). Increasing levels of bile acids activate FXR, in turn inducing SHP. It, in turn, interacts with LRH-1, thereby repressing transcription of CYP7A1 and CYP8B1 (Brendel et al. 2002; Davis et al. 2002; Schoonjans and Auwerx 2002; Frank et al. 2005). The hepatic enzymes cholesterol 7-α hydroxylase (CYP7A1) and sterol 12-α hydroxylase (CYP8B1) are integral components of the neutral pathway through which cholesterol is converted into bile acids (Davis et al. 2002; Bhalla et al. 2004). Thus, any genetic defects in SHP could have direct functional relevance to GBD. Because SHP modulates the transcriptional activity of several nuclear receptors, including HNF-4α, genetic variants in SHP have been examined for association with diabetes- and obesity-related phenotypes (Nishigori et al. 2001; Hung et al. 2003).
Table 6.
Correspondence between the Chromosomal Regions Linked to Clinical GBD (Sets 1 and 2) and the Cholesterol Gallstone Susceptibility Genes (i.e., Lith Loci) Identified by Mouse Models
|
Present Article |
Lith Loci/Mouse Modelsa |
|||||
| Some Positional Candidate Genesb | Locationc | LOD | Set | Lith Locus | Chromosome | Location(cM) |
| TNFR2 (1p36.2) | 1p36.2 | 3.7 | 1 | Lith 8 | 4 | 60.0 |
| SHP (1p36.1); SCP2 (1p32.3) | 1p34.3 | 3.5 | 1 | Lith 8 | 4 | 60.0 |
| APOBd (2p24.1) | 2p24.2 | 2.7 | 2 | Lith 9 | 17 | 54.5 |
| POMCd (2p23.3); ABCG5 (2p21); ABCG8 (2p21) | 2p24.2 | 1.8 | 1 | Lith 9 | 17 | 54.5 |
| NR1I2 (3q13.3) | 3q12.3 | 2.1 | 2 | Lith 14 | 16 | 42.0 |
| LCAT (16q22.1) | 16q23.1 | 1.8 | 1 | Lith 11 | 8 | 58.0 |
| CCKAR (4p15.2); PPARGC1A (4p15.2); LRPAP1 (4p16.3) | 4p15.3 | 1.6 | 1 | Lith 13 | 5 | 30.0 |
| SLC22A1 (6q25.3) | 6q26 | 1.4 | 2 | Lith 3 | 17 | 3.5 |
Lammert et al. 2001, 2002; Paigen and Carey 2002; Lyons et al. 2003; Wittenburg et al. 2003; Mouse Genome Informatics Database.
Near the marker regions of interest in this study and/or the positional candidate genes identified by mouse models; the human cytogenetic band information is provided within parentheses after the gene symbols (UCSC Genome Browser). TNFRSF1B = tumor necrosis factor receptor subfamily, member 1B, or TNFR2; SHP = small heterodimer partner (NROB2 = nuclear receptor subfamily 0, group B, member 2); SCP2 = sterol carrier protein 2 [MIM 184755]; APOB = apolipoprotein B [MIM 107730]; POMC = proopiomelanocortin [MIM 176830]; ABCG5 = ATP-binding cassette, subfamily G, member 5 [MIM 605459]; ABCG8 = ATP-binding cassette, subfamily G, member 8 [MIM 605460]; NR1I2 = nuclear receptor subfamily 1, group I, member 2 (PXR = pregnane X receptor); LCAT = lecithin-cholesterol acyltransferase [MIM 606967]; CCKAR = cholecystokinin receptor; PPARGC1A = peroxisome proliferator-activated receptor-γ, coactivator 1, α [MIM 604517]; LRPAP1 = LDL-related protein-associated protein 1; SLC22A1 = solute carrier family 22 (organic cation transporter), member 1 [MIM 602607].
Genetic locations prioritized by the strength of evidence of linkage in our data (set 1 and set 2).
The Lith 9 location (at 54.5 cM on mouse chromosome 17) is homologous to human chromosome 2p21, and the candidate genes at this location are ABCG5 and ABCG8; APOB and POMC are located at 2–4 cM on mouse chromosome 12, but their homologous regions are 2p24.1 and 2p23.3 on human chromosome 2, respectively.
Several other chromosomal regions across the genome exhibited suggestive or potential evidence of linkage to symptomatic GBD, and some of these findings appear to have relevance to the chromosomal regions in the mouse harboring certain Lith loci (table 6). In the absence of human data to verify our linkage findings, the rich data for Lith loci (including the Lith 8 locus discussed above) appear to be helpful for understanding the potential genetic mechanisms that underlie the GBD phenotype in human populations. Our suggestive linkage finding on chromosome 10p12.2 near marker D10S550 strongly corresponds with the finding of a major susceptibility gene for obesity in a French population (Hager et al. 1998). There is further evidence of a gene or genes on chromosome 10p that influence obesity-related phenotypes in other human populations (e.g., Hinney et al. 2000; Comuzzie et al. 2001; Hsueh et al. 2001b; Lindsay et al. 2003). The evidence of linkage near marker D9S2169 on chromosome 9p24.1 corresponds well with our previous suggestive linkage findings of T2DM and age at diabetes onset on the same region in the Mexican American population (Duggirala et al. 1999a). Also, we previously found evidence of a major gene for HDL cholesterol concentrations at a location very close to this region (Arya et al. 2002; also, see Pajukanta et al. [2003] and Badzioch et al. [2004]). Strong evidence of linkage to serum adiponectin levels was found on chromosome 9p in the Pima Indian population (Lindsay et al. 2003).
The chromosomal region near marker D2S1360 (2p24.2), where we found suggestive evidence of linkage to clinical GBD, has been implicated by various other studies as influencing obesity and lipid phenotypes. For example, in a Mexican American population, Comuzzie et al. (1997) found strong evidence of linkage to leptin levels on chromosome 2p. This is one of the obesity linkage findings with the strongest and most-frequent claims of replication (Barsh et al. 2000; Comuzzie 2002; Loos and Bouchard 2003). Additionally, linkage evidence of such phenotypes as familial combined hyperlipidemia (Pajukanta et al. 2003), LDL cholesterol, and Apo B concentrations (Heijmans et al. 2005) was also found at this chromosomal region. Our finding on chromosome 16q at marker D16S3096 (16q23.1) strongly overlaps with that of a major gene for HDL cholesterol concentrations found on chromosome 16q in a Mexican American population (Mahaney et al. 2003). Evidence was also reported for linkage of LDL particle size (Badzioch et al. 2004) and low HDL cholesterol phenotype (Pajukanta et al. 2003) to similar genetic regions on chromosome 16q.
Our GBD linkage finding near marker D4S403 on chromosome 4p15.33 corresponds well with our previously reported chromosomal region that harbors a major gene for obesity in Mexican Americans (Arya et al. 2004); similar findings have been reported by other studies (Perusse et al. 2001; Deng et al. 2002; Stone et al. 2002). Another Mexican American family study reported that the same marker region harbors a major gene that influences variation in the compound lipid factor or phenotype associated with HDL cholesterol and triglyceride concentrations (Cai et al. 2004). An important positional candidate gene for GBD near the D4S403 region is cholecystokinin A receptor (CCKAR [MIM 118444]) (4p15.2 ). CCKAR plays an important role in mediating gallbladder contraction and in secreting pancreatic enzymes. Several studies have shown that the impaired gallbladder motility could be a result of the defect of the CCKAR (Wang et al. 2004; Ding et al. 2005; Zhu et al. 2005). Because gallbladder hypomotility is an important factor in cholesterol gallstone formation, any defect of the CCKAR gene could relate to our finding on chromosome 4p15.
As reported in table 4, the analyses based on nondiabetic individuals only—albeit with reduced sample sizes—yielded the strongest evidence of linkage to clinical GBD on chromosome 1q and for linkage of total GBD on chromosome 11q. The strongest evidence of linkage to clinical GBD occurred at a location between markers D1S1679 (1q23.3) and D1S1677 (1q23.3). This linkage finding corresponds with previous linkage findings on chromosome 1q that identified susceptibility gene(s) for phenotypes including T2DM, MS, or their related phenotypes (Hanson et al. 1998; Pajukanta et al. 1998, 2003; Elbein et al. 1999; Reed et al. 2001; Broeckel et al. 2002; Xiang et al. 2002; Huertas-Vázquez et al. 2004; Langefeld et al. 2004; Ng et al. 2004; Wiltshire et al. 2004). The highest LOD score for total GBD in nondiabetic individuals occurred at markers D11S2000 (11q22.3) and D11S4464 (11q24.1) on chromosome 11q (table 4). Several studies, including our own, have found evidence of the existence of a locus near marker D11S4464 that influences susceptibility to T2DM and obesity or to their related phenotypes (Hanson et al. 1998; Elbein et al. 1999; Duggirala et al. 2001, 2003a; Atwood et al. 2002; Stone et al. 2002; Arya et al. 2004).
Several studies have examined the nature of associations between GBD and genetic variants in candidate genes. As noted by Katsika et al. (2005), however, polymorphisms in the genes apolipoprotein E (APOE), hepatic phospholipid transporter (ABCB4), and the rate-limiting enzyme of bile salt synthesis (CPY7A1) appear to be consistently associated with GBD (e.g., Juvonen et al. 1993; Bertomeu et al. 1996; Rosmorduc et al. 2003; Jiang et al. 2004). In the present study, we found no evidence of linkage of clinical GBD to the chromosomal region containing the APOE gene. However, there was weak evidence of linkage of clinical GBD to the chromosomal regions harboring the genes ABCB4 on chromosome 7q (near markers D7S3046 and D7S2204; LOD = 0.8 in set 1 and LOD = 1.1 in set 2) and CPY7A1 (near markers D8S1136 and D8S2324; LOD = 0.7 in set 1 and LOD = 0.5 in set 2) on chromosome 8q.
We performed a preliminary linkage analysis of clinical GBD, using data from a subset of the SAFDGS (N=349) and the SAFDS original genome-scan data (Duggirala et al. 2003b), which is different from the CIDR genome-scan data used for the present study. In that preliminary study, we identified a location near marker D11S1984 on chromosome 11p15.5 that significantly influences the clinical GBD. Several mucin genes are located at this chromosomal region. However, we failed to reconfirm the original clinical GBD linkage finding at this 11p chromosomal region in the present study, on the basis of the larger data set (fig. 1), although there was some weak evidence of linkage at the marker D11S1984 (LOD = 1.0), on the basis of the two-point analysis. In addition, we found three chromosomal regions that exhibited suggestive evidence of linkage to clinical GBD, including the marker regions D10S245 (chromosome 10p12.1), D6S1035 (chromosome 6q26), and D8S270 (chromosome 8q13.2-q21.3). Of these findings, in the present study, only the linkage finding on chromosome 10 continues to exhibit suggestive evidence of linkage to clinical GBD, but the evidence of linkage at the other two regions was found to be very weak (D6S1035 [LOD = 0.5] and near D8S270 [LOD = 0.7]). However, the marker D6S1035 region was found to be potentially linked to both clinical and total GBD in the sub–data set containing nondiabetic individuals only. In addition to the new CIDR marker data, a potential explanation of the discrepancies between the above-discussed preliminary findings and current findings is the expanded population (i.e., the sample size of the present study is almost double that of the preliminary study).
Also, some other genetic findings of GBD in the present study need further explanations. Previous studies, including our own, using family data for the genetics of symptomatic GDB suggested that ∼25%–44% of variation in symptomatic GBD is attributable to genetic factors (Duggirala et al. 1999b; Nakeeb et al. 2002; Katsika et al. 2005). Although the present findings add further strength to such observations, the heritability estimated in this study for symptomatic GBD is high (e.g., 64% in the total data) (table 2). However, it should be noted that heritability estimates are population sample–specific and can be influenced by such factors as study population, design, ascertainment criteria, and the covariates considered for the analysis. All of these factors could have influenced the observed heritability of symptomatic GBD in our population. In regard to the differences in linkage profiles across our data sets, overall, the symptomatic GBD appears to be informative for genetic analysis in both total and nondiabetic individuals–only data sets, perhaps because of the severe nature of the symptomatic GBD. There appear to be potential genetic factors that could determine gallstones to become symptomatic after their formation in the gallbladder. Since total GBD is highly heritable in the nondiabetic data set (53%) compared with total GBD in the total sample (26%) and since several potential or suggestive linkage signals are present in the nondiabetic sub–data set with total GBD information, despite the reduced sample size, it appears that some unknown mechanisms (e.g., diabetes duration and gallbladder motility problems) that are unique to the diabetic environment are interacting with or masking the influences of the genetic factors.
In consideration of our present findings of GBD, it is apparent that a complex genetic architecture underlies the phenotypic expression of GBD. As Lee (2004) succinctly states, “the single most important way for the body to get rid of excess cholesterol is the secretion of bile acids and cholesterol into bile.” Our study localized two major susceptibility loci for clinical GBD that could have strong functional relevance to the defects related to the mechanism of supersaturation of bile with cholesterol. In addition, our study provides potential evidence of genetic factors that could influence other abnormalities of the hepatobiliary system, such as hypomotility of the gallbladder and cholesterol nucleation. It is reassuring to note that several of our linkage findings, including the major ones on chromosome 1p, appear to overlap with the positions of the Lith loci that have been reported to contribute to cholesterol gallstone formation in mice. Also evident from our study is the overlapping of some of the present linkage findings with those reported by other studies that relate to various conditions that occur with GBD, including obesity and diabetes.
In summary, we performed a genomewide search to localize susceptibility genes for GBD in Mexican Americans and found strong evidence of the possible existence of two novel susceptibility loci on chromosome 1p that influence variation in clinical GBD. To our knowledge, this is the first report of major genetic determinants of GBD in human populations. Relatively strong and/or potential evidence of linkage to GBD was also found at several genetic locations on chromosomes 1q, 2p, 3q, 4p, 8p, 9p, 10p, 11q, and 16q. Confirmation of our results in other populations would strengthen our linkage findings. We plan to screen the strong positional candidate genes, such as SHP1 and TNFR2, on chromosome 1p, to identify potential functional variant(s) that may relate to our linkage findings. Given the epidemic of obesity in both developed and developing countries, the prevalence of obesity-related comorbidities such as cholelithiasis is expected to become increasingly burdensome. Our findings may pave the way for prevention and treatment of GBD.
Acknowledgments
This study was supported by National Institutes of Health grants DK53889, DK42273, DK47482, and MH59490. We thank the CIDR for performing the new SAFDGS genome scan. We thank Norberto Isaac and Shelley Porter for performing ultrasonograms. We appreciate the nursing and dietetic care provided by the staff of the Frederic C. Bartter General Clinical Research Center at the South Texas Veterans Health Care Systems, Audie Murphy Division. We acknowledge Barbara Chapman, Sherry L. Cummins, Irma Jean Gomez, Rosa M. Pelayo, Margeret de la Garza, Rajeswari Cheruvu, Roy G. Resendez, Allison Mittler, Robin J. Lumpkin, Marcel Fourcaudot, and Paul Streng, for excellent technical support. We thank Drs. Hanna E. Abboud and Ralph A. DeFronzo, for support and encouragement, and Drs. Robin J. Leach and Dawn K. Richardson, for providing help regarding DNA sample/molecular genetic–related issues. We warmly thank the families from SAFDGS for their enthusiasm and cooperation.
Web Resources
URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for genetic map information used for the purpose of comparison)
- Mouse Genome Informatics Database, http://www.informatics.jax.org/ (for mouse map information regarding the Lith loci)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for TNFR2, SHP, ARH, CCKAR, SCP2, APOB, POMC, ABCG5, ABCG8, NR1I2, LCAT, PPARGC1A, LRPAP1, and SLC22A1)
- UCSC Genome Browser, http://www.genome.ucsc.edu/cgi-bin/hgGateway/ (for physical map and cytogenetic band information regarding microsatellite markers and genes)
References
- Al-Kateb H, Bahring S, Hoffmann K, Strauch K, Busjahn A, Nurnberg G, Jouma M, Bautz EK, Dresel HA, Luft FC (2002) Mutation in the ARH gene and a chromosome 13q locus influence cholesterol levels in a new form of digenic-recessive familial hypercholesterolemia. Circ Res 90:951–958 10.1161/01.RES.0000018002.43041.08 [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI (1994) Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet 54:535–543 [PMC free article] [PubMed] [Google Scholar]
- Arya R, Duggirala R, Almasy L, Rainwater DL, Mahaney MC, Cole S, Dyer TD, Williams K, Leach RJ, Hixson JE, MacCluer JW, O’Connell P, Stern MP, Blangero J (2002) Linkage of high-density lipoprotein-cholesterol concentrations to a locus on chromosome 9p in Mexican Americans. Nat Genet 30:102–105 10.1038/ng810 [DOI] [PubMed] [Google Scholar]
- Arya R, Duggirala R, Jenkinson CP, Almasy L, Blangero J, O’Connell P, Stern MP (2004) Evidence of a novel quantitative-trait locus for obesity on chromosome 4p in Mexican Americans. Am J Hum Genet 74:272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R, Duggirala R, Williams JT, Almasy L, Blangero J (2001) Power to localize the major gene for disease liability is increased after accounting for the effects of related quantitative phenotypes. Genet Epidemiol Suppl 21:S774–S778 [DOI] [PubMed] [Google Scholar]
- Atwood LD, Heard-Costa NL, Cupples LA, Jaquish CE, Wilson PW, D’Agostino RB (2002) Genomewide linkage analysis of body mass index across 28 years of the Framingham Heart Study. Am J Hum Genet 71:1044–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badzioch MD, Igo RP Jr, Gagnon F, Brunzell JD, Krauss RM, Motulsky AG, Wijsman EM, Jarvik GP (2004) Low-density lipoprotein particle size loci in familial combined hyperlipidemia: evidence for multiple loci from a genome scan. Arterioscler Thromb Vasc Biol 24:1942–1950 10.1161/01.ATV.0000143499.09575.93 [DOI] [PubMed] [Google Scholar]
- Barsh GS, Farooqi IS, O’Rahilly S (2000) Genetics of body-weight regulation. Nature 404:644–651 [DOI] [PubMed] [Google Scholar]
- Bertomeu A, Ros E, Zambón D, Vela M, Pérez-Ayuso RM, Targarona E, Trías M, Sanllehy C, Casals E (1996) Apolipoprotein E polymorphism and gallstones. Gastroenterology 111:1603–1610 10.1016/S0016-5085(96)70023-9 [DOI] [PubMed] [Google Scholar]
- Bhalla S, Ozalp C, Fang S, Xiang L, Kemper JK (2004) Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha: functional implications in hepatic cholesterol and glucose metabolism. J Biol Chem 279:45139–45147 10.1074/jbc.M405423200 [DOI] [PubMed] [Google Scholar]
- Boehnke M (1991) Allele frequency estimation from data on relatives. Am J Hum Genet 48:22–25 [PMC free article] [PubMed] [Google Scholar]
- Boehnke M, Lange K (1984) Ascertainment and goodness of fit of variance component models for pedigree data. Prog Clin Biol Res 147:173–192 [PubMed] [Google Scholar]
- Boland LL, Folsom AR, Rosamond WD, Atherosclerosis Risk in Communities (ARIC) Study Investigators (2002) Hyperinsulinemia, dyslipidemia, and obesity as risk factors for hospitalized gallbladder disease: a prospective study. Ann Epidemiol 12:131–140 10.1016/S1047-2797(01)00260-5 [DOI] [PubMed] [Google Scholar]
- Bossé Y (2004) Genetic susceptibility to the metabolic syndrome. PhD dissertation, Laval University, Quebec (http://www.theses.ulaval.ca/2004/22151/ch07.html) (accessed January 5, 2006)
- Brendel C, Schoonjans K, Botrugno OA, Treuter E, Auwerx J (2002) The small heterodimer partner interacts with the liver X receptor alpha and represses its transcriptional activity. Mol Endocrinol 16:2065–2076 10.1210/me.2001-0194 [DOI] [PubMed] [Google Scholar]
- Broeckel U, Hengstenberg C, Mayer B, Holmer S, Martin LJ, Comuzzie AG, Blangero J, Nurnberg P, Reis A, Riegger GA, Jacob HJ, Schunkert H (2002) A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet 30:210–214 10.1038/ng827 [DOI] [PubMed] [Google Scholar]
- Burke JP, Duggirala R, Hale DE, Blangero J, Stern MP (2000) Genetic basis of acanthosis nigricans in Mexican Americans and its association with phenotypes related to type 2 diabetes. Hum Genet 106:467–472 10.1007/s004390000274 [DOI] [PubMed] [Google Scholar]
- Cai G, Cole SA, Freeland-Graves JH, MacCluer JW, Blangero J, Comuzzie AG (2004) Principal component for metabolic syndrome risk maps to chromosome 4p in Mexican Americans: the San Antonio Family Heart Study. Hum Biol 76:651–665 10.1353/hub.2005.0001 [DOI] [PubMed] [Google Scholar]
- Carey MC (1993) Pathogenesis of gallstones. Am J Surg 165:410–419 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG (2002) The emerging pattern of the genetic contribution to human obesity. Best Pract Res Clin Endocrinol Metab 16:611–621 10.1053/beem.2002.0224 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Funahashi T, Sonnenberg G, Martin LJ, Jacob HJ, Black AE, Maas D, Takahashi M, Kihara S, Tanaka S, Matsuzawa Y, Blangero J, Cohen D, Kissebah A (2001) The genetic basis of plasma variation in adiponectin, a global endophenotype for obesity and the metabolic syndrome. J Clin Endocrinol Metab 86:4321–4325 10.1210/jc.86.9.4321 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Hixson JE, Almasy L, Mitchell BD, Mahaney MC, Dyer TD, Stern MP, MacCluer JW, Blangero J (1997) A major quantitative trait locus determining serum leptin levels and fat mass is located on human chromosome 2. Nat Genet 15:273–276 10.1038/ng0397-273 [DOI] [PubMed] [Google Scholar]
- Davis RA, Miyake JH, Hui TY, Spann NJ (2002) Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP. J Lipid Res 43:533–543 [PubMed] [Google Scholar]
- Deng HW, Deng H, Liu YJ, Liu YZ, Xu FH, Shen H, Conway T, Li JL, Huang QY, Davies KM, Recker RR (2002) A genomewide linkage scan for quantitative-trait loci for obesity phenotypes. Am J Hum Genet 70:1138–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl AK (1991) Epidemiology of natural history of gallstone disease. Gastroenterol Clin North Am 20:1–19 [PubMed] [Google Scholar]
- ——— (2000) Cholelithiasis and the insulin resistance syndrome. Hepatology 31:528–530 10.1002/hep.510310238 [DOI] [PubMed] [Google Scholar]
- Diehl AK, Schwesinger WH, Holleman DR Jr, Chapman JB, Kurtin WE (1994) Gallstone characteristics in Mexican Americans and non-Hispanic whites. Dig Dis Sci 39:2223–2228 10.1007/BF02090375 [DOI] [PubMed] [Google Scholar]
- Diehl AK, Stern MP (1989) Special health problems of Mexican-Americans: obesity, gallbladder disease, diabetes mellitus, and cardiovascular disease. Adv Intern Med 34:73–96 [PubMed] [Google Scholar]
- Ding X, Lu CY, Mei Y, Liu CA, Shi YJ (2005) Correlation between gene expression of CCK-A receptor and emptying dysfunction of the gallbladder in patients with gallstones and diabetes mellitus. Hepatobiliary Pancreat Dis Int 4:295–298 [PubMed] [Google Scholar]
- Duggirala R, Almasy L, Blangero J, Jenkinson CP, Arya R, DeFronzo RA, Stern MP, O’Connell P, American Diabetes Association GENNID Study Group (2003a) Further evidence for type 2 diabetes susceptibility locus on chromosome 11q. Genet Epidemiol 24:240–242 10.1002/gepi.10233 [DOI] [PubMed] [Google Scholar]
- Duggirala R, Blangero J, Almasy L, Arya R, Dyer TD, Williams KL, Leach RJ, O’Connell P, Stern MP (2001) A major locus for fasting insulin concentrations and insulin resistance on chromosome 6q with strong pleiotropic effects on obesity-related phenotypes in nondiabetic Mexican Americans. Am J Hum Genet 68:1149–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O’Connell P, Stern MP (1999a) Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 64:1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggirala R, Dodd GD, Fowler S, Schneider J, Arya R, Diehl AK, Almasy L, O’Connell P, Stern MP, Blangero J (2003b) Gallbladder disease is influenced by a major locus on chromosome 11p in Mexican Americans. Am J Hum Genet Suppl 73:195 [Google Scholar]
- Duggirala R, Mitchell BD, Blangero J, Stern MP (1999b) Genetic determinants of variation in gallbladder disease in the Mexican-American population. Genet Epidemiol 16:191–204 [DOI] [PubMed] [Google Scholar]
- Duggirala R, Williams JT, Williams-Blangero S, Blangero J (1997) A variance component approach to dichotomous trait linkage analysis using a threshold model. Genet Epidemiol 14:987–992 [DOI] [PubMed] [Google Scholar]
- Dyke B (1996) PEDSYS: a pedigree data management system. Tech rep 2, Population Genetics Laboratory, Department of Genetics, Southwest Foundation for Biomedical Research, San Antonio [Google Scholar]
- Eden ER, Naoumova RP, Burden JJ, McCarthy MI, Soutar AK (2001) Use of homozygosity mapping to identify a region on chromosome 1 bearing a defective gene that causes autosomal recessive homozygous hypercholesterolemia in two unrelated families. Am J Hum Genet 68:653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein SC, Hasstedt SJ (2002) Quantitative trait linkage analysis of lipid-related traits in familial type 2 diabetes: evidence for linkage of triglyceride levels to chromosome 19q. Diabetes 51:528–535 [DOI] [PubMed] [Google Scholar]
- Elbein SC, Hoffman MD, Teng K, Leppert MF, Hasstedt SJ (1999) A genome-wide search for type 2 diabetes susceptibility genes in Utah Caucasians. Diabetes 48:1175–1182 [DOI] [PubMed] [Google Scholar]
- Everhart JE (1993) Contributions of obesity and weight loss to gallstone disease. Ann Intern Med 119:1029–1035 [DOI] [PubMed] [Google Scholar]
- ——— (1995) Digestive diseases and diabetes. In: Diabetes in America, 2nd ed. NIH publication no. 95–1468. National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, pp 457–483 [Google Scholar]
- Everhart JE, Kahre M, Hill M, Maurer KR (1999) Prevalence and ethnic differences in gallbladder disease in the United States. Gastroenterology 117:632–639 10.1016/S0016-5085(99)70456-7 [DOI] [PubMed] [Google Scholar]
- Everhart JE, Yeh F, Lee ET, Hill MC, Fabsitz R, Howard BV, Welty TK (2002) Prevalence of gallbladder disease in American Indian populations: findings from the Strong Heart Study. Hepatology 35:1507–1512 10.1053/jhep.2002.33336 [DOI] [PubMed] [Google Scholar]
- Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (2001) Executive summary of the third report of National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (ATP III). JAMA 285:2486–2497 10.1001/jama.285.19.2486 [DOI] [PubMed] [Google Scholar]
- Fernandez-Real JM, Vendrell J, Ricart W, Broch M, Gutierrez C, Casamitjana R, Oriola J, Richart C (2000) Polymorphism of the tumor necrosis factor-alpha receptor 2 gene is associated with obesity, leptin levels, and insulin resistance in young subjects and diet-treated type 2 diabetic patients. Diabetes Care 23:831–837 [DOI] [PubMed] [Google Scholar]
- Frank C, Makkonen H, Dunlop TW, Matilainen M, Vaisanen S, Carlberg C (2005) Identification of pregnane X receptor binding sites in the regulatory regions of genes involved in bile acid homeostasis. J Mol Biol 346:505–519 10.1016/j.jmb.2004.12.003 [DOI] [PubMed] [Google Scholar]
- Garcia CK, Wilund K, Arca M, Zuliani G, Fellin R, Maioli M, Calandra S, Bertolini S, Cossu F, Grishin N, Barnes R, Cohen JC, Hobbs HH (2001) Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science 292:1394–1398 10.1126/science.1060458 [DOI] [PubMed] [Google Scholar]
- Geurts JM, Janssen RG, van Greevenbroek MM, van der Kallen CJ, Cantor RM, Bu X, Aouizerat BE, Allayee H, Rotter JI, de Bruin TW (2000) Identification of TNFRSF1B as a novel modifier gene in familial combined hyperlipidemia. Hum Mol Genet 9:2067–2074 10.1093/hmg/9.14.2067 [DOI] [PubMed] [Google Scholar]
- Glenn CL, Wang WY, Benjafield AV, Morris BJ (2000) Linkage and association of tumor necrosis factor receptor 2 locus with hypertension, hypercholesterolemia and plasma shed receptor. Hum Mol Genet 9:1943–1949 10.1093/hmg/9.13.1943 [DOI] [PubMed] [Google Scholar]
- Grundy SM (2004) Cholesterol gallstones: a fellow traveler with metabolic syndrome? Am J Clin Nutr 80:1–2 [DOI] [PubMed] [Google Scholar]
- Haffner SM, Diehl AK, Valdez R, Mitchell BD, Hazuda HP, Morales P, Stern MP (1993) Clinical gallbladder disease in NIDDM subjects. Diabetes Care 16:1276–1284 [DOI] [PubMed] [Google Scholar]
- Hager J, Dina C, Francke S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant A, Clement K, Guy-Grand B, Froguel P (1998) A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nat Genet 20:304–308 10.1038/3123 [DOI] [PubMed] [Google Scholar]
- Hall MJ, Lawrence L (1998) Ambulatory surgery in the United States: advance data from vital and health statistics. No 300. National Center for Health Statistics, Hyattsville, MD [Google Scholar]
- Hanis CL, Hewett-Emmeett D, Kubrusly LF, Makland MN, Douglas TC, Mueller WH, Barton SA, Yoshimaru H, Kubrusly DB, Gonzalez R, Schull WJ (1993) An ultrasound survey of gallbladder disease among Mexican Americans in Starr County, Texas: frequencies and risk factors. Ethn Dis 3:32–43 [PubMed] [Google Scholar]
- Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC (1998) An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet 63:1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser ER, Boehnke M (1997) Confirmation of linkage results in affected-sib-pair linkage analysis for complex genetic traits. Am J Hum Genet Suppl 61:A278 [Google Scholar]
- Heath SC (1997) Markov chain Monte Carlo methods for radiation hybrid mapping. J Comput Biol 4:505–515 [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Beekman M, Putter H, Lakenberg N, van der Wijk HJ, Whitfield JB, Posthuma D, Pedersen NL, Martin NG, Boomsma DI, Slagboom PE (2005) Meta-analysis of four new genome scans for lipid parameters and analysis of positional candidates in positive linkage regions. Eur J Hum Genet 13:1143–1153 10.1038/sj.ejhg.5201466 [DOI] [PubMed] [Google Scholar]
- Hillebrandt S, Matern S, Lammert F (2003) Mouse models for genetic dissection of polygenic gastrointestinal diseases. Eur J Clin Invest 33:155–160 10.1046/j.1365-2362.2003.01089.x [DOI] [PubMed] [Google Scholar]
- Hinney A, Ziegler A, Oeffner F, Wedewardt C, Vogel M, Wulftange H, Geller F, Stubing K, Siegfried W, Goldschmidt HP, Remschmidt H, Hebebrand J (2000) Independent confirmation of a major locus for obesity on chromosome 10. J Clin Endocrinol Metab 85:2962–2965 10.1210/jc.85.8.2962 [DOI] [PubMed] [Google Scholar]
- Hopper J, Matthews J (1982) Extensions to multivariate normal models for pedigree analysis. Ann Hum Genet 46:373–383 [DOI] [PubMed] [Google Scholar]
- Hsueh WC, Göring HH, Blangero J, Mitchell BD (2001a) Replication of linkage to quantitative trait loci: variation in location and magnitude of the lod score. Genet Epidemiol Suppl 21:S473–S478 [DOI] [PubMed] [Google Scholar]
- Hsueh WC, Mitchell BD, Schneider JL, St Jean PL, Pollin TI, Ehm MG, Wagner MJ, Burns DK, Sakul H, Bell CJ, Shuldiner AR (2001b) Genome-wide scan of obesity in the Old Order Amish. J Clin Endocrinol Metab 86:1199–1205 10.1210/jc.86.3.1199 [DOI] [PubMed] [Google Scholar]
- Huertas-Vazquez A, del Rincon JP, Canizales-Quinteros S, Riba L, Vega-Hernandez G, Ramirez-Jimenez S, Auron-Gomez M, Gomez-Perez FJ, Aguilar-Salinas CA, Tusie-Luna MT (2004) Contribution of chromosome 1q21-q23 to familial combined hyperlipidemia in Mexican families. Ann Hum Genet 68:419–427 10.1046/j.1529-8817.2003.00116.x [DOI] [PubMed] [Google Scholar]
- Hung CC, Farooqi IS, Ong K, Luan J, Keogh JM, Pembrey M, Yeo GS, Dunger D, Wareham NJ, O’Rahilly S (2003) Contribution of variants in the small heterodimer partner gene to birth weight, adiposity, and insulin levels: mutational analysis and association studies in multiple populations. Diabetes 52:1288–1291 [DOI] [PubMed] [Google Scholar]
- Hunt SC, Hopkins PN, Bulka K, McDermott MT, Thorne TL, Wardell BB, Bowen BR, Ballinger DG, Skolnick MH, Samuels ME (2000) Genetic localization to chromosome 1p32 of the third locus for familial hypercholesterolemia in a Utah kindred. Arterioscler Thromb Vasc Biol 20:1089–1093 [DOI] [PubMed] [Google Scholar]
- Jiang ZY, Han TQ, Suo GJ, Feng DX, Chen S, Cai XX, Jiang ZH, Shang J, Zhang Y, Jiang Y, Zhang SD (2004) Polymorphisms at cholesterol 7α-hydroxylase, apolipoproteins B and E and low density lipoprotein receptor genes in patients with gallbladder stone disease. World J Gastroenterol 10:1508–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvonen T, Kervinen K, Kairalouma MI, Lajunen LHJ, Kesäniemi YA (1993) Gallstone cholesterol content is related to apolipoprotein E polymorphism. Gastroenterology 104:1806–1813 [DOI] [PubMed] [Google Scholar]
- Katsika D, Grijbovski A, Einarsson C, Lammert F, Lichtenstein P, Marschall H-U (2005) Genetic and environmental influences on symptomatic gallstone disease: a Swedish study of 43,141 twin pairs. Hepatology 41:1138–1143 10.1002/hep.20654 [DOI] [PubMed] [Google Scholar]
- Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, Schwarz M (2002) Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell 2:713–720 10.1016/S1534-5807(02)00154-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesaniemi YA, Koskenvuo M, Vuoristo, Mittinen TA (1989) Biliary lipid composition in monozygotic and dizygotic pairs of twins. Gut 30:1750–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanuja B, Cheah YC, Hunt M, Nishina PM, Wang DQ, Chen HW, Billheimer JT, Carey MC, Paigen B (1995) Lith1, a major gene affecting cholesterol gallstone formation among inbred strains of mice. Proc Natl Acad Sci USA 92:7729–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosters A, Jirsa M, Groen AK (2003) Genetic background of cholesterol gallstone disease. Biochim Biophys Acta 1637:1–19 [DOI] [PubMed] [Google Scholar]
- Lammert F, Carey MC, Paigen B (2001) Chromosomal organization of candidate genes involved in cholesterol gallstone formation: a murine gallstone map. Gastroenterology 120:221–238 [DOI] [PubMed] [Google Scholar]
- Lammert F, Wang DQ, Wittenberg H, Bouchard G, Hillebrandt S, Taenzler B, Carey MC, Paigen B (2002) Lith genes control mucin accumulation, cholesterol crystallization, and gallstone formation in A/J and AKR/J inbred mice. Hepatology 36:1145–1154 10.1053/jhep.2002.36821 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- Langefeld CD, Wagenknecht LE, Rotter JI, Williams AH, Hokanson JE, Saad MF, Bowden DW, Haffner S, Norris JM, Rich SS, Mitchell BD (2004) Linkage of the metabolic syndrome to 1q23-q31 in Hispanic families: the Insulin Resistance Atherosclerosis Study Family Study. Diabetes 53:1170–1174 [DOI] [PubMed] [Google Scholar]
- Lawrence L, Hall MJ (1999) 1997 summary: National Hospital Discharge Survey. Adv Data 308:1–16 [PubMed] [Google Scholar]
- Lee S (2004) Gallstones: how do we translate an old story into future therapy? Nat Clin Pract Gastroenterol Hepatol 1:2–3 10.1038/ncpgasthep0017 [DOI] [PubMed] [Google Scholar]
- Leitzmann MF, Rimm EB, Willet WC, Spiegelman D, Grodstein F, Stampfer MJ, Colditz GA (1999) Recreational physical activity and the risk of cholycystectomy in women. N Engl J Med 341:777–784 10.1056/NEJM199909093411101 [DOI] [PubMed] [Google Scholar]
- Lindsay RS, Funahashi T, Krakoff J, Matsuzawa Y, Tanaka S, Kobes S, Bennett PH, Tataranni PA, Knowler WC, Hanson RL (2003) Genome-wide linkage analysis of serum adiponectin in the Pima Indian population. Diabetes 52:2419–2425 [DOI] [PubMed] [Google Scholar]
- Liu YJ, Xu FH, Shen H, Liu YZ, Deng HY, Zhao LJ, Huang QY, Dvornyk V, Conway T, Davies KM, Li JL, Recker RR, Deng HW (2004) A follow-up linkage study for quantitative trait loci contributing to obesity-related phenotypes. J Clin Endocrinol Metab 89:875–882 10.1210/jc.2003-030774 [DOI] [PubMed] [Google Scholar]
- Loos RJ, Bouchard C (2003) Obesity: is it a genetic disorder? J Intern Med 254:401–425 10.1046/j.1365-2796.2003.01242.x [DOI] [PubMed] [Google Scholar]
- Lyons MA, Korstanje R, Li R, Sheehan SM, Walsh KA, Rollins JA, Carey MC, Paigen B, Churchill GA (2005) Single and interacting QTLs for cholesterol gallstones revealed in an intercross between mouse strains NZB and SM. Mamm Genome 16:152–163 10.1007/s00335-004-2446-5 [DOI] [PubMed] [Google Scholar]
- Lyons MA, Wittenburg H, Li R, Walsh KA, Leonard MR, Churchill GA, Carey MC, Paigen B (2003) New quantitative trait loci that contribute to cholesterol gallstone formation detected in an intercross of CAST/Ei and 129S1/SvImJ inbred mice. Physiol Genomics 14:225–239 [DOI] [PubMed] [Google Scholar]
- MacDonald FR, Cooperberg PL, Cohen MM (1981) The WES triad: a specific sonographic sign of gallstones in the contracted gallbladder. Gastrointest Radiol 6:39–41 10.1007/BF01890219 [DOI] [PubMed] [Google Scholar]
- Mahaney MC, Almasy L, Rainwater DL, VandeBerg JL, Cole SA, Hixson JE, Blangero J, MacCluer JW (2003) A quantitative trait locus on chromosome 16q influences variation in plasma HDL-C levels in Mexican Americans. Arterioscler Thromb Vasc Biol 23:339–345 10.1161/01.ATV.0000051406.14162.6A [DOI] [PubMed] [Google Scholar]
- Matise TC, Perlin M, Chakravarti A (1994) Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet 6:384–390 10.1038/ng0494-384 [DOI] [PubMed] [Google Scholar]
- Maurer KJ, Ihrig MM, Rogers AB, Ng V, Bouchard G, Leonard MR, Carey MC, Fox JG (2005) Identification of cholelithogenic enterohepatic helicobacter species and their role in murine cholesterol gallstone formation. Gastroenterology 128:1023–1033 10.1053/j.gastro.2005.01.008 [DOI] [PubMed] [Google Scholar]
- McPeek MS, Sun L (2000) Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet 66:1076–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell NR, Elston RC (1974) Multifactorial qualitative traits: genetic analysis and prediction of recurrence risks. Biometrics 30:41–57 [PubMed] [Google Scholar]
- Méndez-Sánchez N, Cardenas-Vazquez R, Ponciano-Rodriguez G, Uribe M (1996) Pathophysiology of cholesterol gallstone disease. Arch Med Res 27:433–441 [PubMed] [Google Scholar]
- Méndez-Sánchez N, Chavez-Tapia NC, Motola-Kuba D, Sanchez-Lara K, Ponciano-Rodriguez G, Baptista H, Ramos MH, Uribe M (2005) Metabolic syndrome as a risk factor for gallstone disease. World J Gastroenterol 11:1653–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Sánchez N, King-Martinez AC, Ramos MH, Pichardo-Bahena R, Uribe M (2004) The Amerindian’s genes in the Mexican population are associated with development of gallstone disease. Am J Gastroenterol 99:2166–2170 10.1111/j.1572-0241.2004.40159.x [DOI] [PubMed] [Google Scholar]
- Misciagna G, Leoci C, Guerra V, Chiloiro M, Elba S, Petruzzi J, Mossa A, Noviello MR, Coviello A, Minutolo MC, Mangini V, Messa C, Cavallini A, De Michele G, Giorgio I (1996) Epidemiology of cholelithiasis in southern Italy. Part II: risk factors. Eur J Gastroenterol Hepatol 8:585–593 [DOI] [PubMed] [Google Scholar]
- Moschetta A, Bookout AL, Mangelsdorf DJ (2004) Prevention of cholesterol gallstone disease by FXR agonists in a mouse model. Nat Med 10:1352–1358 10.1038/nm1138 [DOI] [PubMed] [Google Scholar]
- Nakeeb A, Comuzzie AG, Martin L, Sonnenberg GE, Swartz-Basile D, Kissebah AH, Pitt HA (2002) Gallstones: genetics versus environment. Ann Surg 235:842–849 10.1097/00000658-200206000-00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MC, So WY, Lam VK, Cockram CS, Bell GI, Cox NJ, Chan JC (2004) Genome-wide scan for metabolic syndrome and related quantitative traits in Hong Kong Chinese and confirmation of a susceptibility locus on chromosome 1q21-q25. Diabetes 53:2676–2683 [DOI] [PubMed] [Google Scholar]
- Nishigori H, Tomura H, Tonooka N, Kanamori M, Yamada S, Sho K, Inoue I, Kikuchi N, Onigata K, Kojima I, Kohama T, Yamagata K, Yang Q, Matsuzawa Y, Miki T, Seino S, Kim MY, Choi HS, Lee YK, Moore DD, Takeda J (2001) Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc Natl Acad Sci USA 98:575–580 10.1073/pnas.021544398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen B, Carey MC (2002) Gallstones. In: King RA, Rotter JI, Motulsky AG (eds) The genetic basis of common diseases, 2nd ed. Oxford University Press, Oxford, United Kingdom, pp 298–335 [Google Scholar]
- Paigen B, Schork NJ, Svenson KL, Cheah YC, Mu JL, Lammert F, Wang DQ, Bouchard G, Carey MC (2000) Quantitative trait loci mapping for cholesterol gallstones in AKR/J and C57L/J strains of mice. Physiol Genomics 4:59–65 [DOI] [PubMed] [Google Scholar]
- Pajukanta P, Allayee H, Krass KL, Kuraishy A, Soro A, Lilja HE, Mar R, Taskinen MR, Nuotio I, Laakso M, Rotter JI, de Bruin TW, Cantor RM, Lusis AJ, Peltonen L (2003) Combined analysis of genome scans of Dutch and Finnish families reveals a susceptibility locus for high-density lipoprotein cholesterol on chromosome 16q. Am J Hum Genet 72:903–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajukanta P, Nuotio I, Terwilliger JD, Porkka KV, Ylitalo K, Pihlajamaki J, Suomalainen AJ, Syvanen AC, Lehtimaki T, Viikari JS, Laakso M, Taskinen MR, Ehnholm C, Peltonen L (1998) Linkage of familial combined hyperlipidaemia to chromosome 1q21-q23. Nat Genet 18:369–373 10.1038/ng0498-369 [DOI] [PubMed] [Google Scholar]
- Perusse L, Chagnon YC, Weisnagel SJ, Rankinen T, Snyder E, Sands J, Bouchard C (2001) The human obesity gene map: the 2000 update. Obes Res 9:135–169 [DOI] [PubMed] [Google Scholar]
- Platte P, Papanicolaou GJ, Johnston J, Klein CM, Doheny KF, Pugh EW, Roy-Gagnon MH, Stunkard AJ, Francomano CA, Wilson AF (2003) A study of linkage and association of body mass index in the Old Order Amish. Am J Med Genet C Semin Med Genet 121:71–80 10.1002/ajmg.c.20005 [DOI] [PubMed] [Google Scholar]
- Pomeranz IS, Shaffer EA (1985) Abnormal gallbladder emptying in a subgroup of patients with gallstone. Gastroenterology 88:787–791 [DOI] [PubMed] [Google Scholar]
- Portincasa P, Moschetta A, Calamita G, Margari A, Palasciano G (2003) Pathobiology of cholesterol gallstone disease: from equilibrium ternary phase diagram to agents preventing cholesterol crystallization and stone formation. Curr Drug Targets Immune Endocr Metabol Disord 3:67–81 [PubMed] [Google Scholar]
- Portincasa P, Stolk MFJ, van Erpecum KJ, Palasciano G, van Berge-Henegouwen GP (1995) Cholesterol gallstone formation in man and potential treatments of the gallbladder motility defect. Scand J Gastroenterol Suppl 212:63–78 [DOI] [PubMed] [Google Scholar]
- Puga I, Lainez B, Fernandez-Real JM, Buxade M, Broch M, Vendrell J, Espel E (2005) A polymorphism in the 3′ untranslated region of the gene for tumor necrosis factor receptor 2 modulates reporter gene expression. Endocrinology 146:2210–2220 10.1210/en.2004-1366 [DOI] [PubMed] [Google Scholar]
- Reed DR, Nanthakumar E, North M, Bell C, Price RA (2001) A genome-wide scan suggests a locus on chromosome 1q21-q23 contributes to normal variation in plasma cholesterol concentration. J Mol Med 79:262–269 10.1007/s001090100212 [DOI] [PubMed] [Google Scholar]
- Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS (1999) Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet 65:876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal TC, Siepel T, Zubler J, Horwitz M (1994) The use of ultrasonography to scan the abdomen of patients presenting for routine physical examinations. J Fam Pract 38:380–385 [PubMed] [Google Scholar]
- Rosmorduc O, Hermelin B, Boelle PY, Parc R, Taboury J, Poupon R (2003) ABCB4 gene mutation-associated cholelithiasis in adults. Gastroenterology 125:452–459 10.1016/S0016-5085(03)00898-9 [DOI] [PubMed] [Google Scholar]
- Ruhl CE, Everhart JE (2000) Association of diabetes, serum insulin, and C-peptide with gallbladder disease. Hepatology 31:299–303 10.1002/hep.510310206 [DOI] [PubMed] [Google Scholar]
- Rybicki FJ (2000) The WES sign. Radiology 214:881–882 [DOI] [PubMed] [Google Scholar]
- Sahi T, Paffenbarger RS Jr, Hsieh CC, Lee IM (1998) Body mass index, cigarette smoking, and other characteristics as predictors of self-reported, physician diagnosed gall bladder disease in male college alumni. Am J Epidemiol 147:644–651 [DOI] [PubMed] [Google Scholar]
- Sandler RS, Everhart JE, Donowitz M, Adams E, Cronin K, Goodman C, Gemmen E, Shah S, Rubin R (2002) The burden of selected digestive diseases in the United States. Gastroenterology 122:1500–1511 10.1053/gast.2002.32978 [DOI] [PubMed] [Google Scholar]
- Sarin SK, Negi VS, Dewan R, Sasan S, Saraya A (1995) High familial prevalence of gallstones in the first-degree relatives of gallstone patients. Hepatology 22:138–141 10.1016/0270-9139(95)90365-8 [DOI] [PubMed] [Google Scholar]
- Schoonjans K, Auwerx J (2002) A sharper image of SHP. Nat Med 8:789–791 10.1038/nm0802-789 [DOI] [PubMed] [Google Scholar]
- Self SG, Liang K-Y (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc 82:605–610 [Google Scholar]
- Silva CP, Pereira-Lima JC, Oliveira AG, Guerra JB, Marques DL, Sarmanho L, Cabral MM, Queiroz DM (2003) Association of the presence of Helicobacter in gallbladder tissue with cholelithiasis and cholecystitis. J Clin Microbiol 41:5615–5618 10.1128/JCM.41.12.5615-5618.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Sobel E, Papp JC, Lange K (2002) Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet 70:496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S, Abkevich V, Hunt SC, Gutin A, Russell DL, Neff CD, Riley R, Frech GC, Hensel CH, Jammulapati S, Potter J, Sexton D, Tran T, Gibbs D, Iliev D, Gress R, Bloomquist B, Amatruda J, Rae PM, Adams TD, Skolnick MH, Shattuck D (2002) A major predisposition locus for severe obesity, at 4p15-p14. Am J Hum Genet 70:1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Lee SP (2001) The role of bacteria in gallstone pathogenesis. Front Biosci 6:E93–E103 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Leitzmann MF, Willett WC, Giovannucci EL (2004) Prospective study of abdominal adiposity and gallstone disease in US men. Am J Clin Nutr 80:38–44 [DOI] [PubMed] [Google Scholar]
- Varret M, Rabes JP, Saint-Jore B, Cenarro A, Marinoni JC, Civeira F, Devillers M, Krempf M, Coulon M, Thiart R, Kotze MJ, Schmidt H, Buzzi JC, Kostner GM, Bertolini S, Pocovi M, Rosa A, Farnier M, Martinez M, Junien C, Boileau C (1999) A third major locus for autosomal dominant hypercholesterolemia maps to 1p34.1-p32. Am J Hum Genet 64:1378–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DQ, Schmitz F, Kopin AS, Carey MC (2004) Targeted disruption of the murine cholecystokinin-1 receptor promotes intestinal cholesterol absorption and susceptibility to cholesterol cholelithiasis. J Clin Invest 114:521–528 10.1172/JCI200416801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss KM, Ferrell RE, Hanis CL (1984a) A New World syndrome of metabolic diseases with genetic and evolutionary basis. Yearbook Phys Anthropol 27:153–178 10.1002/ajpa.1330270508 [DOI] [Google Scholar]
- Weiss KM, Ferrell RE, Hanis CL, Styne PN (1984b) Genetics and epidemiology of gallbladder disease in New World native peoples. Am J Hum Genet 36:1259–1278 [PMC free article] [PubMed] [Google Scholar]
- Wiltshire S, Frayling TM, Groves CJ, Levy JC, Hitman GA, Sampson M, Walker M, Menzel S, Hattersley AT, Cardon LR, McCarthy MI (2004) Evidence from a large UK family collection that genes influencing age of onset of type 2 diabetes map to chromosome 12p and to the MODY3/NIDDM2 locus on 12q24. Diabetes 53:855–860 [DOI] [PubMed] [Google Scholar]
- Wittenburg H, Lyons MA, Li R, Churchill GA, Carey MC, Paigen B (2003) FXR and ABCG5/ABCG8 as determinants of cholesterol gallstone formation from quantitative trait locus mapping in mice. Gastroenterology 125:868–881 10.1016/S0016-5085(03)01053-9 [DOI] [PubMed] [Google Scholar]
- World Health Organization (1999) Definition, diagnosis and classification of diabetes mellitus and its complications: report of a WHO consultation. Part I: diagnosis and classification of diabetes mellitus. World Health Organization, Geneva [Google Scholar]
- Xiang K, Wang Y, Zheng T, Shen K, Jia W, Li J, Lin X, Wu S, Zhang G, Wang S, Lu H (2002) Genome-wide scan search for type 2 diabetes susceptibility loci in Chinese. Diabetes Suppl 51:A262 [Google Scholar]
- Yang CC, Sun SS, Lin CC, Kao A, Lee CC (2002) Evidence of impaired gallbladder function in patients with non-insulin-dependent diabetes mellitus by quantitative cholescintigraphy. J Diabetes Complications 16:347–351 10.1016/S1056-8727(01)00218-5 [DOI] [PubMed] [Google Scholar]
- Zhu J, Han TQ, Chen S, Jiang Y, Zhang SD (2005) Gallbladder motor function, plasma cholecystokinin and cholecystokinin receptor of gallbladder in cholesterol stone patients. World J Gastroenterol 11:1685–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]