Abstract
Mucolipidosis II (MLII; I-cell disease) and mucolipidosis IIIA (MLIIIA; classical pseudo-Hurler polydystrophy) are diseases in which the activity of the uridine diphosphate (UDP)–N-acetylglucosamine:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase (GlcNAc-phosphotransferase) is absent or reduced, respectively. In the absence of mannose phosphorylation, trafficking of lysosomal hydrolases to the lysosome is impaired. In these diseases, mistargeted lysosomal hydrolases are secreted into the blood, resulting in lysosomal deficiency of many hydrolases and a storage-disease phenotype. To determine whether these diseases are caused by mutations in the GlcNAc-phosphotransferase α/β–subunits precursor gene (GNPTAB), we sequenced GNPTAB exons and flanking intronic sequences and measured GlcNAc-phosphotransferase activity in patient fibroblasts. We identified 15 different mutations in GNPTAB from 18 pedigrees with MLII or MLIIIA and demonstrated that these two diseases are allelic. Mutations in both alleles were identified in each case, which demonstrated that GNPTAB mutations are the cause of both diseases. Some pedigrees had identical mutations. One frameshift mutation (truncation at amino acid 1171) predominated and was found in both MLII and MLIIIA. This mutation was found in combination with severe mutations (i.e., mutations preventing the generation of active enzyme) in MLII and with mild mutations (i.e., mutations allowing the generation of active enzyme) in MLIIIA. Some cases of MLII and MLIIIA were the result of mutations that cause aberrant splicing. Substitutions were inside the invariant splice-site sequence in MLII and were outside it in MLIIIA. When the mutations were analyzed along with GlcNAc-phosphotransferase activity, it was possible to confidently distinguish these two clinically related but distinct diseases. We propose criteria for distinguishing these two disorders by a combination of mutation detection and GlcNAc-phosphotransferase activity determination.
Mucolipidosis II (MLII [MIM 252500]), mucolipidosis IIIA (MLIIIA [MIM 252600]), and mucolipidosis IIIC (MLIIIC [MIM 252605]) are three related genetic diseases of lysosomal enzyme trafficking. Alterations in the activity of the uridine diphosphate (UDP)–N-acetylglucosamine:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase (GlcNAc-phosphotransferase [IUBMB accession number EC 2.7.8.17]) have been demonstrated in all three of these diseases. Alterations of GlcNAcphosphotransferase are believed to be the primary genetic defect in all three diseases. GlcNAc-phosphotransferase activity is absent in MLII, reduced in MLIIIA, and altered in MLIIIC (Hasilik et al. 1981; Reitman et al. 1981; Varki et al. 1981, 1982; Waheed et al. 1982b).
GlcNAc-phosphotransferase is essential for the lysosomal trafficking of most lysosomal hydrolases. In higher eukaryotes, most lysosomal hydrolases are targeted to the lysosome via a mannose 6-phosphate (M6P)–dependent pathway. In this pathway, lysosomal enzymes are modified by the phosphorylation of mannose in a two-step reaction. In the first step, GlcNAc-phosphotransferase catalyzes the transfer of GlcNAc 1-phosphate from UDP-GlcNAc to certain terminal or penultimate mannoses on high-mannose–type glycans (Reitman and Kornfeld 1981a, 1981b; Waheed et al. 1982a). In the second step, occurring in the trans-Golgi network, the covering GlcNAc is removed by N-acetylglucosamine-1-phosphodiester α-GlcNAcase (IUBMB accession number EC 3.1.4.45) which has the trivial name “uncovering enzyme” (Varki and Kornfeld 1981; Waheed et al. 1981; Do et al. 2002). The lysosomal enzymes, now modified with M6P, bind to M6P receptors in the trans-Golgi network and are translocated to the endosome and subsequently to the lysosome. The recognition of lysosomal hydrolases by GlcNAc-phosphotransferase is the determining step in lysosomal hydrolase trafficking. Lysosomal hydrolases that are substrates for GlcNAc-phosphotransferase exhibit low Km values, whereas nonlysosomal glycoproteins bearing similar glycans have high Km values (Reitman and Kornfeld 1981a; Waheed et al. 1982a; Bao et al. 1996b). This difference in Km appears to explain the substrate-specific modification in lysosomal targeting.
GlcNAc-phosphotransferase has an α2β2γ2-subunit structure with a molecular mass of 540 kDa (Bao et al. 1996a). Recently, we cloned the cDNA and genomic DNA encoding the α/β–subunits precursor gene (GNPTAB) (Kudo et al. 2005). With this cloning and the previous cloning of the γ-subunit gene (GNPTG) (Raas-Rothschild et al. 2000), we concluded that the enzyme is a product of two genes. As a result, GlcNAc-phosphotransferase is an uncommon exception to the one enzyme–one gene theory of Garrod (1902).
Interestingly, before the subunits and gene structures were determined, substantial evidence for heterogeneity in the mucolipidoses had been described. Varki (1981) identified a variant form of MLIII on the basis of substrate recognition, which was later designated MLIIIC. A series of elegant genetic-complementation studies demonstrated heterogeneity and multiple complementation groups in MLIII (Honey et al. 1982; Shows et al. 1982; Mueller et al. 1983; Little et al. 1986). Three complementation groups were identified in MLIII and were designated A, B, and C, where A is classical pseudo-Hurler polydystrophy, C is the variant form described by Varki (1981), and B is represented by a single patient whose cell line has subsequently been lost. Additional studies demonstrated complementation between MLII and MLIIIC (Mueller et al. 1983).
Total or near-total deficiency of GlcNAc-phosphotransferase results in the nearly complete absence of lysosomal targeting in many cell types and tissues and the secretion of most lysosomal enzymes. This defect is clinically recognized as MLII (or I-cell disease), a disease often resulting in death in the 1st decade of life (Kornfeld and Sly 2001). MLIIIA (pseudo-Hurler polydystrophy) is a milder disease in which GlcNAc-phosphotransferase activity is reduced rather than absent (Kornfeld and Sly 2001). MLIIIC (variant pseudo-Hurler polydystrophy) is clinically indistinguishable from MLIIIA but is characterized by a normal level of GlcNAc 1-phosphate transfer to the small-molecule acceptor α-methylmannoside but a reduced level of transfer to a lysosomal hydrolase substrate (Varki et al. 1981). The link between the γ-subunit and the substrate recognition function was provided by our previous studies demonstrating that mutations in the γ-subunit are the molecular basis for MLIIIC but not MLIIIA (Raas-Rothschild et al. 2000, 2004). This result suggested that the α-subunit and/or β-subunit of the enzyme might contain the catalytic portion of GlcNAc-phosphotransferase. Our recent cloning and expression of the α/β–subunits precursor cDNA also supported the localization of the catalytic domain on these two subunits. Since the catalytic activity is absent or decreased in MLII and MLIIIA with all substrates, we speculated that these patients might have mutations in the α- and/or β-subunit. Our recent cloning of GNPTAB allowed us to analyze the mutations in patients with MLII or MLIIIA, to confirm this speculation. Tiede et al. (2005) also cloned the cDNA for GNPTAB and identified seven mutations from six patients with MLII. One of the patients in their study has a mutation identical to one found by us.
In this study, we sequenced all the exons of GNPTAB in 18 patients with MLII or MLIIIA and, when available, their relatives. Mutations in both alleles were identified in all patients, which demonstrates that both MLII and MLIIIA are caused by mutations in GNPTAB. In addition, GlcNAc-phosphotransferase activity was measured in patient fibroblasts, and selected mutant enzymes were expressed and characterized. The results of these studies allowed us to demonstrate that these diseases are allelic, to define the molecular basis of these diseases, and to propose molecular criteria to distinguish them. One of us (M.K.) recently reported a mutation in GNPTAB in a patient with MLIIIA (Steet et al. 2005) that is not included here. Part of the work of the present study was presented at the Joint Meeting of the Society for Glycobiology and the Japanese Society of Carbohydrate Research (Kudo et al. 2004).
Material and Methods
Cell Lines
Fibroblasts or lymphoblasts were derived from subjects with clinically diagnosed MLII and MLIII. Clinical information is shown in table 1. Cell lines from pedigrees GM03066, GM00080, GM00081, GM02013, GM02014, GM02274, GM02687, GM02660, GM01586, GM01589, GM01590, GM02046, GM02047, GM02273, GM02558, GM02559, GM01494, GM07255, GM07256, GM00113, GM01759, GM02065, GM02425, GM03112, GM03685, and GM03392 that have mucolipidosis, as well as normal human fibroblast line GM05565, were obtained from the National Institute of General Medical Sciences (NIGMS) Human Genetic Cells Repository at the Coriell Institute for Medical Research (Camden, NJ). Cell line F1515 was a kind gift from Dr. A. Raas-Rothschild (Hadassah University Hospital, Israel). Cell line F2954 was obtained from Dr. Miko Stewart (Children’s Memorial Hospital, Chicago). Cell lines GM1006 (LT) and C.M. were obtained from Dr. Thomas B. Shows (Roswell Park Cancer Institute, Buffalo, NY). Cell line R.S. was obtained from Dr. Catherine M. Bollard (Texas Children’s Hospital, Houston). Two of the MLII cell lines (GM02273 and GM03112) could not be recovered after freezing. It is well known that fibroblasts from patients with mucolipidosis are frequently difficult to recover after freezing. This work was performed with the approval of the Institutional Research Board of the University of Oklahoma Health Sciences Center. Although it is not current standard practice, patient initials have been used in some cases, to allow comparison with previously published results.
Table 1.
Summary of Clinical Background, GlcNAc-Phosphotransferase Activity, and Identified Mutations
| MucolipidosisClassificationand Individuala | Family | Relationto Proband | Ageb | Sex | Racec | GlcNAcPhosphotransferaseActivityd(%) | Allele 1e | Allele 2e |
| MLII: | ||||||||
| GM03066 | 34 | (Proband) | 23 fwk | Female | White | 1 | FS288X | FS546X |
| GM00080 | 34 | Mother | 16 years | Female | White | 66 | WT | FS546X |
| GM00081 | 34 | Father | 23 years | Male | White | 96 | FS288X | WT |
| GM02013 | 36 | (Proband) | 2 wk | Male | White | <.1 | FS1081X | FS1172X |
| GM02014 | 36 | Mother | 18 years | Female | White | 58 | FS1081X | WT |
| GM02273 | 38 | (Proband) | 2 years | Male | White | … | … | … |
| GM02274 | 38 | Mother | … | Female | White | 66 | FS1172X | WT |
| 469 Proband | 469 | (Proband) | … | … | … | … | … | … |
| GM02687 | 469 | Father | 26 years | Male | White | 82 | FS588X | WT |
| GM02660 | 469 | Mother | 23 years | Female | … | 80 | WT | FS1172X |
| GM01586 | 1908 | (Proband) | 5 wk | Male | White | <.1 | FS1172X | FS1172X |
| GM01589 | 1908 | Father | … | Male | White | 94 | FS1172X | WT |
| GM01590 | 1908 | Mother | … | Female | White | 78 | FS1172X | WT |
| GM03112 | 1909 | (Proband) | 21 fwk | Female | White | … | … | … |
| GM02046 | 1909 | Mother | 25 years | Female | White | 41 | FS737X | WT |
| GM02047 | 1909 | Father | 26 years | Male | White | 52 | WT | FS1172X |
| F2954 | … | … | … | … | … | .3 | *FS1085X (type 1) | FS1172X |
| C.M. | … | … | … | … | … | <.1 | FS211X (type 1) | FS1172X |
| R.S. | … | … | 2 years | Male | … | .0 | FS211X (type 1) | FS1172X |
| MLIIIA: | ||||||||
| GM02558 | 501 | Sister | 9 years | Female | Black | 3 | Q278X | *FS1085X (type 2) |
| GM02559 | 501 | (Proband) | 7 years | Female | Black | 3 | Q278X | *FS1085X (type 2) |
| 501 Brother | 501 | Brother | … | Male | … | … | … | … |
| GM01494 | 1012 | (Proband) | 9 years | Male | White | 14 | K4Q | FS1172X |
| GM07255 | 1012 | Father | 53 years | Male | White | 78 | K4Q | WT |
| GM07256 | 1012 | Mother | 51 years | Female | White | 70 | WT | FS1172X |
| GM00113 | … | (Proband) | 2 years | Female | White | 12 | K4Q | K4Q |
| GM01759 | … | (Proband) | 13 years | Male | White | .8 | *FS211X (type 2) | FS1172X |
| GM02065 | … | (Proband) | 9 years | Male | White | 2 | FS745X | *FS1085X (type 2) |
| GM02425 | … | (Proband) | 15 years | Male | White | 1 | *FS211X (type 2) | FS1172X |
| GM03685 | … | (Proband) | 15 years | Male | White | 1 | *FS1085X (type 2) | FS1172X |
| Reclassified as MLIIIAf: | ||||||||
| GM1006 (LT) | … | … | … | … | … | 5.1 | FS1172X | *FS1202X |
| F1515 | … | … | … | … | … | 4.4 | R1205X | R1205X |
| MLIIIC: | ||||||||
| GM03392 | 610 | Sister | 13 years | Female | White | 122 | WT | WT |
Clinically affected individuals are listed in bold italics.
Age at time of tissue collection. In the case of fetal samples, an estimated gestational age is provided in fetal weeks (fwk) from conception.
Two of the cell lines were classified as Caucasian for race and Arab for ethnicity by the NIGMS Human Genetic Cells Repository, from which we obtained the cell lines.
GlcNAc-phosphotransferase activity was measured as specific activity (pmol/mg/h) in cell lysate and is shown as a percentage of the activity in normal fibroblasts (GM05565).
Mutations are named on the basis of the resulting protein products, which are described in table 5. An asterisk (*) before the mutation indicates that the frameshift is caused by aberrant splicing. WT = wild type.
Reclassified from MLII to MLIIIA.
Culture of Cell Lines
Fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 20% defined fetal bovine serum (FBS) supplemented with 2 mM l-glutamine in 5% CO2/95% air at 37°C until confluent and were harvested using trypsin. Lymphoblasts transformed with Epstein-Barr virus (GM02687 and GM02660) were cultured in DMEM with 15% heat-inactivated FBS supplemented with 2 mM l-glutamine in 5% CO2/95% air at 37°C until the density of the cells was 1–1.4×106 cells/mL and were harvested by centrifugation. Cell pellets were stored at −80°C until used for GlcNAc-phosphotransferase activity assay, genomic DNA preparation, or mRNA preparation.
GlcNAc-Phosphotransferase Enzymatic Activity in Patient Cells
Approximately 3×106 cells were incubated in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, and 2 mM MgCl2) on ice for 45 min, were sonicated for 20 min in an ice-bath sonicator (Ultrasonics Quantrex 90H [L&R Manufacturing]), and then were clarified by centrifugation at 40,000 g for 20 min. GlcNAc-phosphotransferase activity was determined by measuring the transfer of GlcNAc-phosphate from [β-32P]UDP-GlcNAc to α-methylmannoside, as described elsewhere (Reitman et al. 1984). [β-32P]UDP-GlcNAc was synthesized using an improved procedure modified from that of Reitman et al. (1984) In brief, [32P]glucosamine 6-phosphate was synthesized by incubating 200 nmol glucosamine, 100 nmol ATP, and 5.0 mCi [γ-32P]ATP (7,500 Ci/mmol from MP Biomedicals) with 2 U of yeast hexokinase (Sigma) in 200 mM Tris-HCl (pH 7.4) and 20 mM MgCl2 for 20 min at 37°C. After inactivation of hexokinase by boiling for 5 min, 100 nmol glucose 1,6-diphosphate, 200 nmol acetyl coenzyme A, and 200 nmol uridine triphosphate were added to the reaction, together with 7.5 μg recombinant Escherichia coli glmM (phosphoglucosamine mutase), 50 μg recombinant E. coli glmU (pyrophosphorylase and acetyltransferase), and 5 U of inorganic pyrophosphatase from baker’s yeast (Sigma), and were incubated for 30 min at 37°C. The [β-32P]UDP-GlcNAc thus synthesized was purified by chromatography on a Microsorb-MV amino high-performance liquid chromatography column (250×4.6 mm [Varian]) with a linear gradient of 5 mM potassium phosphate buffer (pH 3.0) to 1 M potassium phosphate buffer (pH 4.0). The [β-32P]UDP-GlcNAc peak was identified by GlcNAc-phosphotransferase assay and cochromatography with authentic UDP-GlcNAc. Purity of the [β-32P]UDP-GlcNAc thus synthesized and purified is >95%, and a very low nonspecific background is exhibited in the GlcNAc-phosphotransferase assay. This low background is helpful in assaying MLII fibroblasts. One unit of GlcNAc-phosphotransferase activity is defined as 1 pmol of GlcNAc-phosphate transferred to α-methylmannoside per h in a reaction containing 150 μM UDP-GlcNAc and 100 mM α-methylmannoside. Protein concentration was determined by Bradford assay (Pierce).
PCR Amplification of Exons in GNPTAB
PCR primers to amplify the exons in GNPTAB and sequencing primers were designed on the basis of the genomic sequence of GNPTAB in BAC 14951 (GenBank accession number AC005409) and are listed in table 2. Genomic DNA was prepared from cells by use of DNeasy Tissue Kits (Qiagen). The individual exons or groups of exons, along with the flanking intronic sequences, were amplified by PCR. Primers and PCR conditions are provided in table 3. PCR was performed in a GeneAmp PCR system 2400 thermocycler (Perkin Elmer) by use of the Expand High Fidelity PCR system (Roche), except for exon 1, for which the GC-rich PCR system (Roche) was used. PCR products were separated from the primers with a QIAquick PCR Purification Kit (Qiagen) and were sequenced using the primers listed in table 3.
Table 2.
Oligonucleotide Sequences of Primers Used for PCR Amplification and Sequencing of GNPTAB
| Primer ID | Oligonucleotide Sequence(5′→3′) |
| 1076 | cctggtgaactcattcttcaagacc |
| 1077 | gctaaagtgaacacatcagatgggc |
| 1078 | cataatctctgggtttaaaccctgtg |
| 1082 | aaccctccccagtgcagtgaagc |
| 1083 | aagctctatctttggagttgg |
| 1084 | gctgttttgcttctctttgtgc |
| 1085 | tgctgctttaatagctctcagaatg |
| 1086 | atggaccacaagaaaagaatcacac |
| 1087 | cgtccgtcgccggagctgcaatg |
| 1088 | ggcaaaaccccgtctctaataatg |
| 1089 | gtgatgatgcagtcctgtagtc |
| 1092 | aggactccatctttaaaagtctgc |
| 1103 | gtggtaggcagtaagtgaatac |
| 1105 | ctctaagcaaaccgtcttgg |
| 1106 | cacaaaatcctgtgtaccc |
| 1107 | ctcacctctctgtaagtccc |
| 1109 | gatttgctaagtgacttccacgc |
| 1118 | ctgcctccttcctaatctgtcc |
| 1119 | caatccctttttgaggttaaggcc |
| 1120 | cgctcagtaagaacggtcaacg |
| 1121 | gtttggttaagtctatagcacatgttc |
| 1122 | cacccagtccagaactgtctttc |
| 1123 | gctaaggtaaatctgcttggtcc |
| 1125 | caagtggaaaaccatccacc |
| 1126 | tccaagtcagccttgctgag |
| 1128 | cttcctgggctctccttgttg |
| 1129 | tgctcctaatgaagagttcgtgac |
| 1132 | gtttgcttgtgttcagaactaagg |
| 1134 | tgtgagccactgcgcctgaccag |
| 1135 | ccacagtcattacttacaatgcgc |
| 1136 | cctgctgctagttctgaagtgc |
| 1137 | attgccctagtttcagctctgatg |
| 1138 | gctccaacactgcctgcactg |
| 1139 | cccatagctaaaaggccatctacc |
| 1140 | gtatacactcacccacacacatgc |
| 1141 | gaagtcctctctcctgcctgg |
| 1142 | ctgaagacactgtgctcactcaa |
| 1162 | gggattacaggtgtgagccacc |
| 1163 | tgcctacttcagcagcacatatac |
| 1164 | cctgagcatgagaaagaatgagg |
| 1215 | cttacctccagagagcatagaatc |
| 1216 | tcctatctctcactcaaccac |
| 1219 | ggctcttagaaagtttgatgcac |
| 1233 | ggcggtgaaggggtgatgctgttc |
| 1257 | gcatcacatcacaaagacatctc |
| 1259 | ccacaaaacagcatggtactgg |
| 1349 | acatactgtatcggggcatc |
Table 3.
PCR and Sequencing Primers and PCR Conditions for Amplification of GNPTAB
|
PCR Primera |
||||||
| Exon(s) | Forward | Reverse | AnnealingTemperature(°C) | ElongationTime(s) | PCR ProductSize(bp) | Sequencing Primersa |
| 1 | 1087 | 1088 | 60 | 45 | 386 | 1088, 1233, 1349 |
| 2 | 1076 | 1077 | 60 | 45 | 536 | 1103, 1107 |
| 3 and 4 | 1078 | 1082 | 60 | 90 | 1,832 | 1078, 1105, 1162, 1106 |
| 5 | 1118 | 1119 | 60 | 45 | 737 | 1083, 1084 |
| 6 and 7 | 1085 | 1086 | 58.5 | 45 | 646 | 1085, 1086 |
| 8, 9, and 10 | 1089 | 1109 | 58 | 60 | 1,433 | 1092, 1107 |
| 11 | 1120 | 1121 | 60 | 45 | 424 | 1120, 1121 |
| 12 | 1122 | 1123 | 60 | 45 | 522 | 1123, 1122 |
| 13 | 1163 | 1164 | 60 | 60 | 1,345 | 1163, 1125, 1126, 1128, 1164 |
| 14 and 15 | 1129 | 1134 | 60.5 | 45 | 829 | 1129, 1132 |
| 16 | 1135 | 1136 | 60.5 | 45 | 267 | 1136, 1135 |
| 17 and 18 | 1215 | 1216 | 60 | 45 | 708 | 1215, 1216 |
| 19 | 1139 | 1140 | 60 | 45 | 436 | 1139, 1140 |
| 20 | 1141 | 1142 | 60 | 45 | 717 | 1141, 1142 |
| 21 | 1219 | 1259 | 58.5 | 45 | 297 | 1219, 1257 |
Primer sequences are listed in table 2.
RT-PCR Amplification of mRNA
RT-PCR was used to amplify mRNA from selected cell lines. Total RNA was prepared from fibroblasts by using an RNeasy mini kit and QIA shredder (Qiagen). First-strand cDNA was synthesized from ∼3 μg of total RNA with oligo(dT) or random hexamer primers by use of a First-Strand cDNA synthesis kit (Amersham Pharmacia). cDNA corresponding to exons 3–7 was amplified with primers 5′-cacctgggtgaatggcacagatc-3′ and 5′-gcattagcactaatccagggac-3′. cDNA corresponding to exons 16–19 was amplified with 5′-gcttcctgctgatatcacgcgagc-3′ and 5′-gtggcaatgttgtcattcaggc-3′. cDNA corresponding to exons 16–21 was amplified with 5′-gcttcctgctgatatcacgcgagc-3′ and 5′-gcatcacatcacaaagacatctc-3′. PCR parameters were as follows: 30 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and elongation at 72°C for 45 s. The PCR products from GM01759, GM02425, and GM1006 (LT) were separated from primers by use of QIAquick PCR Purification Kit and were sequenced with the same primers used for PCR. The PCR products from F2954, GM02558, and GM03685 were gel purified and sequenced with primers 1288 and 1289 (table 2).
Expression Plasmids for Mutants
Expression plasmids encoding mutant α/β–subunits precursors—K4Q, D190V, L1168fsX1172 (FS1172X), or W1201fsX1202 (FS1202X)—were constructed by replacing the restriction-enzyme–flanked region in the wild-type sequence in pcDNA6/V5/His-A. Mutant sequences were generated by PCR with the primers described below and with the wild-type expression plasmid as the template. The K4Q expression plasmid was constructed by replacing the 5′-terminal NheI-XhoI fragment with an NheI/XhoI-cleaved PCR fragment amplified with 5′-cgtggcgggctagccaccatgctgttccagctcctgcagagacaaacc-3′ and 5′-ctaaaggtaggcaagtggctc-3′ (Q4 is introduced by underlined nucleotides). The D190V plasmid was constructed by replacing the 5′-terminal NheI-XhoI fragment with an NheI/XhoI-cleaved PCR product amplified by sequential PCR. The first two reactions were performed with primers T7 (5′-taatacgactcactataggg-3′) and 5′-aaccttggtactgtcaaaaacaacaact-3′ and with primers 5′-ccaaggttgttgaagatgcccactctg-3′ and 5′-ctaaaggtaggcaagtggctc-3′ (V190 is introduced by underlined nucleotides). The second PCR was performed with primers T7 and 5′-ctaaaggtaggcaagtggctc-3′ with the use of a mixture of the products from the first reactions as template. Expression plasmids for FS1172X and FS1202X were constructed by replacing the 3′-terminal EcoRV-XbaI fragment with EcoRV/XbaI-cleaved PCR fragments amplified with primers 5′-gcttcctgctgatatcacgcagc-3′ and 5′-catggatttctagaggagcccttgaacagccttcactgtctgagcatc-3′ (for FS1172X) or primers 5′-tttgtctctctagactcaacattcctgcagctcatgcatatga-3′ (for FS1202X) (stop codons are underlined).
Expression of the Mutant α/β–Subunits Precursor cDNA in 293T Cells
The human embryonic kidney cell line 293T was grown in DMEM with 10% (v/v) FBS at 37°C and 5% CO2/95% air. Cells were transfected with empty vector or with vector containing cDNAs encoding wild-type or mutant α/β–subunits precursor, by use of FuGene 6 (Roche). Cells were harvested at 72 h after transfection, and GlcNAc-phosphotransferase activity was measured as described for mucolipidosis cell lines.
Results
Source and Characterization of Cell Lines
Fibroblast or lymphoblast cell lines from 15 patients with a diagnosis of MLII or MLIIIA (and, when possible, their relatives) were obtained from repositories or clinicians as described in the “Material and Methods” section. Cells from NIGMS have a family number assigned when several cell lines are available from a family. There are no data available about any possible genetic relationships between the pedigrees. In three pedigrees with MLII, the cells from the proband were not available; however, cells from both parents were available in two of the three pedigrees. GlcNAc-phosphotransferase activity in cell lysates was measured by an assay for the transfer of [32P]GlcNAc-phosphate to α-methylmannoside as described in the “Material and Methods” section. Activity was expressed as a percentage of the specific activity in the normal fibroblast line (table 1). In this assay, patients with MLII lacked detectable activity, whereas patients with MLIIIA had reduced but detectable activity (Varki et al. 1982; Mueller et al. 1985). In table 1, patients and available relatives are listed along with available clinical information, measured GlcNAc-phosphotransferase activity, and mutations identified by this study. Specific activity of the normal fibroblast lysate was 80–110 pmol/mg/h, comparable to that reported elsewhere (Varki et al. 1982).
Strategy for the Identification of Mutations in GNPTAB
To identify as many mutations as possible, including those that altered splicing, we chose to amplify and sequence all 21 exons and their flanking intronic sequences, as described in the “Material and Methods” section. The PCR products were sequenced and compared with the corresponding GNPTAB sequence in BAC 14951. Mutations were identified in the uncloned PCR product as a second sequence along with the wild-type sequence. For some mutations, additional experiments, such as RT-PCR and expression of mutant cDNAs, were performed to confirm the effect of mutations. The genomic sequence for GNPTAB is numbered starting from the first base in the initiation codon in BAC 14951. The cDNA is numbered according to the cDNA sequence reported elsewhere (GenBank accession number AY687932), and amino acids are numbered starting from the initiator methionine. Mutations are described in accordance with the nomenclature suggested by den Dunnen and Antonarakis (2000). Polymorphisms identified in GNPTAB during mutation analysis are listed in table 4.
Table 4.
Polymorphisms in GNPTAB
| Genomic DNAa | cDNAb | Location |
| del−42_−40CGG | del108_110CGG | Exon 1, 5′ UTR |
| 27A→G | 191A→G | Exon 1 (silent) |
| 33801A→G | 282−117A→G | Intron 1 |
| del42226_42228GTG | IVS4+94_96GTG | Intron 4 |
| 63255A→G | IVS10−166G→A | Intron 10 |
| 65373G→C | IVS12−3G→C | Intron 12 |
| 65696A→G | 2096A→G | Exon 13 (silent) |
| 69559T→C | IVS+5T→C | Intron 15 |
| 73124G→A | IVS17+14G→A | Intron 17 |
| 73347T→C | IVS17−25T→C | Intron 17 |
Mutations Identified in Patients with MLII and Their Parents
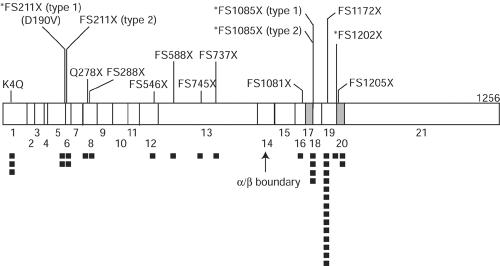
Eight unique mutations were found in the nine pedigrees with clinically diagnosed MLII (table 1). These mutations are listed in table 5, and their locations in the cDNA are shown diagrammatically in fig. 1. Two mutant alleles were found in each patient or family, except for family 38, for which only the maternal cell line was available. Only one patient with MLII was homozygous for a mutation. When parental samples were available, each parent carried a mutant allele in common with the patient. One mutation (FS1172X) was predominant and was found in all the pedigrees with MLII except family 34.
Table 5.
Mutations Identified in GNPTAB
| MucolipidosisClassificationand Mutationa | cDNA Sequenceb | Genomic Sequencec | Location | Consequence | Patients and Families with Mutation |
| MLII: | |||||
| FS211X (type 1) | 773_776delCAGA | g.50096_50100delCAGA | Exon 6 | Frameshift T206fsX211 | C.M., R.S. |
| FS288X | 1012delA | g.59600delA | Exon 8 | Frameshift T284fsX288 | Family 34 |
| FS546X | 1744delC | g.64558delC | Exon 12 | Frameshift C528fsX546 | Family 34 |
| FS588X | 1902_1905tripTATAd | g.65502_65505tripTATA | Exon 13 | Frameshift S581fsX588 | Family 469 |
| FS737X | 2352T→AAA | g.65952delTinsAAA | Exon 13 | Frameshift L730fsX737 | Family 1909 |
| FS1081X | 3395_3398dupCTAC | g.70633_70636dupCTAC | Exon 16 | Frameshift Y1079fsX1081 | Family 36 |
| FS1172X | 3665_3666delTC | g.77209_77210delTC | Exon 19 | Frameshift L1168fsX1172 | Families 36, 38, 469, 1908, 1909; C.M., R.S., F2954 |
| *FS1085X (type 1) | IVS17+1G→A | g.73111G→A | Intron 17 | Skipped exon 17; P1084fsX1085 | F2954 |
| MLIIIA: | |||||
| K4Q | 174A→C | g.10A→C | Exon 1 | Missense K4Q | Family 1202, GM00113 |
| *FS211X (type 2) | 733A→T | g.44667A→T | Exon 5 | 4-nt del from 3′ end of exon 5; D190fsX211/missense D190V | GM01759, GM02425 |
| Q278X | 996C→T | g.59584C→T | Exon 8 | Stop codon Q278X | Family 501 |
| FS745X | 2215_2219delACTCA | g.65815_65819delACTCA | Exon 13 | Frameshift S685fsX745 | GM02065 |
| FS1172X | 3665_3666delTC | g.77209_77210delTC | Exon 19 | Frameshift L1168fsX1172 | Family 1012, GM01759, GM02425, GM03685, GM1006 (LT) |
| R1205X | 3777C→T | g.81501C→T | Exon 20 | Stop codon R1205X | F1515 |
| *FS1085X (type 2) | IVS17+6T→G | g.73116T→G | Intron 17 | Skipped exon 17; P1084fsX1085 | Family 501, GM02065, GM03685 |
| FS1202X | IVS19−1G→A | g.81490G→A | Intron 19 | Skipped exon 20; W1201fsX1202 | GM1006 (LT) |
Figure 1.
Location of mutations on the GlcNAc-phosphotransferase α/β–subunits precursor cDNA. The vertical lines indicate positions of mutations. Numbers below the boxes are exon numbers. Exons that are deleted as a result of splice-site mutations are shaded. An asterisk (*) before the name of a mutation indicates that the frameshift is caused by aberrant splicing. Blackened squares under the cDNA show the frequency of the mutant alleles in this study.
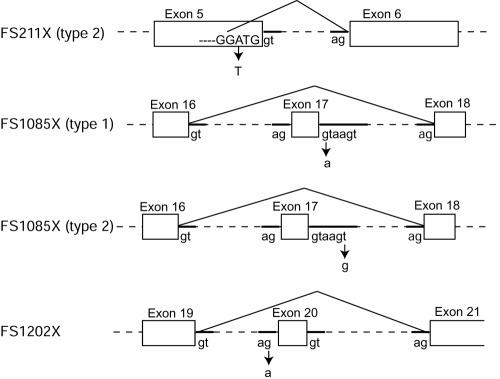
All the mutations, except for the one in F2954, were insertions or deletions of nucleotide(s) causing frameshift and truncation of the protein product. These frameshift mutations were named after the location of the introduced stop codons (table 5). One allele in F2954 carried a substitution at the exon 17–intron 17 junction (IVS17+1G→A) in the invariant (GT…AG) splice-site sequence (Breathnach and Chambon 1981), whereas the other allele carried FS1172X. To determine the outcome of this substitution, cDNA corresponding to exons 16–21 was amplified by RT-PCR by use of RNA from F2954 fibroblasts as template and was sequenced. Two distinct products were found at equal abundance, and each product contained a single mutation. The FS1172X allele contained a 2-nt deletion in the cDNA, as expected. The IVS17+1G→A allele led to aberrant splicing with deletion of exon 17 (fig. 2), resulting in truncation at amino acid 1084—namely, *FS1085X (type 1) (an asterisk [*] indicates that the frameshift is caused by aberrant splicing).
Figure 2.
Structure of mutations causing aberrant splicing. Mutations and the resulting mRNA splicing that cause F211X (type 2), FS1085X (type 1), FS1085X (type 2), and FS1202X are shown. Nucleotide substitutions are indicated (arrows). RNA was analyzed by RT-PCR with oligo(dT) or random hexamer primers, and the amplified products were sequenced.
Five of the mutations, FS211X (type 1), FS288X, FS546X, FS588X, and FS737X, truncate the protein in the α-subunit, yielding truncated α-subunit and no β-subunit. The rest of the mutations, FS1081X, FS1172X, and *FS1085X (type 2), truncate the protein in the β-subunit, yielding intact α-subunit and truncated β-subunit (fig. 1). To examine the effect of truncation on the GlcNAc-phosphotransferase activity, the expression plasmid for the longest product (FS1172X; 1171-aa product) was constructed. Transient transfection of 293T cells with the FS1172X plasmid did not increase GlcNAc-phosphotransferase activity, compared with that of mock-transfected cells. The absence of additional activity in the cells transfected with FS1172X plasmid was consistent with the absence of activity in the fibroblast GM01586, which was homozygous for the FS1172X mutation. Since the other mutations generate proteins shorter than FS1172X, the protein from these mutations is likely to be inactive as well. The lack of GlcNAc-phosphotransferase activity from mutations found in patients with MLII is consistent with the very low activity (⩽1% of normal fibroblasts) detected in fibroblasts of patients with MLII.
Mutations Identified in Patients with MLIIIA and Their Parents
Six unique mutations were found in the seven pedigrees with clinically diagnosed MLIIIA (table 1). The mutations are listed in table 5, and their locations in the cDNA are shown diagrammatically in fig. 1. Two mutant alleles were found in each patient or family. Only one patient with MLIIIA was homozygous for the mutation. When parental samples were available, each parent carried a mutant allele in common with the patient.
Four of the mutations were single-nucleotide substitutions, and the other two were nucleotide deletions. Each single-nucleotide substitution affects the protein product in different ways, whereas the nucleotide deletions cause a frameshift and yield inactive enzyme in the same way as mutations found in MLII.
The first substitution, c.174A→C, causes the amino acid substitution K4Q. This mutation was homozygous in GM00113 and heterozygous in GM01494. The effect of the K4Q substitution was examined by transient transfection of 293T cells with K4Q plasmid. Transfection resulted in ∼20%–40% activity when compared with cells transfected with the wild-type sequence. We conclude that the K4Q substitution reduced but did not eliminate GlcNAc-phosphotransferase activity. GlcNAc-phosphotransferase activity in GM00113 fibroblast lysate, homozygous for K4Q, was 12% (table 1), which is consistent with K4Q being a mild mutation. It is interesting to note that the diagnosis had been made by the time the patient was 2 years old, although the GM00113 fibroblasts have very high activity (12%). GlcNAc-phosphotransferase activity in GM01494 fibroblast lysate, heterozygous for K4Q, was 14% of normal fibroblasts (see data for MLIIIA in table 1). Since the other mutant allele was the null FS1172X, the product of the K4Q mutation could be directly observed and was reasonably consistent with K4Q being a mild mutation. The reported age at which fibroblasts were obtained, 9 years, is also consistent with the relatively high enzyme activity.
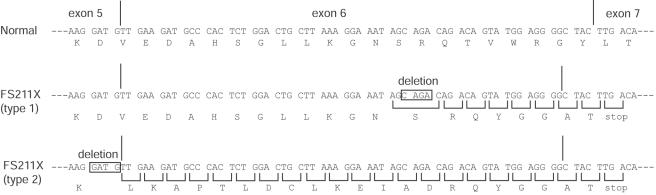
The second substitution, c.733A→T, is located 3 nt upstream from the 3′-end of exon 5 (table 5). This mutation was heterozygous in GM01759 and GM02425. The effect of this substitution is complex. This nucleotide substitution should change amino acid Asp190 (GAT) to Val (GTT). However, when a cDNA encoding the D190V substitution was expressed in 293T cells, it produced 86% of GlcNAc-phosphotransferase activity. Therefore, the substitution of valine for aspartic acid at position 190 does not significantly affect the enzyme activity. To investigate potential other effects of the c.733A→T substitution, the RNA was analyzed. RT-PCR amplification of exons 3–7 from GM01759 and GM02425 RNA revealed two RNA sequences in both cell lines. The mRNA from the c.733A→T allele contained a 4-nt deletion resulting in FS211X (type 2) at the 3′-end of the exon 5 sequence (figs. 2 and 3). Sequencing the cDNA product revealed that a new splice site was generated between G44665 and G44666 instead of the original site between G44669 and G44670 (fig. 2). Even though the original splice site is unaltered, the new site appears to be preferred. Since the other allele in GM01759 and GM02425 is FS1172X, GlcNAc-phosphotransferase activity in these patients is from the c.733A→T allele. GlcNAc-phosphotransferase activity of GM01759 and GM02425 fibroblasts was 0.8% and 1%, respectively, which is consistent with the lack of full-length enzyme. Fibroblasts from probands GM01759 and GM02425 were collected when the patients were aged 13 and 15 years, respectively, which strongly suggests that the diagnosis of MLIIIA is correct. The long clinical course and very low fibroblast enzyme activity may suggest that splicing is especially disrupted in these fibroblasts.
Figure 3.
Comparison of frameshift mutations that cause termination at amino acid 211. Deletions in the cDNA sequence change the reading frame in FS211X (type 1) and FS211X (type 2). Nucleotides deleted from the cDNA are boxed. The wild-type reading frame is indicated by spacing; the abnormal reading frame, by brackets.
The third substitution, c.996C→T, is a nonsense mutation truncating the enzyme at amino acid 277—namely, Q278X. This mutation probably results in enzyme with no activity. This mutation was heterozygous in the proband GM02558 and affected sister GM02559.
The fourth substitution, IVS17+6T→G, is located at the junction of exon 17 and intron 17. This mutation was found in GM02558, GM02559, GM02065, and GM03685. Although the substitution is outside the invariant sequence, it alters splicing. RT-PCR amplification of cDNA containing exons 16–21 from GM02558 mRNA revealed two sequences; one was wild type derived from the Q278X allele, and the other contained a deletion of exon 17 (fig. 2). Skipping of exon 17 results in a frameshift at amino acid 1084 and termination at amino acid 1085 (FS1085X, type 2). The same result was previously observed in a patient with MLII (F2954), caused by IVS17+1G→A (FS1085X, type 1). Although mRNA sequences derived from these two mutations are the same (fig. 2), the GlcNAc-phosphotransferase activity and clinical outcomes are different. Fibroblasts with FS1085 (type 2) exhibited 1%–3% GlcNAc-phosphotransferase activity, whereas F2954 fibroblasts with FS1085X (type 1) exhibited <0.1% activity (table 1). Since, in these fibroblasts, the other alleles are null alleles, the observed difference is the difference between FS1085X types 1 and 2. This difference may be because of the location of the substitutions. It seems probable that the substitution outside the invariant splice site may occasionally allow the correct splicing, although we could not demonstrate the presence of a product with the wild-type sequence in fibroblasts.
The final two mutations are deletions of nucleotides. One is FS1172X, which was also found in patients with MLII. The other is FS745X, which truncates the protein in the α-subunit. These two frameshift mutations and the nonsense mutation Q278X are thought to not yield active enzyme. These three null mutations were always found heterozygous with other mutations that yield active enzyme in patients with MLIIIA. At least one of the alleles in each patient with MLIIIA is K4Q, *FS211X (type 2), or *FS1085X (type 2), which allow generation of some active enzyme.
Mutations Identified in Patients with Reclassified MLIIIA
Patients GM1006 and F1515 were given the clinical diagnosis of MLII. On the basis of the GlcNAc-phosphotransferase activity and mutations identified in these patients (table 1), we propose that they should be classified as having MLIIIA. Mutations found in these patients are listed in table 5 with those of the other patients with MLIIIA, and the locations in the cDNA are shown diagrammatically in fig. 1.
In patient GM1006, one allele carried the common FS1172X mutation, and the other carried a substitution at the intron 19–exon 20 junction, IVS19−1G→A. The substitution was in the invariant (GT…AG) splice-site sequence. A region containing exons 16–21 of mRNA from GM1006 was amplified by RT-PCR, which demonstrated that the intron 19–exon 20 splice-site mutation led to aberrant splicing, resulting in a deletion of exon 20 (fig. 2). This causes a frameshift and truncates the protein at amino acid 1201 (FS1202X). Because of the proximity of the two mutations found in this patient (exon 19 and intron 19), it was possible to demonstrate that these two mutations were always found on different transcripts. Nearly 100% of the transcripts from patient GM1006 contained the exon 20 deletion, suggesting that the mRNA containing the 2-nt deletion in exon 19 is poorly expressed or unstable. This is in contrast to the result for patient F2954, who is heterozygous for FS1172X mutation, in which both transcripts were expressed at a similar level as the transcript in normal human fibroblasts. Fibroblasts from proband GM1006 expressed 5.1% GlcNAc-phosphotransferase activity, which is well above the maximum typically observed in patients with MLII (<1%), suggesting that the classification should be MLIIIA. Since one allele carries the FS1172X null mutation, this unexpectedly high activity should be from FS1202X deleting the final 55 aa. Transient transfection of 293T cells with FS1172X or FS1202X plasmids resulted in 0% and 2.8% increase in activity, respectively, compared with cells transfected with the wild-type sequence. Therefore, the observed activity in patient GM1006 resulted from the FS1202X allele. This enzyme activity in fibroblasts could be the result of normal transcript from leaky splicing at the original or alternative splice sites, although we could not demonstrate the presence of the wild-type sequence by RT-PCR.
Patient F1515 appeared to be homozygous for R1205X caused by a substitution in exon 20; however, since a family study was not possible, a genomic deletion could not be excluded. F1515 fibroblasts expressed 4.4% GlcNAc-phosphotransferase activity (table 1), suggesting that the correct diagnosis is MLIIIA. Since we demonstrated that truncation at amino acid 1201 retains 2.8% of wild-type GlcNAc-phosphotransferase activity when expressed in 293T cells, we believe R1205X is likely to retain a small amount of enzyme activity. This mutation was also reported by Tiede et al. (2005), who found the mutation in a patient with MLII.
Patient with MLIIIC
To confirm that MLIIIC is not caused by GNPTAB mutations, DNA from GM03392 (MLIIIC) fibroblasts was sequenced. No mutations in GNPTAB were found. This result supports the hypothesis that the MLIIIC phenotype is exclusively caused by mutations in the γ-subunit gene. Fibroblast GlcNAc-phosphotransferase activity was 122% of normal, consistent with the MLIIIC diagnosis (table 1).
Discussion
In a previous study, we demonstrated that mutations in the γ-subunit of GlcNAc-phosphotransferase were the cause of MLIIIC (Raas-Rothschild et al. 2000). This was subsequently expanded by Raas-Rothschild et al. (2004). To understand the relationship between MLII and MLIIIA, this study involved sequencing of GNPTAB from pedigrees with MLII or MLIIIA, as well as measuring GlcNAc-phosphotransferase activity in their fibroblasts by use of a specific and sensitive assay. These studies allowed us to elucidate the complex molecular genetics of these disorders.
Nine pedigrees with MLII and nine with MLIIIA were analyzed in this study. Two mutant alleles were identified in 17 of the 18 pedigrees. In the remaining family (family 38), only a maternal cell line was available, and, consequently, only the maternal mutant allele could be identified. The identification of two mutant GNPTAB alleles in each of the 17 adequate pedigrees strongly suggests that the MLII or MLIIIA phenotypes are only caused by GNPTAB mutations. Since both pedigrees with MLII and pedigrees with MLIIIA always had GNPTAB mutations, this confirmed that these two diseases are allelic, which is consistent with the fact that GlcNAc-phosphotransferase activity is affected in both diseases and that MLII and MLIIIA are not complemented by each other, as discussed below. This study provides the criteria to distinguish between MLII and MLIIIA on the basis of the location and type of the mutations.
In the 18 pedigrees with MLII or MLIIIA, 15 different mutations were identified, including 2 nonsense mutations, 8 frameshift mutations (insertions and/or deletions), 1 missense mutation, and 4 mutations that affect splicing. Mutations causing aberrant splicing were confirmed by RT-PCR. The missense mutation and some of the frameshift mutations near the 3′-terminus of the cDNA were tested for GlcNAc-phosphotransferase activity after transient expression in 293T cells, to confirm the effect on enzyme activity. Five of the 15 different mutations were found in multiple patients or pedigrees. The remaining 10 mutations were found in only a single patient or family. The most common mutation was the FS1172X null mutation found in 13 of 18 families, including 9 pedigrees with MLII and 4 pedigrees with MLIIIA. This is the only mutation found in both MLII and MLIIIA. FS1172X was always caused by the same 2-nt deletion in exon 19. Since the mutation was a deletion of CT from a repeating CTCT sequence, this may be the result of a mismatch-repair error. This frameshift causes truncation of the enzyme at amino acid 1171 in the β-subunit and appears to be a null allele. When a cDNA with this mutation was expressed, no activity could be demonstrated. Additionally, fibroblasts from patient GM01586, who is homozygous for this mutation, exhibited the very low value of <0.1% GlcNAc-phosphotransferase activity. Since FS1172X is the most common mutant allele identified in both MLII and MLIIA in our study (13 of 32 cell lines), it is surprising that only a single homozygote (GM01586) was identified. This might be the result of adverse selection or our small sample size. In the remaining 11 pedigrees with this allele, there was never evidence of enzyme activity from this allele. The second frequent mutation was FS1085X (type 2), shared by three pedigrees with MLIIIA. The remaining three shared mutations, FS211X (type 1), FS211X (type 2), and K4Q, were found in two pedigrees each.
Given the multiple mutations and the presence of the same mutation in pedigrees with MLII and MLIIA, how can these two diseases be distinguished molecularly? In the 11 pedigrees listed by the repositories or clinicians as having MLII, the following mutations were found. Nine pedigrees with MLII shared the common FS1172X allele, which was homozygous in one case (family 1908) and present in a single maternal sample in another case (family 38). In six of the remaining seven pedigrees, the FS1172X was paired with a second mutation: FS211X (type 1) (two cases), FS588X, FS737X, FS1081X, or FS1085X (type 1). These are all due to insertion or deletion of nucleotide(s), except for the FS1085X (type 1) that is a substitution in the invariant splice-site (GT…AG) sequence. None of these mutations were predicted to generate active GlcNAc-phosphotransferase. In the only MLII pedigree that lacks the FS1172X mutation (family 34), the proband was heterozygous for FS288X and FS546X. Unexpectedly, no amino acid substitutions were found as a cause of MLII. All fibroblasts or lymphoblasts from patients with MLII lacked GlcNAc-phosphotransferase activity (⩽1%) and had mutations that precluded the production of mRNA coding for full-length or active GlcNAc-phosphotransferase. We propose that these criteria—very low GlcNAc-phosphotransferase activity and mutations yielding truncated proteins—should define the MLII diagnosis. We believe that the two patients (GM1006 and F1515) clinically diagnosed with MLII were misclassified, and the proper classification is MLIIIA. This is based on enzyme activity (∼5% of normal), which we believe to be incompatible with a diagnosis of MLII.
In the nine pedigrees with MLIIIA, including the two reclassified cases, the mutations determined were qualitatively different. Although the common FS1172X null mutation was found in five pedigrees, and additionally two patients were heterozygous for Q278X or FS745X, these null mutations were always paired with a mild mutation, such as an amino acid substitution, truncation of the enzyme near the carboxyl-terminus, or a leaky splice-site mutation. Two patients were homozygous for a mutation: K4Q or R1205X. Fibroblast GlcNAc-phosphotransferase activity in patients with MLIII varied from 0.8% to 14% of normal and overlapped with the values found for patients with MLII (⩽1%). When the patients’ ages were available, all except one were in the range from 7 to 15 years. Given the age range, it is unlikely that any of these cases represent misclassified MLII. There does not appear to be any obvious correlation between age and fibroblast GlcNAc-phosphotransferase activity. We propose that MLIIIA is defined by the presence of at least one mild mutation that can produce active GlcNAc-phosphotransferase.
Genetic heterogeneity within the mucolipidoses has been investigated by cell fusion studies (heterokaryon analysis) (Honey et al. 1982; Shows et al. 1982; Mueller et al. 1983; Little et al. 1986). These studies demonstrated several important features about the mucolipidoses, including the presence of multiple complementation groups. On the basis of this technique, MLIII was divided into MLIIIA, MLIIIB, and MLIIIC, confirming the previous identification of a variant form of MLIII by Varki et al. (1981) that was based on enzyme activity with different substrates. Group B is represented by a single patient whose cell line was subsequently lost. Group A represents classical MLIII, and group C represents the variant form. In this study, we demonstrated that MLIIIA is caused by mutations in GNPTAB on human chromosome 12. We previously showed that MLIIIC was caused by mutations in the γ-subunit gene on chromosome 16 (Raas-Rothschild et al. 2000, 2004); therefore, the molecular basis for the complementation is obvious. Mueller et al. (1983) also demonstrated complementation between MLII and MLIIIC but not between MLII and MLIIIA nor between MLII and MLIIIIB. Again, these results are expected because we now know that MLII and MLIIIA are allelic. Less understandable is the complementation within MLII reported by Shows et al. (1982). This complementation was only observed when the fusion partner was fibroblasts from GM1006 (LT). To investigate this observation, we obtained the GM1006 (LT) cell line from Dr. Shows. Our analyses strongly suggested that this patient should be classified as having MLIIIA, not MLII, on the basis of the GlcNAc-phosphotransferase activity (5.2% of normal) and the FS1202X mutation that was active on transfection. Since we reclassified patient GM1006 (LT), we conclude that complementation within MLII was not demonstrated by Shows et al. (1982).
Since we now know the α- and β-subunits are derived from one gene, intragenic complementation in MLII or MLIIIA is possible in principal. For example, if one allele carries an α-subunit mutation and the other carries a β-subunit mutation, active GlcNAc-phosphotransferase might be formed from the active subunits. In our study, the only patient that might demonstrate intragenic complementation is patient GM01494. In GM01494 cells, an active α-subunit (amino acids 1–928) might be provided by the FS1172X allele, and an active β-subunit (amino acids 929–1256), by the K4Q allele. However, GlcNAc-phosphotransferase activity of fibroblast GM01494 was only 14% of normal. This is only slightly higher than the average of the activity in GM00113 (K4Q/K4Q) (12%) and GM01586 (FS1272X/FS1272X) (<0.1%), which suggests that other factors, such as unstable mRNA, may prevent complementation in this case. Additionally, persons with fully complementing mutations would not be expected to display symptoms that would identify them as candidates for GNPTAB sequencing.
Our study proved that MLII and MLIIIA are allelic on the basis of gene sequence, and we proposed criteria to distinguish them. MLII is defined by two severe mutations, whereas MLIIIA is defined by at least one mild mutation that can produce active GlcNAc-phosphotransferase. This approach allowed us to reclassify as MLIIIA two cases that were previously misdiagnosed as MLII, and it provided a molecular basis to understand a large body of complementation data. The identification of 15 different mutations, including 5 common mutations, provides a basis for screening, carrier testing, prenatal diagnosis, genetic counseling, and prediction of prognosis.
Acknowledgments
We thank the NIGMS Human Genetic Cells Repository at the Coriell Institute for Medical Research (Camden, NJ), Dr. A. Raas-Rothschild (Hadassah University Hospital, Israel), Dr. Thomas B. Shows (Roswell Park Cancer Institute, Buffalo, NY), Dr. Miko Stewart (Children’s Memorial Hospital, Chicago), and Dr. Catherine M. Bollard (Texas Children’s Hospital, Houston), for providing cell lines from patients with MLII and MLIIIA. We thank Ms. Courtney P. Kerbo, for culturing patient cells, and the sequencing core facility at the Oklahoma Medical Research Foundation, for sequencing. We also thank Dr. Anil D’Souza for initial efforts at detecting mutations in patients with mucolipidosis.
Footnotes
Presented in part at the 1998 Annual Meeting of The American Society of Human Genetics, Denver, October 27–31 [abstract 74], and at the Joint Meeting of the Society for Glycobiology and the Japanese Society of Carbohydrate Research, Honolulu, November 17–20 [abstract 16].
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BAC 14951 containing GNPTAB [accession number AC005409], cDNA encoding GlcNAc-phosphotransferase α/β–subunit precursor [accession number AY687932], and the following exons and the flanking intronic sequences: exon 1 [accession number BV677459], exon 2 [accession number BV677460], exons 3 and 4 [accession number BV677461], exon 5 [accession number BV677462], exons 6 and 7 [accession number BV677463], exons 8, 9, and 10 [accession number BV677464], exon 11 [accession number BV677465], exon 12 [accession number BV677466], exon 13 [accession number BV677467], exons 14 and 15 [accession number BV677468], exon 16 [accession number BV677469], exons 17 and 18 [accession number BV677470], exon 19 [accession number BV677471], exon 20 [accession number BV677472], and exon 21 [accession number BV677463])
- IUBMB (International Union of Biochemistry and Molecular Biology) Enzyme Nomenclature, http://www.chem.qmul.ac.uk/iubmb/enzyme/ (for accession numbers EC 2.7.8.17 and EC 3.1.4.45)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MLII, MLIIIA, and MLIIIC)
References
- Bao M, Booth JL, Elmendorf BJ, Canfield WM (1996a) Bovine UDP-N-acetylglucosamine: lysosomal enzyme N-acetylglucosamine-1-phosphotransferase. I. Purification and subunit structure. J Biol Chem 271:31437–31445 10.1074/jbc.271.49.31437 [DOI] [PubMed] [Google Scholar]
- ——— (1996b) Bovine UDP-N-acetylglucosamine: lysosomal enzyme N-acetyglucosamine-1-phosphotransferase. II. Enzymatic characterization and identification of the catalytic subunit. J Biol Chem 271:31446–31451 10.1074/jbc.271.49.31446 [DOI] [PubMed] [Google Scholar]
- Breathnach R, Chambon P (1981) Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem 50:349–383 10.1146/annurev.bi.50.070181.002025 [DOI] [PubMed] [Google Scholar]
- Canfield WM, Bao M, Pan J, D’Souza A, Brewer K, Pan H, Roe B, Raas-Rothschild A (1998) Mucolipidosis II and mucolipidosis IIIA are caused by mutations in the GlcNAc-phosphotransferase α/β gene on chromosome 12p. Am J Hum Genet Suppl 63:A15 [Google Scholar]
- den Dunnen JT, Antonarakis SE (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15:7–12 [DOI] [PubMed] [Google Scholar]
- Do H, Lee W, Ghosh P, Hollowell T, Canfield WM, Kornfeld R (2002) Human mannose 6-phosphate-uncovering enzyme is synthesized as a proenzyme that is activated by the endoprotease furin. J Biol Chem 277:29737–29744 10.1074/jbc.M202369200 [DOI] [PubMed] [Google Scholar]
- Garrod AE (1902) The incidence of alkaptonuria: a study in chemical individuality. Lancet 2:1611–1620 [PMC free article] [PubMed] [Google Scholar]
- Hasilik A, Waheed A, von Figura K (1981) Enzymatic phosphorylation of lysosomal enzymes in the presence of UDP-N-acetylglucosamine: absence of the activity in I-cell fibroblasts. Biochem Biophys Res Commun 98:761–767 10.1016/0006-291X(81)91177-3 [DOI] [PubMed] [Google Scholar]
- Honey NK, Mueller OT, Little LE, Miller AL, Shows TB (1982) Mucolipidosis III is genetically heterogeneous. Proc Natl Acad Sci USA 79:7420–7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld S, Sly WS (2001) I-cell disease and pseudo-Hurler polydystrophy: disorders of lysosomal enzyme phosphorylation and localization. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease. Vol III. McGraw-Hill, New York, pp 3469–3505 [Google Scholar]
- Kudo M, Bao M, D’Souza A, Ying F, Pan H, Roe BA, Canfield WM (2005) The α- and β-subunits of the human UDP-N-acetylglucosamine:lysosomal enzyme phosphotransferase are encoded by a single cDNA. J Biol Chem 280:36141–36149 10.1074/jbc.M509008200 [DOI] [PubMed] [Google Scholar]
- Kudo M, Kerbo CP, Canfield WM (2004) Mutations in the GlcNAc-phosphotransferase α-/β-subunits precursor gene are the molecular basis of both mucolipidosis II and IIIA. Glycobiology 14:1057–1058 [Google Scholar]
- Little LE, Mueller OT, Honey NK, Shows TB, Miller AL (1986) Heterogeneity of N-acetylglucosamine 1-phosphotransferase within mucolipidosis III. J Biol Chem 261:733–738 [PubMed] [Google Scholar]
- Mueller OT, Honey NK, Little LE (1983) Mucolipidosis II and III: the genetic relationships between two disorders of lysosomal enzyme biosynthesis. J Clin Invest 72:1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller OT, Little LE, Miller AL, Lozzio CB, Shows TB (1985) I-cell disease and pseudo-Hurler polydystrophy: heterozygote detection and characteristics of the altered N-acetyl-glucosamine-phosphotransferase in genetic variants. Clin Chim Acta 150:175–183 10.1016/0009-8981(85)90242-6 [DOI] [PubMed] [Google Scholar]
- Raas-Rothschild A, Bargal R, Goldman O, Ben-Asher E, Groener JE, Toutain A, Stemmer E, Ben-Neriah Z, Flusser H, Beemer FA, Penttinen M, Olender T, Rein AJ, Bach G, Zeigler M (2004) Genomic organisation of the UDP-N-acetylglucosamine-1-phosphotransferase gamma subunit (GNPTAG) and its mutation in mucolipidosis III. J Med Genet 41:e52 10.1136/jmg.2003.015222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raas-Rothschild A, Cormier-Daire V, Bao M, Genin E, Salomon R, Brewer K, Zeigler M, Mandel H, Toth S, Roe B, Munnich A, Canfield WM (2000) Molecular basis of variant pseudo-Hurler polydystrophy (mucolipidosis IIIC). J Clin Invest 105:673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman ML, Kornfeld S (1981a) Lysosomal enzyme targeting: N-acetylglucosaminylphosphotransferase selectively phosphorylates native lysosomal enzymes. J Biol Chem 256:11977–11980 [PubMed] [Google Scholar]
- ——— (1981b) UDP-N-acetylglucosamine:glycoprotein N-acetylglucosamine-1-phosphotransferase. Proposed enzyme for the phosphorylation of the high mannose oligosaccharide units of lysosomal enzymes. J Biol Chem 256:4275–4281 [PubMed] [Google Scholar]
- Reitman ML, Lang L, Kornfeld S (1984) UDP-N-acetylglucosamine: lysosomal enzyme N-acetylglucosamine-1-phosphotransferase. Methods Enzymol 107:163–172 [DOI] [PubMed] [Google Scholar]
- Reitman ML, Varki A, Kornfeld S (1981) Fibroblasts from patients with I-cell disease and pseudo-Hurler polydystrophy are deficient in uridine 5′-diphosphate-N-acetylglucosamine:glycoprotein N-acetylglucosaminylphosphotransferase activity. J Clin Invest 67:1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shows TB, Mueller OT, Honey NK, Wright CE, Miller AL (1982) Genetic heterogeneity of I-cell disease is demonstrated by complementation of lysosomal enzyme processing mutants. Am J Med Genet 12:343–353 10.1002/ajmg.1320120312 [DOI] [PubMed] [Google Scholar]
- Steet RA, Hullin R, Kudo M, Martinelli M, Bosshard NU, Schaffner T, Kornfeld S, Steinmann B (2005) A splicing mutation in the α/β GlcNAc-1-phosphotransferase gene results in an adult onset form of mucolipidosis III associated with sensory neuropathy and cardiomyopathy. Am J Med Genet A 132:369–375 [DOI] [PubMed] [Google Scholar]
- Tiede S, Storch S, Lubke T, Henrissat B, Bargal R, Raas-Rothschild A, Braulke T (2005) Mucolipidosis II is caused by mutations in GNPTA encoding the α/β GlcNAc-1-phosphotransferase. Nat Med 11:1109–1112 10.1038/nm1305 [DOI] [PubMed] [Google Scholar]
- Varki A, Kornfeld S (1981) Purification and characterization of rat liver α-N-acetylglucosaminyl phosphodiesterase. J Biol Chem 256:9937–9943 [PubMed] [Google Scholar]
- Varki AP, Reitman ML, Kornfeld S (1981) Identification of a variant of mucolipidosis III (pseudo-Hurler polydystrophy): a catalytically active N-acetylglucosaminylphosphotransferase that fails to phosphorylate lysosomal enzymes. Proc Natl Acad Sci USA 78:7773–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Reitman ML, Vannier A, Kornfeld S, Grubb JH, Sly WS (1982) Demonstration of the heterozygous state for I-cell disease and pseudo-Hurler polydystrophy by assay of N-acetylglucosaminylphosphotransferase in white blood cells and fibroblasts. Am J Hum Genet 34:717–729 [PMC free article] [PubMed] [Google Scholar]
- Waheed A, Hasilik A, von Figura K (1981) Processing of the phosphorylated recognition marker in lysosomal enzymes: characterization and partial purification of a microsomal α-N-acetylglucosaminylphosphodiesterase. J Biol Chem 256:5717–5721 [PubMed] [Google Scholar]
- Waheed A, Hasilik A, von Figura K (1982a) UDP-N-acetylglucosamine:lysosomal enzyme precursor N-acetylglucosamine-1-phosphotransferase. J Biol Chem 257:12322–12331 [PubMed] [Google Scholar]
- ——— (1982b) Deficiency of UDP-N-acetylglucosamine:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase in organs of I-cell patients. Biochem Biophys Res Commun 105:1052–1058 10.1016/0006-291X(82)91076-2 [DOI] [PubMed] [Google Scholar]