Summary
During a normal cell cycle, chromosomes are exposed to many biochemical reactions that require specific types of DNA movement. Separation forces move replicated chromosomes into separate sister cell compartments during cell division, and the contemporaneous acts of DNA replication, RNA transcription and cotranscriptional translation of membrane proteins cause specific regions of DNA to twist, writhe and expand or contract. Recent experiments indicate that a dynamic and stochastic mechanism creates supercoil DNA domains soon after DNA replication. Domain structure is subsequently reorganized by RNA transcription. Examples of transcription-dependent chromosome remodelling are also emerging from eukaryotic cell systems.
Introduction
A working model of the bacterial chromosome has been a long-standing goal of molecular biology. Once the outlines of replication and transcription were revealed in the late 1960s, the physical implications of Watson and Crick’s structure raised basic questions about chromosome mechanics (Maaloe et al., 1966). Resolving many of the most basic questions about chromosome behaviour proved to be a daunting challenge [see the review by Higgins et al. (2005)]. Recent discoveries in Escherichia coli (Postow et al., 2004), Salmonella typhimurium (Deng et al., 2004; Stein et al., 2005), Caulobacter crescentus (Viollier et al., 2004; Gitai et al., 2005) and Bacillus subtilis (Britton et al., 1998; Lemon and Grossman, 1998; Dworkin and Losick, 2002; Lindow et al., 2002; Wu and Errington, 2003; 2004) move us closer to a composite view of dynamic chromosome behaviour, and make it easier to test and understand the 270-odd sequenced bacterial genomes in the current NCBI database.
If fully extended, the 4.6 million base pair E. coli chromosome would stretch over 1 mm. How does a 2-μm-long bacterium condense DNA 1000-fold and still allow the dynamic DNA movement necessary for RNA transcription and DNA replication to proceed without everything getting snarled (Holmes and Cozzarelli, 2000)? A partial solution to the packing problem is negative supercoiling, which causes the double helix to adopt a branched and plectonemic or interwound structure. Cells treated with lysozyme and mild ionic detergent release non-viscous ‘nucleoids’1 that can be analysed by sedimentation through sucrose gradients and by electron microscopy (Worcel and Burgi, 1972; Giorno et al., 1975; Kavenoff and Ryder, 1976). The unfolded structure reveals an interwound DNA conformation. Sensitive and non-disruptive topological tests of plasmid DNA structure that are produced by recombination with the lambda integrase confirmed the presence of interwound DNA in vivo (Bliska and Cozzarelli, 1987).
The chromosome superhelix density for mid-log phase cultures of E. coli is about σ= −0.06, where σ= ΔLk/Lk0 (Bauer et al., 1980; Drlica and Rouviere-Yaniv, 1987; Pettijohn, 1996). This value results from a dynamic equilibrium established by competing activities of DNA gyrase, which introduces negative supercoils, and two enzymes that remove negative supercoils – TopoI (Menzel and Gellert, 1983) and Topo IV (Zechiedrich et al., 2000). The supercoil density of chromosomal DNA must exist in a window of ±20% of the normal mean value for cell growth (Drlica, 1992). Hypo-supercoiling creates problems in segregation (Steck and Drlica, 1984; Hiraga et al., 1989; Holmes and Cozzarelli, 2000; Sawitzke and Austin, 2000) and leads to poor chromosome function that is either toxic or lethal (Zechiedrich et al., 1997). Hyper-supercoiling promotes the formation of Z-DNA at positions with long alternating GC or AT base pairs, causes the extrusion of cruciforms at inverted repeats, and stabilizes other unusual structures such as intramolecular triplexes (H-DNA), intermolecular triplexes and R-loops. These unusual structures impede RNA polymerase, slow or stall DNA replication forks, and create substrates for endonucleases and Holliday junction resolving enzymes (Higgins and Vologodskii, 2004).
In addition to compacting DNA, negative supercoils are dynamic. The slithering and branching of the interwound strands allow DNA to act like a chaperone, promoting the long-range assembly and disassembly of protein–DNA complexes (Higgins et al., 2005). Furthermore, negative supercoils facilitate the transition from duplex- to single-stranded conformation, a state through which DNA replication, transcription and recombination all proceed (Funnell et al., 1986; Artsimovitch, 2005; Kuzminov and Stahl, 2005).
Domain numerology
A supercoil domain can be defined as the segment of DNA relaxed by the introduction of a single- or double-strand break (Pettijohn, 1996; Postow et al., 2004). In vivo, single-strand breaks occur through a variety of mechanisms (Strauss, 2005). These include chemical damage from oxygen radicals, alkylation and depurination by chemicals generated by endogenous and exogenous mechanisms, attack from cellular endonucleases and the process of semi-discontinuous DNA replication. Because supercoiled DNA strands rotate rapidly (Oram et al., 1994), large chromosomes are partitioned into small units to shield the genome from catastrophic negative superhelical loss. Early DNase I nicking studies of E. coli nucleoids by Worcel and Burgi showed that multiple breaks were required to relax a chromosome fully in vitro; the estimated number of domains was 4–30 (Worcel and Burgi, 1972). Using 3H-tri-methylpsoralen binding as a probe of supercoil structure in vivo, Sinden and Pettijohn treated cells with X-rays to introduce breaks and estimated that E. coli contained 40 domains per genome equivalent (Sinden and Pettijohn, 1981). Recent studies indicate the presence of 400 domains [for an explanation of why the numbers differ, see the review by Higgins et al. (2005)].
A method developed in 1996 permitted the examination of domains in defined segments of the large bacterial chromosome using site-specific recombination as an assay. This test involved less structural perturbation than either physical extraction of DNA or X-irradiation (Higgins et al., 1996). S. typhimurium chromosomes marked with pairs of directly repeated res sites spaced at different intervals were exposed to the controlled induction of Tn3 or γδ resolvases. Unlike recombination catalysed by λ Int, phage P1 Cre, or yeast 2μ Flp recombinases, which pair sites by random DNA collisions, the resolvase system relies on negative supercoiling to precisely align two 140 bp res sites (Stark and Boocock, 1995). Two res sites can recombine and mark a chromosome with a deletion of non-essential genes if and only if they reside in the same supercoil domain (Fig. 1A). Three conclusions from this study challenged the conventional notions of chromosome structure. First, a distance penalty for recombination efficiency approximated first order decay. This suggested cell-to-cell variation in domain location. Second, the slope of the first order decay revealed that resolution efficiency decreased by 50% for each 15–17 kb distance separating a pair of res sites (a term called the 1/2D). Assuming that two points in a chromosome form a loop (see below) and that barrier placement has a Poisson distribution, the mean domain size was estimated to be 20 kb, which leads to an estimate three to four times larger than the 40 domain structure predicted by previous methods (Higgins et al., 1996). Third, domains appeared and disappeared over time. Surprisingly, chromosome movement was more dynamic in stationary-phase cells than in exponential log phase cells (Higgins et al., 1996; Staczek and Higgins, 1998; Higgins, 1999).
Fig. 1. Assays measuring supercoil domain structure in Salmonella and E. coli.
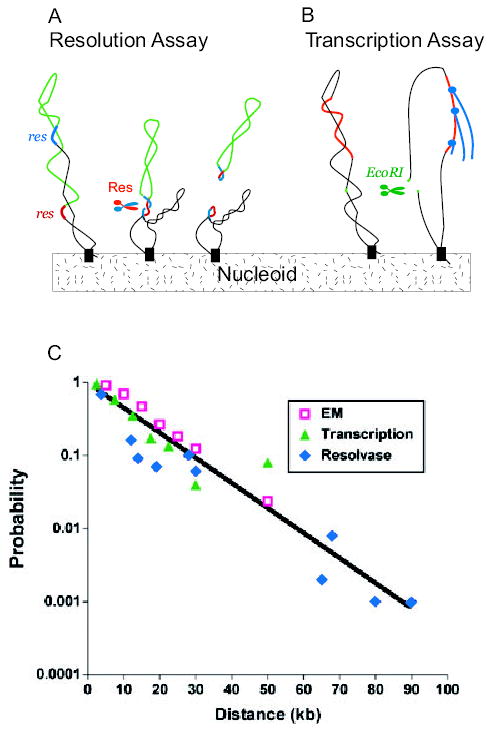
A. A set of Salmonella strains were made with a pair of directly repeated 140 bp Tn3 res sites (red and blue DNA) separated by different segments of bacterial DNA (green). After expressing the γδ site-specific recombinases (Res), a deletion forms if and only if the res sites reside in a common supercoil domain (Stein et al., 2005).
B. Escherichia coli cells contain 300 supercoil responsive genes (SSGs) whose expression is rapidly increased or decreased by a loss of negative supercoiling (Peter et al., 2004). Expression of EcoRI causes double-strand breaks and loss of supercoiling from domains with a restriction site. Changes in RNA levels measured with whole genome microarrays measure supercoil diffusion when data are combined with distance from the promoter to restriction sites in the genome (Postow et al., 2004).
C. The probability of supercoil loop detection as a function of DNA length in kb is plotted for resolution assays (blue diamonds) SSG transcription data (green triangles) and loop sizes measured from EM images (red squares). (Lisa Postow provided the transcription data and plot.)
Recently, Cozzarelli’s laboratory devised a new method for enumerating domains in E. coli (Postow et al., 2004). Negative supercoiling is a product of transcription as well as a factor that can modulate increased or decreased output from specific promoters (McGovern et al., 1994; Willenbrock and Ussery, 2004; Travers and Muskhelishvili, 2005). The Postow method takes advantage of 306 genes that are widely distributed in the genome and which respond rapidly and reliably to supercoil relaxation. After DNA relaxation, a third of the supercoil-sensitive genes (SSGs) become induced for transcription and two-thirds respond with transcriptional repression (Peter et al., 2004). By controlling the in vivo expression of type II restriction enzymes, Postow made double-strand breaks at precise chromosomal locations. The distance from a SwaI site to the promoter of an SSG was combined with microarray expression patterns before and after chromosomal cleavage. The best-fit model of the E. coli chromosome using SSG transcription data consisted of variable loops, random barrier positions and a 10 kb mean domain size. This model predicts ~450 domains per genome equivalent (Postow et al., 2004).
Support for a 400-domain chromosome came from two sources. First, tracings of EM images of gently lysed nucleoids showed that loops ranged from 2 to 66 kb and the best-fit model supported variable loops with an 11 kb mean (Postow et al., 2004).
Second, Postow’s domain estimates in E. coli agreed with new results in S. typhimurium, which developed from studies aimed at understanding how domains change over time (Scheirer and Higgins, 2001; Deng et al., 2004). Stein et al. discovered that once the WT Tn3 resolvase is produced, it remains active inside cells for more than 3 h. Because short-lived domains would be invisible to such a long lasting enzyme, Stein worked out a method to modulate the time-span of cell exposure to γδ resolvase. An 11-amino-acid extension was added to the C-terminus of Res protein (Res-SsrA). The extension corresponds to a sequence that is normally appended to proteins by the SsrA or Tm RNA trans-translation system (Stein et al., 2005). The tag had no effect on the enzyme’s catalytic efficiency or mechanism, but it made the resolvase an efficient substrate for ClpXP degradation (Keiler et al., 1996; Hayes et al., 2002a,b). By modifying selected residues in the tag (Flynn et al., 2001), Stein generated five resolvases that persist for periods varying from 5 min to several hours. Over a 100 kb chromosomal segment, domain structure changed over a 3 h window. Assayed for a 10 min period in exponential growth, the mean domain size was 11–13 kb. Thus, like E. coli, Salmonella has ~400 domains per genome equivalent, and the agreement between three different techniques measuring domain sizes in two species of bacteria is striking (Fig. 1C).
When do supercoil domains form? The process of DNA replication requires all DNA binding proteins and all topological impediments to unwinding of the helix by the DnaB helicase to be removed to allow efficient DNA synthesis. DNA replication proceeds at the rate of 800 bp per second per fork, and supercoils behind the fork are lost because of discontinuities in strand synthesis (Hingorani and O’Donnell, 2005). Peter showed that domains must reform rapidly after DNA replication. He used the 306 super-coil reporter SSGs mentioned above to measure how fast domains form after replication (Peter, 2000). Because there was no significant transcription change in the SSGs following replication, Peter concluded that domains reformed so rapidly that RNA polymerase had no time to synthesize significant levels of RNA before supercoil density was restored. The post-replication period is undoubtedly the time when most of the 400 stochastic supercoil domains are established.
Domains remodelled by transcription
In 2001, Scheirer reported that transcription from a phage promoter caused the appearance of a supercoil diffusion barrier in a Mu prophage (Scheirer and Higgins, 2001). To follow up, Deng created test intervals at several locations in the Salmonella chromosome (Deng et al., 2004). The impact of transcription was tested using a module derived from Tn10. Constitutive transcription of tetA, the gene for an integral plasma membrane tetracycline efflux pump, created region-specific barriers to γδ recombination and reduced resolution by 20-fold in a 14 kb interval at several chromosome locations. In plasmid studies, transcription anchors DNA to the cell membrane by cotranscriptional translation, and this attachment enhanced topological effects on DNA (Lynch and Wang, 1993). However, in the Salmonella chromosome, two cytoplasmic proteins, β-galactosidase (β-gal) and aminoglycoside-3′-O-phospho-transferase, the product of the Tn5 kan gene, reduced resolution as much as transcription of tetA. Thus, membrane attachment was not required to generate supercoil barriers in a wild-type (WT) Salmonella chromosome.
How does transcription remodel chromosome structure? X-ray crystal structures of RNA polymerase show that DNA enters RNA polymerase through one channel and RNA emerges from another (Perederina et al., 2004; Geszvain and Landick, 2005). Thus, transcription would either remove any previously established DNA attachments or move them along the DNA and out of the path of RNA polymerase. This would generate an impediment free zone for RNA synthesis and possibly pile up barriers near the transcription terminus. What does a transcription-generated domain look like? Two rules suggest a loop. First, any gene positioned between a pair of res sites always inhibits resolution once the gene is highly transcribed. Second, when a pair of res sites lies downstream of a highly transcribed gene, resolution is inhibited only when one res site is within 1 kb of the transcription terminator sequence of the highly transcribed gene (S. Deng and N.P. Higgins, unpubl. data). A loop that includes the transcribed sequence plus a little extra DNA (average about 500 bp) is the simplest model that explains this pattern.
A bonus in the Salmonella transcription experiments came from discovering the time-course for the formation and dissolution of a transcription domain. Using a short-lived resolvase discussed above, cells were induced to transcribe tetA by adding chlorotetracycline (CLT). The maximum impact of the transcription on inhibiting resolution required 20 min of cell growth. Conversely, when maximally induced cultures were washed free of CLT, barriers to resolution disappeared over a 20 min interval. This shows that domain formation and dissolution does not coincide with transcription; transcription domains persist for a length of time roughly equal to the time for a new wave of replication to reach dichotomously replicating chromosomes. However, once high transcription established a domain, it persisted, representing a type of topological memory.
Is transcription memory functionally useful?
New methods allow the analysis in variation of expression at the single cell level (Rosenfeld et al., 2005). The mean lac operon expression profile of growing cultures spans a several hundred-fold range. However, all cells do not respond alike, and in most individual cells (Fig. 2A), lac expression behaves like a switch that modulates between low and high expression states (Novick and Weiner, 1957). Ozbudak found cell populations to behave hysteretically over a broad range of inducer levels (Ozbudak et al., 2004). With E. coli cultures started from cells that were un-induced for lac expression (Fig. 2B, bottom panel), growth for 20 h in different inducer concentrations yielded reproducible bi-stable cell populations with high or low GFP expression. With cell cultures initiated from a maximally induced cell population (Fig. 2B, top panel), the same inducer concentrations resulted in higher mean cellular expression levels and bi-stable populations containing much higher frequencies of high expression cells. Hysteresis shows that bacterial cultures ‘remember’ the initial growth conditions. One mechanism contributing to hysterisis involves the LacY permease, which concentrates inducer by actively transporting it into cells. However, the formation of a stable free-running transcription domain may also contribute to efficient transcription persistence in cells that achieve induction.
Fig. 2. Hysteresis and bistability in single cell transcription profiles.
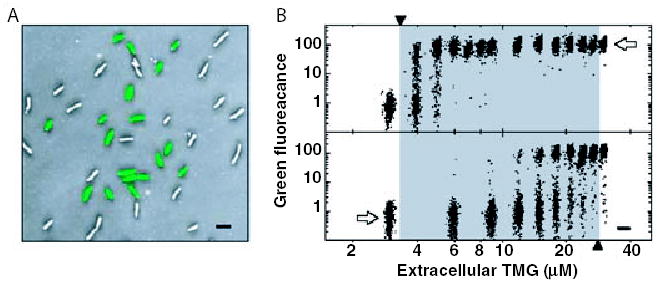
A. Overlayed green fluorescence and inverted phase-contrast images of cells that are initially un-induced for lac expression, then grown for 20 h in 18 μM thio-methylgalactoside (TMG), a non-metabolizable lactose analogue. The cells show a bimodal distribution of lac expression, with induced cells having over one hundred times the green fluorescence of uninduced cells.
B. A series of cell populations, initially un-induced (lower panel) or fully induced (upper panel) for lac expression, were grown 20 h in media containing various amounts of TMG. Scatter plots show log (green fluorescence) for about 1000 cells in each population. Each scatter plot is centred at a point indicating the underlying TMG concentration (Ozbudak et al., 2004). Figure reproduced with permission from the Nature Publishing Group.
Transcription memory may contribute to the expression of operons that lack a repressor control system. The bgl operon has no repressor but changes expression in response to DNA structure and TopoI and gyrase activity (DiNardo et al., 1982; Manna et al., 2001). There are also interesting genes like hipA (Moyed and Bertrand, 1983; Balaban et al., 2004), which cause a variable fraction of cells to stop growth and thereby survive exposure to antibiotics. Transcription memory could provide the epistatic mechanism to generate lineages with different expression states as minor cellular subpopulations.
Eukaryotic cells also move chromosome proteins with RNA polymerase and mark genes with transcription memory signals. One example in budding yeast is cohesin. Following DNA replication, chromosomes are decorated with cohesin, which has three important roles in cell division. In mitotic yeast cells, cohesin maintains a contact between sister chromatids after DNA synthesis, and it promotes attachment of chromosomes to the spindle. In meiosis, cohesin produces cohesion between sister chromatids that is necessary for efficient homologous recombination (Ross and Cohen-Fix, 2004). New studies using ChIP-on-chip technology showed that cohesins are dispersed by RNA polymerase from specific loading sites to positions near the 3′ end of transcribed genes (Glynn et al., 2004; Lengronne et al., 2004; Ross and Cohen-Fix, 2004). Another example is transcription-mediated nucleosome replacement. Histones deposited during DNA replication are naturally stable. However, if a Drosophila gene is expressed after replication, nucleosomes containing the histone H3 subunit disappear and are replaced with nucleosomes containing the histone variant H3.3 (Schwartz and Ahman, 2005). Third, the condensin proteins, closely related in structure to cohesins, mediate dosage compensation of sex chromosomes and serve as bookmarks for the inducible expression of heatshock genes (Xing et al., 2005).
Transcription neighbourhoods
How many transcription domains exist in the average bacterial cell? Deng tested the relationship between transcription activity and resolution efficiency in Salmonella (Deng et al., 2004). Using a strain with lacZ regulated from the tetA promoter with a WT TetR repressor, un-induced cultures gave 15 units of β-galactosidase activity and the recombination efficiency (1/2D) was comparable to previously analysed stochastic neighbourhoods with a mean 12 kb domain. Cells incubated with 5 μg ml−1 CLT generated >600 units of β-gal and the resolution efficiency of this region fell 10-fold. The measured RNA:DNA ratio of lacZ message for cells expressing 15 and 600 units of β-gal was 0.45 and 15 respectively (Wei et al., 2001). With the microarray scale it was possible to predict transcription effects genome-wide using RNA:DNA ratios for genes in Salmonella or E. coli.
Most (~70%) of the >4200 genes with measurable expression in E. coli or Salmonella give an RNA/DNA signal of less than one (Wei et al., 2001; Bernstein et al., 2002). In fact, all organisms for which RNA abundance has been studied including yeast, mouse and humans show the same genome-wide transcript structure; >70% of all eukaryotic protein genes contain steady-state RNA levels of less than one molecule per cell (Kuznetsov et al., 2002; Blewitt et al., 2004). In Gram-negative bacteria, most mRNAs turn over with a half-life of 3–4 min, so the transcription rate of 70% of the chromosome must be less than one message per 3 min, including 30% of the known essential genes. For 27% of E. coli genes, the steady-state RNA:DNA ratio is less than four, which would hinder resolution by less than 30%. Thus, 97% of bacterial genes have little or no transcriptional impact on domain structure. This confirms results obtained by French using electron microscopy to locate transcribing RNA polymerase molecules in E. coli DNA. The images from this study showed long stretches of DNA with an occasional single polymerase and very rare zones with ‘Christmas trees’ of multiple polymerases with short branches at the operon start and long branches near the operon 3′ end (French and Miller, 1989).
Only about 110–130 (2.5%) of E. coli genes located at 50 positions would inhibit resolution of a 14 kb interval by twofold or more, and only 25 protein encoding genes at about 15 locations have mRNA:DNA ratios of >10, which inhibits resolution by fivefold or more. High transcription zones in bacterial chromosomes are spaced at intervals (Allen et al., 2003; Lobner-Olesen et al., 2003; Jeong et al., 2004; Manna et al., 2004) and if the stable RNA genes, which include seven ribosomal operons and several tRNA gene clusters, are included, only ≈30 sites would show region-specific domains with fivefold or greater effects on resolution. Specific regions in Salmonella with highly transcribed non-essential genes were tested for agreement between the steady-state microarray RNA:DNA ratio and the resulting impact on resolution. The results validated microarray expression data for predicting where region-specific domains can be found [for specific examples see supplementary data in the study by Deng et al. (2004)].
Domain barrier elements
With a 400-domain chromosome and transcription causing formation of region-specific domains, two questions arise. First, what makes a domain? One hypothesis is that barriers are simple knots and tangles in DNA (Higgins et al., 1996), but the stability of domains made by transcription suggests that proteins are involved. A set of small structural proteins plus a handful of nucleotide cofactor-driven enzymes play key roles in chromosome behaviour. Small abundant ‘nucleoid-associated proteins’ (NAPs) act as architectural components that shape DNA. These proteins are often present at concentrations exceeding the level needed to saturate their known high-affinity sites, and they contribute to global chromosome structure by binding DNA ‘non-specifically’ (Crozat et al., 2005; Johnson et al., 2005). These proteins include HU, IHF, H-NS, StpA, FIS, LRP, SeqA and DPS. Each protein was originally discovered through its ability to condense DNA or to stimulate complex DNA transactions like transcription, transposition and site-specific recombination. Johnson et al. recently reviewed the biochemical properties as well as structural models of X-ray DNA-cocrystals for most of these proteins (Johnson et al., 2005).
Other than RNA and DNA polymerases and the type I and type II topoisomerases mentioned above, three catalytic enzymes channel chromosome motion in living cells. A three-protein complex of MukB, MukE and MukF (Muk-BEF) (Yamazoe et al., 1999) is a member of a highly conserved structural maintenance of chromosome (SMC) proteins. This family includes condensins and cohesins. MreB is an actin-like protein involved in moving chromosomal DNA after replication (van den Ent et al., 2001), and FtsK is an ATP-driven DNA conveyer (Aussel et al., 2002; Pease et al., 2005).
What are the most likely proteins for controlling formation of loop domains? Four proteins warrant special mention: DNA gyrase, MukBEF, FIS and HNS. DNA gyrase re-establishes a proper value of σ and it also controls the mean domain number. Certain gyrase hypomorphs have more than twice the WT number of domains (Staczek and Higgins, 1998). Stein confirmed this result with short-lived resolvases and showed that some gyrase hypomorphs limit resolution to segments much shorter than 12 kb (Stein et al., 2005). The topological impact of severe gyrase hypomorphs is most dramatic at the terminus of DNA replication. This can be seen as a large effect on Tn3 resolution in the region that lies adjacent to the dif site (Z. Pang, R. Chen and N.P. Higgins, submitted). Microarray studies also show strong effects of gyrase mutants on transcription profiles near the terminus (Jeong et al., 2004).
The E. coli and Salmonella MukBEF complex is a prime candidate for stabilizing loop domains in chromosomes. SMC proteins are conserved across the bacteria, archaea and eukaryotic kingdoms and this family includes cohesins and condensins. SMC proteins have globular amino- and carboxyl-terminal domains separated by long coiled-coil regions with a central flexible hinge (Jessberger and Gasser, 1998). SMC proteins form antiparallel, interwound dimers that hydrolyse ATP and bind DNA at each end (Hirano and Hirano, 1998; Hirano et al., 2001). Although not enough MukB is present in E. coli cells to account for 400 domains, this protein could account for the transcription-dependent domains. The movement of RNA polymerase would reduce negative supercoiling ahead of RNA polymerase, thereby creating a high affinity site for gyrase (Higgins and Cozzarelli, 1982; Moulin et al., 2005). A complex involving both MukBEF and gyrase could stabilize a loop and facilitate transcription by relieving the supercoil flux generated by high level transcription. In other organisms, SMC complexes like MukBEF are involved in chromosome condensation, dosage compensation of transcription, DNA repair and recombination (Cobbe and Heck, 2000; Holmes and Cozzarelli, 2000; Hirano, 2002).
Two NAPs make DNA loops in vitro. Electron microscopy studies show that H-NS and FIS both stabilize branched loops through cooperative interactions when mixed with plasmid DNA at appropriate concentrations (Dame et al., 2000; Schneider et al., 2001). Interestingly, both proteins have been implicated in global chromosome structure in several assays. H-NS influences both the frequency (Falconi et al., 1991) and the location (Swingle et al., 2004) of transposon insertions in the bacterial chromosome. H-NS is present at 10 000–20 000 copies per cell (Azam et al., 1999), and deletions of hns alter expression of about 5% of E. coli proteins (Hommais et al., 2001). FIS also has intriguing properties. In early log phase, FIS is abundant (30 000 molecules per cell), and it is the only NAP that disappears when cells enter stationary phase (<1000 molecules per cell). As domain barriers decrease in stationary phase, this is an intriguing correlation. FIS has also been implicated in the regulation of both the gyrA and the gyrB genes (Schneider et al., 1997, 1999; Keane and Dorman, 2003)
Finally, Hardy devised a genetic screen to find chromosomal domain barrier genes (Hardy and Cozzarelli, 2005). Modules with supercoil-responsive promoters were introduced into the E. coli chromosome at two locations. One promoter was linked to β-lactamase and the other controlled β-gal expression. Chromosome supercoiling relies on domains to maintain the mean chromosomal value of σ, but plasmids are single molecules without domain boundaries. Mutations were sought that alter chromosome supercoiling while leaving the plasmid σ-values at the normal mean value. Transposon mutagenesis uncovered five non-essential genes that appear to contribute to chromosome domain structure. The screen/selection uncovered H-NS and Fis mutations, which are in the category of expected genes (see above), plus three unexpected genes. These ‘new’ domain candidates are tktA, a transketolase, pgm, phosphoglucomutase and dskA, a multicopy suppressor of dnaK and mukB knockouts. Deletions of each of these genes showed reproducible effects on the supercoil-responsive reporters in the chromosome but did not change the superhelical density of pBR322-derived plasmids in the same cell. How these new genes alter chromosomal DNA structure has yet to be explained.
Conclusions
Some scientists have been reluctant to accept the notion that variable domain organization is good design for chromosomal DNA. Nonetheless, stochastic events in cell development are not rare (Rosenfeld et al., 2005), and variation is an inherent aspect of regulated gene expression in all tissues and organisms (Blewitt et al., 2004). The key property of bacterial chromosome compaction is pliability. Replication takes place within a DNA ‘factory’ and temporal rules for initiation are predictable for different growth rates. Transcription changes in response to internal and external environment, and during cotranscriptional translation, insertion of a protein into the membrane or periplasm will tether its gene to the cell membrane. As most of the DNA is rarely transcribed, workable units with variable divisions are acceptable most of the time. For the 3% of DNA with very active transcription, what is made during replication can be reworked to accommodate efficient gene transcription, even though life in the fast lane requires both RNA and DNA synthesis to occur simultaneously at different locations and at 10-fold different rates. Thus, a bacterial chromosome requires a statistical view in lieu of the highly ordered X-ray crystal structures of proteins that do the biochemical work on a chromosome.
Above the stochastic 10 kb domain level, long range order is beginning to come into focus. Using fluorescent in situ hybridization or fluorescent DNA binding proteins with specific DNA binding sites, specific genetic regions have been located and tracked in growing cell populations (Glaser et al., 1997; Gordon et al., 1997; Niki and Hiraga, 1998; Lemon and Grossman, 2000; Niki et al., 2000; Roos et al., 2001; Li et al., 2002; 2003; Lau et al., 2003) and by using random collision site-specific reactions catalysed by the phage λ Int and Xis proteins with attR and attL sites placed in the genome as unique locations, Garcia-Russell showed that some segments of chromosome do not often make intimate contact with each other (Garcia-Russell et al., 2004). In a similar study of E. coli with many endpoints, Valens et al. found a distance rule for lambda attL X attR recombination that indicated colocalization of very large regions (Valens et al., 2004). Furthermore, an amazing structure was revealed in C. crescentus using ‘tagged’ chromosomes with tet- and lac-operator modules bound with TetR-cyan fluorescent protein and LacI-yellow fluorescent protein (Lau et al., 2003). Foci were resolved in 112 different strains, using thousands of images for each strain (Viollier et al., 2004). Every tagged region moved to a replication factory and then to a post-replication position in sister cells that reflected genetic linkage to the origin or terminus of the chromosome. More recently, using chromosomes marked at two sites close to the dif site, Wang et al. demonstrated an intriguing genome organization in E. coli (Wang et al., 2005). For slow growing cells, sites separated by 150 kb move independently to the centrally located replication factory, and then to a unique location of the replicated nucleoids. Moreover, sites near the terminus in different replichores moved to opposite edges the nucleoid. These studies show that the left and right replichores, which have either near-continuous or discontinuous synthetic patterns, also have different traffic patterns in living cells. High fidelity chromosome positioning has been equated to geologic stratification or stabilization after deposition. For the chromosome, stability may be underpinned by numerous 10 kb domains (Breier and Cozzarelli, 2004). Whatever accounts for 400 domains and the longer range order in bacterial genomes, combinatorial mutation schemes will be required to pin down domain size phenotypes and more unpredictable players like tktA and pgm seem likely to emerge.
Acknowledgments
We thank Nick Cozzarelli and Christine Hardy for sharing data before publication. Lisa Postow generated the graph in Fig. 1C, and David Sherratt provided prepublication data on nucleoid structure in E. coli. We thank members of the Higgins lab for comments and criticism of the manuscript. Work in the NPH lab was supported by grants from the National Institutes of Health – GM33143 and the National Science Foundation – MCB9122048.
Footnotes
The term nucleoid is also used in reference to a subcellular region of a bacterium that stains with DNA-binding drugs like DAPI (Bohrmann et al., 1991; Cunha et al., 2001).
Note added in proof
In the brief time since this manuscript was submitted, two papers appeared with a focus on the long-range organization of the E. coli chromosome. 1. Bates, D., and Kleckner, N. (2005) Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121: 899–911. 2. Elmore, S., Muller, M., Vischer, N., Odijk, T., and Woldringh, C. (2005) Single-particle tracking of oriC-GFP fluorescent spots during chromosome segregation in Escherichia coli. J Struct Biol 136: 53–66.
References
- Allen TE, Herrgard MJ, Liu M, Qiu Y, Glasner JD, Blattner FR, Palsson BO. Genome-scale analysis of the uses of the Escherichia coli genome: model-driven analysis of heterogeneous data sets. J Bacteriol. 2003;185:6392–6399. doi: 10.1128/JB.185.21.6392-6399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch, I. (2005) Control of transcription termination and antitermination. In The Bacterial Chromosome Higgins, N.P. (ed.). Washington, DC: American Society for Microbiology Press, pp. 311–326.
- Aussel L, Barre FX, Aryoy M, Stasiak A, Stasiak AZ, Sherratt DJ. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Bauer WR, Crick FHC, White JH. Super-coiled DNA. Sci Am. 1980;243:100–113. [PubMed] [Google Scholar]
- Bernstein JA, Khodursky AB, Lin PH, Lin-Chao SL, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA. 2002;99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt ME, Chong S, Whitelaw E. How the mouse got its spots. Trends Genet. 2004;20:550–554. doi: 10.1016/j.tig.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Bliska JB, Cozzarelli NR. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987;194:205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Bohrmann B, Villiger W, Johansen R, Kellenberger E. Coralline shape of the bacterial nucleoid after cryofixation. J Bacteriol. 1991;173:3149–3158. doi: 10.1128/jb.173.10.3149-3158.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier AM, Cozzarelli NR. Linear ordering and dynamic segregation of the bacterial chromosome. Proc Natl Acad Sci USA. 2004;101:9175–9176. doi: 10.1073/pnas.0403722101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbe N, Heck MM. Review: SMC’s in the world of chromosome biology-from prokaryotes to higher eukaryotes. J Struct Biol. 2000;129:123–143. doi: 10.1006/jsbi.2000.4255. [DOI] [PubMed] [Google Scholar]
- Crozat E, Philippe N, Lenski RE, Geiselmann J, Schneider D. Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics. 2005;169:523–532. doi: 10.1534/genetics.104.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha S, Woldringh CL, Odijk T. Polymer-mediated compaction and internal dynamics of isolated Escherichia coli nucleoids. J Struct Biol. 2001;136:53–66. doi: 10.1006/jsbi.2001.4420. [DOI] [PubMed] [Google Scholar]
- Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Stein RA, Higgins NP. Transcription-induced barriers to supercoil diffusion in the Salmonella typhimurium chromosome. Proc Natl Acad Sci USA. 2004;101:3398–3403. doi: 10.1073/pnas.0307550101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S, Voelkel KA, Sternglanz R, Reynolds AE, Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- Drlica K. Control of bacterial supercoiling. Mol Microbiol. 1992;6:425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- Drlica K, Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin J, Losick R. Does RNA polymerase help drive chromosome segregation in bacteria? Proc Natl Acad Sci USA. 2002;99:14089–14094. doi: 10.1073/pnas.182539899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- Falconi M, McGovern V, Gualerzi C, Hillyard D, Higgins NP. Mutations altering chromosomal protein H-NS induce mini-Mu transposition. New Biol. 1991;3:615–625. [PubMed] [Google Scholar]
- Flynn JM, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci USA. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Miller OL. Transcription mapping of the Escherichia coli chromosome by electron microscopy. J Bacteriol. 1989;171:4207–4216. doi: 10.1128/jb.171.8.4207-4216.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell BE, Baker TA, Kornberg A. Complete enzymatic replication of plasmids containing the origin of the Escherichia coli chromosome. J Biol Chem. 1986;261:5616–5624. [PubMed] [Google Scholar]
- Garcia-Russell N, Harmon TG, Le TQ, Amaladas NH, Mathewson RD, Segall AM. Unequal access of chromosomal regions to each other in Salmonella: probing chromosome structure with phage 1 integrase-mediated long-range rearrangements. Mol Microbiol. 2004;52:329–344. doi: 10.1111/j.1365-2958.2004.03976.x. [DOI] [PubMed] [Google Scholar]
- Geszvain, K., and Landick, R. (2005) The structure of bacterial RNA polymerase. In The Bacterial Chromosome Higgins, N.P. (ed.). Washington, DC: American Society for Microbiology Press, pp. 283–296.
- Giorno R, Hecht RM, Pettijohn D. Analysis by isopycnic centrifugation of isolated nucleoids of Escherichia coli. Nucleic Acids Res. 1975;2:1559–1567. doi: 10.1093/nar/2.9.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Glaser P, Sharpe ME, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:e259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon GS, Sitnikov D, Webb CD, Teleman A, Straight ARL, Murray AW, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- Hardy, C., and Cozzarelli, N.R. (2005) A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol doi:10.1111/j.1365-2958.2005.04799.x. [DOI] [PubMed]
- Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem. 2002a;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc Natl Acad Sci USA. 2002b;99:3440–3445. doi: 10.1073/pnas.052707199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, N.P. (1999) DNA supercoiling and its consequences for chromosome structure and function. In Organization of the Prokaryotic Genome, Vol. 1. Charlebois, R.L. (ed.). Washington, DC: ASM, pp. 189–202.
- Higgins NP, Cozzarelli NR. The binding of gyrase to DNA: analysis by retention to nitrocellulose filters. Nucleic Acids Res. 1982;10:6833–6847. doi: 10.1093/nar/10.21.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, N.P., and Vologodskii, A. (2004) Topological behavior of plasmid DNA. In Plasmid Biology Phillips, G., and Funnell, B. (eds). Washington, DC: American Society for Microbiology Press, pp. 181–201.
- Higgins NP, Yang X, Fu Q, Roth JR. Surveying a supercoil domain by using the γδ resolution system in Salmonella typhimurium. J Bacteriol. 1996;178:2825–2835. doi: 10.1128/jb.178.10.2825-2835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, N.P., Deng, S., Pang, Z., Stein, R., Champion, K., and Manna, D. (2005) Domain behavior and supercoil dynamics in bacterial chromosomes. In The Bacterial Chromosome Higgins, N.P. (ed.). Washington, DC: American Society for Microbiology Press, pp. 133–153.
- Hingorani, M.M., and O’Donnell, M. (2005) DNA elongation. In The Bacterial Chromosome Higgins, N.P. (ed.). Washington, DC: American Society for Microbiology Press, pp. 193–216.
- Hiraga S, Niki H, Ogura T, Ichinose D, Mori H, Ezaki B, Jaffe A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 2002;16:399–414. doi: 10.1101/gad.955102. [DOI] [PubMed] [Google Scholar]
- Hirano M, Hirano T. ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J. 1998;17:7139–7148. doi: 10.1093/emboj/17.23.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Anderson DE, Erickson HP, Hirano T. Bimodal activation of SMC ATPase by intra- and inter-milecular interactions. EMBO J. 2001;20:3238–3250. doi: 10.1093/emboj/20.12.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes VF, Cozzarelli NR. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc Natl Acad Sci USA. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, et al. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- Jeong KS, Ahn J, Khodursky AB. Spatial patterns of transcriptional activity in the chromosome of Escherichia coli. Genome Biol. 2004;5:R86. doi: 10.1186/gb-2004-5-11-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger RFC, Gasser SM. Chromosome dynamics: the SMC protein family. Curr Opin Genet Dev. 1998;8:254–259. doi: 10.1016/s0959-437x(98)80149-4. [DOI] [PubMed] [Google Scholar]
- Johnson, R.C., Johnson, L.M., Schmidt, J.W., and Gardner, J.F. (2005) The major nucleoid proteins in the structure and function of the E. coli chromosome. In The Bacterial Chromosome Higgins, N.P. (ed.). Washington, DC: American Society for Microbiology Press, pp. 65–132.
- Kavenoff R, Ryder O. Electron microscopy of membrane-associated folded chromosomes of Escherichia coli. Chromosoma. 1976;55:13–25. doi: 10.1007/BF00288323. [DOI] [PubMed] [Google Scholar]
- Keane OM, Dorman CJ. The gyr genes of Salmonella enterica serovar Typhimurium are repressed by the factor for inversion stimulation, Fis. Mol Genet Genomics. 2003;270:56–65. doi: 10.1007/s00438-003-0896-1. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PRH, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kuzminov, A., and Stahl, F.W. (2005) Overview of homologous recombination and repair machines. In The Bacterial Chromosome Higgins, N.P. (ed.). Washington, DC: American Society for Microbiology Press, pp. 349–368.
- Kuznetsov VA, Knott GD, Bonner RF. General statistics of stochastic process of gene expression in eukaryotic cells. Genetics. 2002;161:1321–1332. doi: 10.1093/genetics/161.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD. Movement of replicating DNA through a stationary replisome. Mol Cell. 2000;6:1321–1330. doi: 10.1016/s1097-2765(00)00130-1. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Shihori Yokobayashi S, Kelly GP, Itoh T, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sergueev K, Austin S. The segregation of the Escherichia coli origin and terminus of replication. Mol Microbiol. 2002;46:985–995. doi: 10.1046/j.1365-2958.2002.03234.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Youngren B, Sergueev K, Austin S. Segregation of the Escherichia coli chromosome terminus. EMBO J. 2003;50:825–834. doi: 10.1046/j.1365-2958.2003.03746.x. [DOI] [PubMed] [Google Scholar]
- Lindow JC, Tritton RA, Grossman AD. Structural maintenance of chromosomes protein of Bacillus subtilis affects supercoiling in vivo. J Bacteriol. 2002;184:5317–5322. doi: 10.1128/JB.184.19.5317-5322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobner-Olesen A, Marinus MG, Hansen FG. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc Natl Acad Sci USA. 2003;100:4672–4677. doi: 10.1073/pnas.0538053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch AS, Wang JC. Anchoring of DNA to the bacterial cytoplasmic membrane through cotranscriptional synthesis of polypeptides encoding membrane proteins or proteins for export: a mechanism of plasmid hypernegative supercoiling in mutants deficient in DNA topoisomerase I. J Bacteriol. 1993;175:1645–1655. doi: 10.1128/jb.175.6.1645-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaloe, O., Schaechter, M., and Kjeldgaard, N.O. (1966) The bacterial nucleus. In Control of Macromolecular Synthesis Maaloe, O., and Kjeldgaard, N.O. (eds). New York and Amsterdam: W.A. Benjamin, pp. 188–197.
- McGovern V, Higgins NP, Chiz S, Jaworski A. H-NS over-expression induces an artificial stationary phase by silencing global transcription. Biochimie. 1994;76:1030–1040. doi: 10.1016/0300-9084(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Manna D, Wang X, Higgins NP. Mu and Is1 transposition exhibits strong orientation bias at the E. coli bgl locus. J Bacteriol. 2001;183:3328–3335. doi: 10.1128/JB.183.11.3328-3335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna D, Breier AM, Higgins NP. Microarray analysis of transposition targets in Escherichia coli: the impact of transcription. Proc Natl Acad Sci USA. 2004;101:9780–9785. doi: 10.1073/pnas.0400745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R, Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA super-coiling. Cell. 1983;34:105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Moulin L, Rahmouni AR, Boccard F. Topological insulators inhibit diffusion of transcriptioninduced positive supercoils in the chromosome of Escherichia coli. Mol Microbiol. 2005;55:601–610. doi: 10.1111/j.1365-2958.2004.04411.x. [DOI] [PubMed] [Google Scholar]
- Moyed HS, Bertrand KP. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Hiraga S. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 1998;12:1036–1045. doi: 10.1101/gad.12.7.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram M, Shipstone E, Halford SE. Synapsis by Tn3 resolvase: speed and dependence on DNA super-coiling. Biochem Soc Trans. 1994;22:303. doi: 10.1042/bst022303s. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Lim HN, Shraiman BI, van Oudenaarden A. Multistability in the lactose utilization network of Escherichia coli. Nature. 2004;427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- Pease PJ, Levy O, Cost GJ, Gore J, Ptacin JL, Sherratt D, et al. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel – structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Peter, B.J. (2000) The structure of replicating DNA in Escherichia coli PhD Thesis. University of California, Berkeley, CA, USA.
- Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettijohn, D.E. (1996) The nucleoid. In Escherichia coli and Salmonella, Vol. 1. Neidhardt, F.C. (ed.). Washington, DC: American Society for Microbiology Press, pp. 158–166.
- Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos M, Van Geel A, Aarsman M, Veuskens J, Woldringh CL, Nanninga N. The replicated ftsQAZ and minB chromosomal regions of Escherichia coli segregate on average in line with ncleoid movement. Mol Microbiol. 2001;39:633–640. doi: 10.1046/j.1365-2958.2001.02263.x. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JM, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- Ross KE, Cohen-Fix O. Cohesins slip sliding away. Nature. 2004;430:520–521. doi: 10.1038/430520b. [DOI] [PubMed] [Google Scholar]
- Sawitzke JA, Austin S. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc Natl Acad Sci USA. 2000;97:1671–1676. doi: 10.1073/pnas.030528397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheirer K, Higgins NP. Transcription induces a supercoil domain barrier in bacteriophage Mu. Biochimie. 2001;83:155–159. doi: 10.1016/s0300-9084(00)01215-3. [DOI] [PubMed] [Google Scholar]
- Schneider R, Travers A, Muskhelishvili G. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol Microbiol. 1997;26:519–530. doi: 10.1046/j.1365-2958.1997.5951971.x. [DOI] [PubMed] [Google Scholar]
- Schneider R, Travers A, Kutateladze T, Muskhelishvili G. A DNA architectural protein couples cellular physiology and DNa topology in Escherichia coli. Mol Microbiol. 1999;24:953–964. doi: 10.1046/j.1365-2958.1999.01656.x. [DOI] [PubMed] [Google Scholar]
- Schneider R, Lurz R, Luder G, Tolksdorf C, Travers A, Muskhelishvili G. An acrhitectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res. 2001;29:5107–5114. doi: 10.1093/nar/29.24.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BE, Ahman K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden RR, Pettijohn DE. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc Natl Acad Sci USA. 1981;78:224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staczek P, Higgins NP. DNA gyrase and topoisomerase IV modulate chromosome domain size in vivo. Mol Microbiol. 1998;29:1435–1448. doi: 10.1046/j.1365-2958.1998.01025.x. [DOI] [PubMed] [Google Scholar]
- Stark, M.W., and Boocock, M.R. (1995) Topological selectivity in site-specific recombination. In Mobile Genetic Elements, Vol. 58. Sherratt, D. (ed.). Oxford: IRL Press, p. 179.
- Steck TR, Drlica K. Bacterial chromosome segregation: evidence for DNA gyrase involvement in decatenation. Cell. 1984;36:1081–1088. doi: 10.1016/0092-8674(84)90058-8. [DOI] [PubMed] [Google Scholar]
- Stein R, Deng S, Higgins NP. Measuring chromosome dynamics on different timescales using resolvases with varying half-lives. Mol Microbiol. 2005;56:1049–1061. doi: 10.1111/j.1365-2958.2005.04588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, B.S. (2005) Excision repair and bypass. In The Bacterial Chromosome Higgins, N.P. (ed.). Washington, DC: American Society for Microbiology Press, pp. 431–447.
- Swingle B, O’Carroll M, Haniford D, Derbyshire KM. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol Microbiol. 2004;52:1055–1067. doi: 10.1111/j.1365-2958.2004.04051.x. [DOI] [PubMed] [Google Scholar]
- Travers A, Muskhelishvili G. DNA supercoiling – a global transcriptional regulator for enterobacterial growth? Nat Rev Microbiol. 2005;3:150–160. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.J., Possoz, C., and Sherratt, D. (2005) Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev (in press). [DOI] [PMC free article] [PubMed]
- Wei Y, Lee JM, Richmond C, Blattner FR, Rafalski JA, LaRossa RA. High density microarray-mediated gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbrock H, Ussery DW. Chromatin architecture and gene expression in Escherichia coli. Genome Biol. 2004;5:252.251–252.255. doi: 10.1186/gb-2004-5-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A, Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972;71:127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol Microbiol. 2003;49:1463–1475. doi: 10.1046/j.1365-2958.2003.03643.x. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, et al. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- Yamazoe M, Onogi T, sunako Y, Niki H, Yamanaka K, Ichimura T, Hiraga S. Complex formation of MukB, MukE, and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 1999;18:5873–5884. doi: 10.1093/emboj/18.21.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich EL, Khodursky AB, Cozzarelli NR. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich EL, Arkady BK, Bachellier S, Chen D, Lilley DM, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA super-coiling in Escherichia coli. J Biol Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]


