Abstract
Telomeres play a central role in cellular senescence and cancer pathobiology and are associated with age-related diseases such as atherosclerosis and dementia. Telomere length varies between individuals of the same age, is influenced by DNA-damaging factors such as oxidative stress, and is heritable. We performed a quantitative-trait linkage analysis using an ∼10-cM genomewide map for mean leukocyte terminal-restriction fragment (TRF) lengths measured by Southern blotting, in 2,050 unselected women aged 18–80 years, comprising 1,025 complete dizygotic twin pairs. Heritability of mean batch-adjusted TRF was 36% (95% confidence interval [CI] 18%–48%), with a large common environmental effect of 49% (95% CI 40%–58%). Significant linkage was observed on chromosome 14 (LOD 3.9) at 14q23.2, and suggestive linkage at 10q26.13 (LOD 2.4) and 3p26.1 (LOD 2.7). This is the first report of loci, mapped in a sample of healthy individuals, that influence mean telomere variation in humans.
The telomere is a complex DNA-protein structure that caps the end of eukaryotic chromosomes and is responsible for maintaining chromosome integrity, the architecture of the nucleus and chromosomes, and the complete replication of chromosome termini (Flint et al. 1997; Strachan and Read 2003). Human telomeres contain TTAGGG repeats, which are hypothesized to act as a molecular clock by effectively counting the number of cell divisions (Harley 1997). The progressive telomere shortening with each replicative cycle is one of the pathways that leads to replicative senescence of cultured somatic cells (Harley et al. 1990; Bodnar et al. 1998; Vaziri and Benchimol 1998). White blood cell (WBC) telomere length is inversely related to the blood donor’s age, which suggests that telomere attrition occurs with cell division not only in vitro but also in vivo (Harley 1997; Takubo et al. 2002; Gardner et al. 2005; Valdes et al. 2005). Yet WBC telomere length is highly variable at birth (Okuda et al. 2002) and during adult life (Valdes et al. 2005). Monogenic diseases due to mutations in the catalytic subunit of telomerase (Harley 2005; Marrone et al. 2005) and aplastic anemias (Yamaguchi et al. 2005) have been identified, but a host of other mutations and other genes may account for variation in WBC telomere length in the general population.
In the present study, we performed a quantitative-trait linkage analysis of WBC mean terminal-restriction fragment (TRF) length, measured by Southern blotting (Benetos et al. 2001), for 2,050 adult female DZ twins comprising 1,025 twin pairs. The mean age (±SD) for the DZ twins was 47.8±12.4 years, with an age range of 18–80 years at the time of sample collection. For these cross-sectional data, we have demonstrated elsewhere a linear age-related decrease in TRF length (±SE) of ∼27±1.5 bp per year (Valdes et al. 2005). This figure is consistent with estimates that WBC telomeres reduce by ∼15–40 bp per year after accounting for sample age range (Slagboom et al. 1994; Frenck et al. 1998; Benetos et al. 2001; Brouilette et al. 2003; Vasa-Nicotera et al. 2005).
Twins were identified from the St. Thomas’ United Kingdom adult twin registry, were invited to participate in the study, and were measured for an extensive range of clinical phenotypes related to cardiovascular disease, obesity, diabetes, and osteoporosis. Both twins attended the clinic together for the collection of clinical data, which included age, height, and weight. General medical, gynecological, and lifestyle questionnaires were completed at interview, and blood samples were taken for DNA extraction. These twins have been shown to be comparable to age-matched singleton populations for most age-related traits (Andrew et al. 2001).
Human telomere length is highly variable at birth and thereafter, among individuals and chromosomes (Harley et al. 1990; Okuda et al. 2002). Telomere length is difficult to measure without experimental error, and this can be attributed, in part, to different methods used (Slagboom et al. 1994; Cawthon 2002). Therefore, to control for laboratory experimental batch effects for TRF lengths, each twin pair was randomly assigned to a gel batch with respect to age and zygosity. Southern analysis was performed on DNA from each twin, was electrophoresed in adjacent lanes, and subsequently was referred to the nearest molecular weight ladder. Each twin DNA sample pair was resolved in duplicate on two gels.
The samples were digested overnight with restriction enzymes HinfI (0.5 U/μL) and RsaI (0.5 U/μL) (Boehringer Mannheim). DNA samples and DNA ladders were resolved on a 0.5% agarose gel (20×20 cm) at 50 volts (GNA-200 Pharmacia Biotech). After 16 h, the DNA was depurinated for 30 min in 0.25N HCl, was denatured for 30 min in 0.5 mol/liter NaOH/1.5 mol/liter NaCl, and was neutralized for 30 min in 0.5 mol/liter Tris, pH 8/1.5 mol/liter NaCl. The DNA was transferred for 1 h to a positively charged nylon membrane (Boehringer Mannheim) by use of a vacuum blotter (Appligene [Oncor]). The membranes were hybridized at 65°C with the telomeric probe digoxigenin 3′-end labeled 5′-(CCTAAA)3 overnight in 5× saline sodium citrate (SSC) and 0.1% Sarkosyl, 0.02% SDS, and 1% blocking reagent and were washed three times at room temperature in 2× SSC, 0.1% SDS, each for 15 min, and once in 2× SSC for 15 min. The probe was detected by the digoxigenin luminescent detection procedure and was exposed on x-ray film.
The mean TRF length was calculated as TRF=ΣODi/Σ(ODi/MWi), where ODi is optical density at a given position in the lane and MWi is molecular weight at that position (Okuda et al. 2000). This formula accounts for the fact that longer telomeres bind more labeled probes and consequently appear darker on the x-ray film. The mean of the duplicates measured in different gels for each sample was used for data analysis. The intraclass correlation for duplicate TRF length measurements by use of different gels was 0.97 for both MZ and DZ twins.
Previous reports have suggested that telomere length is subject to genetic control (Slagboom et al. 1994; Jeanclos et al. 2000; Strachan and Read 2003; Vasa-Nicotera et al. 2005). For the present study, TRF length was also measured in an additional 55 MZ twins pairs to provide an approximate heritability estimate for these data, defined as the proportion of total TRF variance attributable to genetic factors.
TRF-length heritability was estimated using structural equation modeling that was implemented using Mx software applied to MZ and DZ twins (Neale and Maes 1991). Laboratory batch effects, in which DNA samples were processed in groups of up to 28 at a time, were associated with TRF (square of the correlation coefficient, R2=19% for a simple linear regression) and with donor age (R2=15%). Residuals for TRF length were obtained by regressing TRF length on batch categories (and other covariates for the heritability analysis), by use of the regression cluster option in Stata to account for correlation within twin pairs.
By use of a classic twin model (Neale and Maes 1991), including age as a covariate for both MZ and DZ twin pairs, the best-fitting model for batch-adjusted TRF-length scores provided a narrow heritability estimate of 36% (95% CI 18%–48%) attributable to additive polygenic effects and a large shared familial effect of 49% (95% CI 40%–58%). The intraclass correlation for TRF length (±SD)—adjusted for age, smoking status (never, current, or former smoker), and BMI—was 0.84±0.035 for MZ twin pairs and was 0.67±0.015 for DZ twins. Heritability estimates differed surprisingly little between those based on raw TRF-length scores (data not shown), residuals of TRF length adjusted for batch effects only, and batch-adjusted TRF-length scores that included covariates (table 1).
Table 1.
TRF Length Heritability Model Estimates[Note]
|
Heritability Estimates |
||||||||||||
| A |
C/D |
E |
Fit Statistics |
|||||||||
| Estimate and Model | a2 | 95% CI | c2 | d2 | 95% CI | e2 | 95% CI | No. ofEstimatedParameters | −2 LL | Δ χ2 | Δ df | P |
| TRF Length: | ||||||||||||
| ACE | .33 | .17–.44 | .53 | … | .50–.62 | .14 | .10–.21 | 6 | 3,323 | … | … | … |
| ADE | .91 | .89–.93 | … | 0 | .00–.02 | .09 | .07–.11 | 6 | 3,486 | … | … | … |
| CE | … | .71 | … | .70–.74 | .29 | .26–.32 | 5 | 3,334 | 11 | 1 | .001 | |
| AE | .91 | .89–.93 | … | … | … | .09 | .07–.11 | 5 | 3,486 | 163 | 1 | 0 |
| TRF Length and covariates: | ||||||||||||
| ACE | .36 | .18–.48 | .49 | … | .40–.58 | .16 | .11–.24 | 6 | 3,088 | … | … | … |
| ADE | .9 | .87–.92 | … | 0 | .00–.02 | .1 | .08–.13 | 6 | 3,195 | … | … | … |
| CE | … | … | .67 | … | .64–.7 | .33 | .30–.36 | 5 | 3,099 | 11 | 1 | .001 |
| AE | .9 | .87–.92 | … | … | … | .1 | .08–.13 | 5 | 3,195 | 107 | 1 | 0 |
Note.— All models include age as a covariate and use the residuals of raw TRF length, adjusted for experimental batch effects. Additional covariates are BMI and smoking history (never, current, or former smoker). A = additive polygenic effects; C = familial common environment; D = dominance effects; E = specific individual effects and measurement error. A, C, D, and E are variance components terms; a, c, d, and e refer to the standard deviation. Model-fit statistics for models CE and AE are presented as submodels of ACE. The best-fit models are highlighted in bold italics.
Although the heritability for these data appears to be ostensibly low, on closer inspection, the estimate is probably comparable with those from previous reports, since these did not explicitly model potentially important common environment effects. For example, familial and sib-pair correlations will overestimate heritability when shared environmental effects exist, since such estimates confound shared environmental and genetic factors (Jeanclos et al. 2000; Vasa-Nicotera et al. 2005). A previous twin heritability estimate for WBC TRF length has also been calculated, in which batch effects accounted for up to 42% of total TRF length variance (Slagboom et al. 1994). By contrast, ∼19% of TRF length variance was estimated to be attributable to laboratory batch effects with the use of our methods. Using a total of 115 MZ and DZ male and female twin pairs aged 2–63 years, Slagboom et al. (1994) observed a narrow heritability estimate of 78%, after accounting for the variance attributable to batch and cohort effects. The study did not test for evidence of common environment and additive polygenic effects in the same model. If we similarly model batch and age effects and exclude common environment effects (table 1), we also obtain a high heritability estimate of ∼90%. However, the best-fitting model for these data includes additive polygenic and common environment effects. Given the inherent difficulty of the precise measurement of telomere length, a likely explanation for discrepancy in reported heritability estimates is simply that common environmental effects have not been estimated in previous models. This is important, since, by design, shared sample protocol and the random allocation of twin-pair samples to the same laboratory gel batch will ensure that the estimate of “common environment” for TRF length includes any unmeasured experimental error shared between siblings.
The large common environment we observe for these data is also likely to genuinely reflect environmental factors that influence telomere-length variation shared between siblings, in addition to including systematic experimental error shared by twin-pair samples that are not accounted for by recorded batch categories. For the narrow purpose of gene mapping, the answer to what causes common environmental variation in telomere length does not strictly matter as long as it is explicitly modeled. What is important for gene mapping is that there be a significant heritable component and (for improved power) a large residual correlation (Allison et al. 1998; Sham et al. 2000).
Genome scans were performed using DNA extracted from venous blood samples of the study subjects. Scans involved the use of standard fluorescence-based genotyping methodologies (Pritchard et al. 1995) for the analysis of 737 highly polymorphic microsatellite markers from the ABI Prism linkage mapping set (Applied Biosystems) and Genethon Genetic Linkage Map (Dib et al. 1996). Gemini Genomics conducted the genotyping, with 365 core microsatelites genotyped for 600–1,300 DZ sib pairs and 372 markers providing gap fills. As a result, the 737 markers provided coverage of ∼1 marker at least every 10 cM for all sib pairs. Twin zygosity and family relationships were rigorously investigated, and discrepant pairs were discarded from further analyses. The estimated genotyping error rate was <1%, with use of duplicate blind controls for 2%–5% of all the samples genotyped.
Map positions and ordering of all marker loci were determined from the Genethon Genetic Linkage Maps and Haldane map function. Approximate support intervals were generated using a 2-LOD drop approach. Individuals in the study were considered to be randomly ascertained, because sampling was not based on subject TRF length. Twin pairs formed independent families, and no additional sibs were considered in the analysis.
Two multipoint genomewide variance-components linkage analyses were performed—the first for batch-adjusted TRF length and the second for batch-adjusted TRF length including the covariates of donor age and BMI. BMI was included as a covariate of TRF length, since BMI is negatively correlated with TRF length (−0.11) and, with smoking status, is observed to be an important covariate for these data (Valdes et al. 2005). The phenotypic correlation between TRF length and BMI appeared to be primarily accounted for by shared environmental factors, with no evidence of a genetic correlation between the two variables for these data (genetic correlation coefficient rg=-0.03; 95% CI −0.58 to 0.43). Smoking history was not included as a covariate in the genome scan, since this was not significant after adjusting for all other covariates.
Since common environment effects were important for all heritability models considered, even after accounting for batch effects, common environment was modeled as a random effect in the linkage analyses. The genomewide linkage scan was implemented using QTDT (Abecasis et al. 2000) rather than Merlin (Abecasis et al. 2002), since the former has an option to use MZ phenotypes to estimate the common environment variance components, in addition to specifying measured covariates as fixed effects. The TRF length adjusted for covariates resulted in a perfect Gaussian distribution, with a kurtosis of 2.95, and therefore fulfilled the assumption of trait normality for variance-components linkage analysis.
In addition, we also implemented a genomewide scan, using a modified version of the Haseman-Elston method in a generalized linear model (Barber et al. 2004), in which the square of the sibling differences was regressed on estimated identical-by-descent (IBD) status at each locus. By modeling the difference in TRF length between siblings, any shared measurement error was effectively removed.
Empirical P values for the three observed loci were obtained by running 1,000 genomewide scans at 10-cM intervals, by use of multipoint IBD genotype data (a total of 362,000 tests) in which the phenotype was randomly shuffled before each scan. The routine was programmed in Stata (StatCorp), and the tests of linkage were implemented as a modified version of the Haseman-Elston method in a generalized linear model (Barber et al. 2004).
Female twin pairs had mean TRF length (±SD) of 7.1±0.69 Kbp (ranging from 5.1 to 9.4), mean BMI of 25.3±4.8 kg/m2 (ranging from 13.9 to 48.2), and a prevalence of 47% reporting never having smoked. A genomewide scan provided significant evidence of linkage (table 2) at 14q23.2 for both batch-adjusted TRF length (LOD 3.9) and batch-adjusted TRF length with covariates (LOD 3.4). Evidence of suggestive linkage was also observed at 10q26.13 (LOD 2.1–2.4) and 3p26.1 (LOD 2.7). Linkage analysis using optimal Haseman-Elston methods implemented using GLM and robust to deviations from normality (Barber et al. 2004) gave the same results for chromosomes 14 and 10, but the peak LOD score at 25 cM on chromosome 3 dropped to a LOD score of 1.4.
Table 2.
Multipoint Genomewide Linkage Results for the Mean TRF Length with Use of QTDT[Note]
|
TRF Length Peak LOD Score |
||||||
| Chromosome | No. of Pairs | Marker | Length(cM) | Approximate 2-LOD interval | With No Covariates | With Age and BMIas Covariates |
| 3 | 946 | D3S1304 | 20 | 0–50 | 2.7 | 2.69 |
| 10 | 946 | D10S587 | 160 | 105–182 | 2.4 | 2.1 |
| 14 | 947 | D14S63 | 60 | 40–85 | 3.9 | 3.4 |
Note.— The variance-components model includes a random effect for common environment, estimated including MZ phenotypes. Genomewide linkage analyses were conducted twice, once for batch-adjusted TRF length only and once for batch-adjusted TRF length with covariates age and BMI.
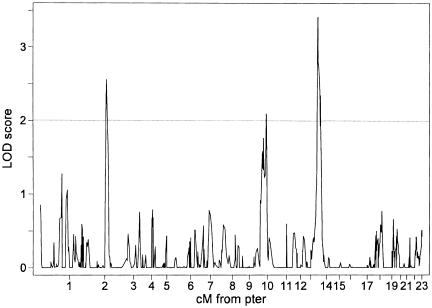
Permutation tests provided empirical counts of 24, 108, and 9,688, from a total of 362,000 tests in which results were obtained that were similar to or more extreme than those originally observed on chromosomes 14, 10, and 3, respectively. The counts are equivalent to pointwise (and genomewide) P values of .000066 (.024), .00029 (.11), and .027 (1) and suggest that the evidence of linkage to chromosome 14 is highly significant, to chromosome 10 is suggestive, and to chromosome 3 is not significant. Apart from these three loci, no evidence of linkage for these data was observed anywhere in the genome at a threshold of LOD 1.4 or above (fig. 1), including previous reports of possible linkage on chromosomes 12 (Vasa-Nicotera et al. 2005) and X (Nawrot et al. 2004).
Figure 1.
Chromosomal LOD-score results for batch-adjusted TRF length, including a random effect for common environment and covariates BMI and donor age.
Vasa-Nicotera et al. (2005) reported a maximum two-point LOD score of 3.21 to D12S345 (at 54.4 cM) for 173 families of primarily male siblings. We found little evidence of two-point linkage to D12S345 for our data at P=.52 (LOD 0.09). Although the sample size for this single genotype for these data was low (n=180 female sib pairs), our data should provide ∼90% power (at α=0.05, equivalent to LOD 0.9 and under the assumption of a recombination fraction of zero) to detect a large QTL effect such as the one reported that accounts for, say, 20% or more of the variation in telomere length. (We chose a smaller QTL heritability estimate for the power calculation than that cited at 49% by Vasa-Nicotera and colleagues [2005], because such estimates are known to be upwardly biased, with the QTL effect size and test statistic completely confounded in gene-detection experiments, such as a novel genome scan [Göring et al. 2001]). The maximum multipoint LOD scores we observed on chromosome 12 for 946 unaffected female sib pairs were LOD 0.65 (at 0 cM) and LOD 0.53 (at 90 cM). However, our study cannot rule out the presence of loci on chromosome 12 with modest or small effects (e.g., a QTL with a heritability of <15%).
The lack of replication between samples could be due to a lack of power to detect small effects and/or different recruitment strategies between studies. Vasa-Nicotera et al. (2005) used a random subsample of primarily male subjects from the British Heart Foundation (BHF) Family Heart Study, with probands originally selected for experience of heart disease (such as a heart attack, angina, or coronary bypass surgery) before age 65 years. Given that telomere length tends to be shorter among subjects with heart disease than among unaffected individuals, such an ascertainment scheme may possibly result in expression of different genes that influence telomere dynamics. By contrast, the data used for the present study are all unselected, healthy sisters. Since mean age-adjusted TRF length is, on average, longer in women than in men (Jeanclos et al. 2000; Benetos et al. 2001; Nawrot et al. 2004; Vasa-Nicotera et al. 2005), this may also reflect the fact that aspects of telomere biology and gene expression are dependent on sex-specific factors such as estrogen that, in turn, might result in different linkage results for men and women. The telomere linkage results for both the BHF family and the twin data still require independent replication.
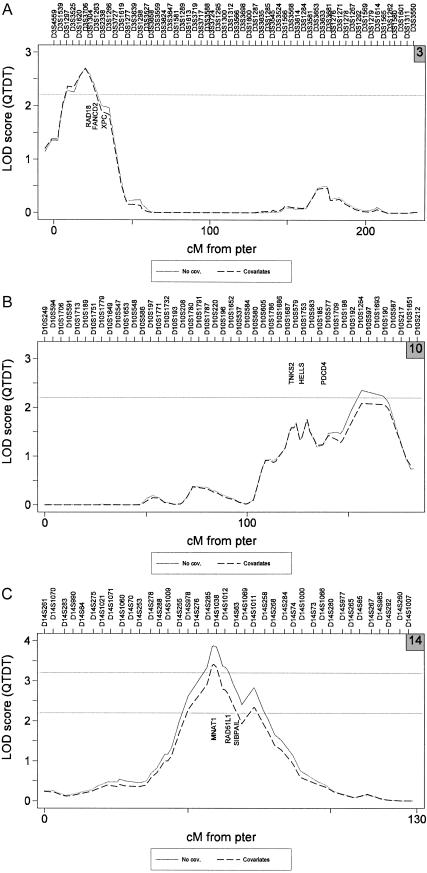
The three linkage loci observed in the present study (fig. 2) have been examined in preliminary detail by use of Genesniffer, a bioinformatics computer-assisted search of publicly available databases, to look for potential candidates. We identified 459 known or predicted genes in the 14q23.2 linkage region, 340 genes at 10q26.13, and 128 at 3p26.1. We used Genesniffer to exclude over half of these genes, since they showed no evidence of relationship with telomere dynamics, according to our automated search criteria using information from the National Center for Biotechnology Information Gene, Online Mendelian Inheritance in Man, and PubMed databases. Nine plausible candidates were identified (fig. 2) by manually searching the remaining Genesniffer pages and references, partly guided by a simple weighted-scoring system, for any evidence that the remaining identified genes in the linkage regions might play a role in telomere biology.
Figure 2.
LOD-score results for batch-adjusted TRF length, analyzed with and without covariates BMI and donor age. Both genome scans included a random effect for common environment. For graph labeling purposes, marker positions are evenly spaced and are accurate only for order. Potential candidate genes for each linkage peak include chromosome 3 (RAD18 = postreplication repair protein; FANCD2 = Fanconi anemia complementation group D2 protein; XPC = xeroderma pigmentosum, complementation group C) (A), chromosome 10 (TNKS2 = tankyrase, TRF1-interacting ankyrin-related; HELLS = helicase, lymphoid-specific; PDCD4 = programmed cell death 4) (B), and chromosome 14 (MNAT1 = ménage à trois 1; RAD51L1 = RAD51-like 1; SIPA1L1 = signal-induced proliferation-associated 1 like 1) (C).
In conclusion, this is the first study, to our knowledge, to identify potential genetic loci that influence telomere length in a large sample of unselected individuals. Work is now under way to identify important genes that contribute to human telomere dynamics.
Acknowledgments
We thank all the twins for participating in this study and Kate Elliott for her expert bioinformatics assistance in writing and running Genesniffer scripts on our behalf. This study was funded by Wellcome Trust Project Grant 074951. A.A.’s telomere research is supported by National Institutes of Health grants AG021593 and AG020132 and by The Healthcare Foundation of New Jersey.
References
- Abecasis GR, Cardon LR, Cookson WOC (2000) A general test of association for quantitative traits in nuclear families. Am J Hum Genet 66:279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Allison DB, Thiel B, St Jean P, Elston RC, Infante MC, Schork NJ (1998) Multiple phenotype modeling in gene-mapping studies of quantitative traits: power advantages. Am J Hum Genet 63:1190–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew T, Hart DJ, Snieder H, de Lange M, Spector TD, MacGregor AJ (2001) Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res 4:464–477 10.1375/1369052012803 [DOI] [PubMed] [Google Scholar]
- Barber MJ, Cordell HJ, MacGregor AJ, Andrew T (2004) Gamma regression improves Haseman-Elston and variance components linkage analysis for sib-pairs. Genet Epidemiol 26:97–107 10.1002/gepi.10299 [DOI] [PubMed] [Google Scholar]
- Benetos A, Okuda K, Lajemi M, Kimura M, Thomas F, Skurnick J, Labat C, Bean K, Aviv A (2001) Telomere length as an indicator of biological aging: the gender effect and relation with pulse pressure and pulse wave velocity. Hypertension 37:381–385 [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE (1998) Extension of life-span by introduction of telomerase into normal human cells. Science 279:349–352 10.1126/science.279.5349.349 [DOI] [PubMed] [Google Scholar]
- Brouilette S, Singh RK, Thompson JR, Goodall AH, Samani NJ (2003) White cell telomere length and risk of premature myocardial infarction. Arterioscler Thromb Vasc Biol 23:842–846 10.1161/01.ATV.0000067426.96344.32 [DOI] [PubMed] [Google Scholar]
- Cawthon RM (2002) Telomere measurement by quantitative PCR. Nucleic Acids Res 30:e47 10.1093/nar/30.10.e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 10.1038/380152a0 [DOI] [PubMed] [Google Scholar]
- Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett NA, King A, Higgs DR (1997) The relationship between chromosome structure and function at a human telomeric region. Nat Genet 15:252–257 10.1038/ng0397-252 [DOI] [PubMed] [Google Scholar]
- Frenck RW Jr, Blackburn EH, Shannon KM (1998) The rate of telomere sequence loss in human leukocytes varies with age. Proc Natl Acad Sci USA 95:5607–5610 10.1073/pnas.95.10.5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JP, Li S, Srinivasan SR, Chen W, Kimura M, Lu X, Berenson GS, Aviv A (2005) Rise in insulin resistance is associated with escalated telomere attrition. Circulation 111:2171–2177 10.1161/01.CIR.0000163550.70487.0B [DOI] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD, Blangero J (2001) Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 69:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB (1997) Human ageing and telomeres. Ciba Found Symp 211:129–139 [DOI] [PubMed] [Google Scholar]
- ——— (2005) Telomerase therapeutics for degenerative diseases. Curr Mol Med 5:205–211 10.2174/1566524053586671 [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW (1990) Telomeres shorten during ageing of human fibroblasts. Nature 345:458–460 10.1038/345458a0 [DOI] [PubMed] [Google Scholar]
- Jeanclos E, Schork NJ, Kyvik KO, Kimura M, Skurnick JH, Aviv A (2000) Telomere length inversely correlates with pulse pressure and is highly familial. Hypertension 36:195–200 [DOI] [PubMed] [Google Scholar]
- Marrone A, Walne A, Dokal I (2005) Dyskeratosis congenita: telomerase, telomeres and anticipation. Curr Opin Genet Dev 15:249–257 10.1016/j.gde.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Nawrot TS, Staessen JA, Gardner JP, Aviv A (2004) Telomere length and possible link to X chromosome. Lancet 363:507–510 10.1016/S0140-6736(04)15535-9 [DOI] [PubMed] [Google Scholar]
- Neale MC, Maes HHM (1991) Methodology for genetic studies of twins and families. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- Okuda K, Bardeguez A, Gardner JP, Rodriguez P, Ganesh V, Kimura M, Skurnick J, Awad G, Aviv A (2002) Telomere length in the newborn. Pediatr Res 52:377–381 10.1203/01.PDR.0000022341.72856.72 [DOI] [PubMed] [Google Scholar]
- Okuda K, Khan MY, Skurnick J, Kimura M, Aviv H, Aviv A (2000) Telomere attrition of the human abdominal aorta: relationships with age and atherosclerosis. Atherosclerosis 152:391–398 10.1016/S0021-9150(99)00482-7 [DOI] [PubMed] [Google Scholar]
- Pritchard LE, Kawaguchi Y, Reed PW, Copeman JB, Davies JL, Barnett AH, Bain SC, Todd JA (1995) Analysis of the CD3 gene region and type 1 diabetes: application of fluorescence-based technology to linkage disequilibrium mapping. Hum Mol Genet 4:197–202 [DOI] [PubMed] [Google Scholar]
- Sham PC, Cherny SS, Purcell S, Hewitt JK (2000) Power of linkage versus association analysis of quantitative traits, by use of variance-components models, for sibship data. Am J Hum Genet 66:1616–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagboom PE, Droog S, Boomsma DI (1994) Genetic determination of telomere size in humans: a twin study of three age groups. Am J Hum Genet 55:876–882 [PMC free article] [PubMed] [Google Scholar]
- Strachan T, Read AP (2003) Human molecular genetics, 3rd ed. Garland Science/Taylor and Francis Group, Oxford, United Kingdom [Google Scholar]
- Takubo K, Izumiyama-Shimomura N, Honma N, Sawabe M, Arai T, Kato M, Oshimura M, Nakamura K (2002) Telomere lengths are characteristic in each human individual. Exp Gerontol 37:523–531 10.1016/S0531-5565(01)00218-2 [DOI] [PubMed] [Google Scholar]
- Valdes AM, Andrew T, Gardner JP, Kimura M, Oelsner E, Cherkas LF, Aviv A, Spector TD (2005) Obesity, cigarette smoking, and telomere length in women. Lancet 366:662–664 10.1016/S0140-6736(05)66630-5 [DOI] [PubMed] [Google Scholar]
- Vasa-Nicotera M, Brouilette S, Mangino M, Thompson JR, Braund P, Clemitson J-R, Mason A, Bodycote CL, Raleigh SM, Louis E, Samani NJ (2005) Mapping of a major locus that determines telomere length in humans. Am J Hum Genet 76:147–151 (erratum 76:373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H and Benchimol S (1998) Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol 8:279–282 10.1016/S0960-9822(98)70109-5 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS (2005) Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med 352:1413–1424 10.1056/NEJMoa042980 [DOI] [PubMed] [Google Scholar]