Abstract
One of the phenotypes of mice with targeted disruption of the uncoupling protein-2 gene (Ucp2−/−) is greater macrophage phagocytic activity and free radical production, resulting in a striking resistance to infectious microorganisms. In this study, the molecular mechanisms of this enhanced immune response were investigated. We found that levels of nitric oxide measured in either plasma or isolated macrophages from Ucp2−/− mice are significantly elevated in response to bacterial lipopolysaccharide challenge compared with similarly treated Ucp2+/+ mice. Likewise, expression of inducible nitric-oxide synthase and inflammatory cytokines is higher in Ucp2−/− mice in vivo and in vitro. Key steps in the activation cascade of nuclear factor (NF)-κB, including IκB kinase and nuclear translocation of NF-κB subunits, are all remarkably enhanced in Ucp2−/− mice, most notably even under basal conditions. The elevated basal activity of IκB kinase in macrophages from Ucp2−/− mice can be blocked by cell-permeable inhibitors of superoxide and hydrogen peroxide generation, but not by a specific inhibitor for inducible nitric-oxide synthase. Isolated mitochondria from Ucp2−/− cells produced more superoxide/hydrogen peroxide. We conclude that mitochrondrially derived reactive oxygen from Ucp2−/− cells constitutively activates NF-κB, resulting in a “primed” state to both potentiate and amplify the inflammatory response upon subsequent stimulation.
Uncoupling protein (UCP)1-2 is a mitochondrial inner membrane carrier protein that was discovered through its homology to the brown fat UCP1 (1). Whereas UCP1 has been clearly established as the molecular mediator of non-shivering thermogenesis (reviewed in Ref. 2), the function of UCP2 remains somewhat of an enigma. Several features of UCP2 led us to initially propose that it was a bona fide uncoupling protein involved in the dissipation of excess metabolic fuel as heat. These aspects of UCP2 included its structural resemblance to UCP1, its ability to uncouple respiration in model assay systems, and a chromosomal location to a region with genetic linkage to obesity and hyperinsulinemia (1, 3). However, although Ucp2 mRNA is expressed in a broad array of tissues in humans and rodent models (3, 4), including metabolically important organs, it exists in minute amounts compared with the level of UCP1 in brown fat. Moreover, it is present in cell types such as pancreatic β-cells, lymphocytes, and neurons that are not typically associated with thermogenesis (5–7). These and other features of UCP2 (8), together with the critical finding that targeted disruption of the Ucp2 gene did not result in obesity, cold sensitivity, or demonstrable differences in coupling efficiency in isolated mitochondria (9–11), strongly suggested a functionally distinct role for this protein.
One of the phenotypes of Ucp2−/− mice was a striking resistance to infectious microorganisms associated with greater macrophage phagocytic activity and free radical production (9). We showed that macrophages from Ucp2−/− mice produced reactive oxygen species (ROS) using the nitro blue tetrazolium reduction assay and hypothesized that UCP2 plays a role in host immune defense by regulating the production of ROS. A broad definition of ROS can include reactive oxygen as well as nitrogen species, and macrophages are known to generate reactive nitrogen species (RNS) such as nitric oxide (NO) in addition to superoxide. However, the nitro blue tetrazolium assay cannot distinguish between the ROS and RNS generated (12) or the mechanisms responsible for it. In the present study we explored the molecular basis of this amplified immune response in Ucp2−/− mice using lipopolysaccharide (LPS), a membrane glycolipid of Gram-negative bacteria that is a well-established activator of both the “reactive oxygen” and “reactive nitrogen” generating pathways (13, 14). We show that in response to LPS challenge, Ucp2−/− mice produce large amounts of nitric oxide and cytokines both in vivo and from isolated macrophages in vitro as compared with wild-type mice.
As one of the major transcription factors responsible for increasing the expression of iNOS and inflammatory cytokines, we find that in spleen or isolated macrophages of Ucp2−/− mice, the NF-κB system is activated at several steps in its pathway (importantly, even under basal conditions). NF-κB is most frequently composed of two DNA-binding subunits, p50 and p65, which exist as an inactive form in the cytoplasm bound to an inhibitory protein (IκB). Almost all of the signals that lead to activation of NF-κB converge on IκB kinase (IKK). Activation of IKK leads to the phosphorylation of IκBs, which targets them for ubiquitination and degradation, thus allowing the NF-κB dimer to translocate to the nucleus to regulate target gene expression (15). In the present study we show that IKK activity is elevated in spleen as well as isolated macrophages from Ucp2−/− mice, even under basal conditions, and isolated mitochondria from Ucp2−/− spleen produce significantly more hydrogen peroxide as compared with wild-type mice. Together with the finding that cell-permeable preparations of the ROS inhibitors superoxide dismutase and catalase could block the basal elevation of IKK activity in isolated macrophages, our results support the idea that deficiency of UCP2 results in greater release from mitochondria (16), leading in turn to persistent activation of NF-κB and exacerbation of subsequent inflammatory challenges.
EXPERIMENTAL PROCEDURES
Animals and Sample Collection
Generation of Ucp2−/− mice was described previously (9). Twelve-week-old male mice were used that had been backcrossed from the mixed C57Bl/6J-129Sv/J strain background to C57Bl/6J mice for 10 generations. All animal experiments were approved by the Institutional Animal Care and Use Committees of Duke University and CIIT Centers for Health Research. LPS (Sigma) was administered to mice at a dose of 4 μg/g or 15 μg/g body weight (bw) (17) in 100 μl of PBS by intraperitoneal injection or given PBS vehicle. Blood samples were collected 6 or 16 h later under pentobarbital anesthesia (50 μg/g bw). Plasma was separated in Ficoll-Paque Plus (Amersham Biosciences), and plasma protein was quantified by Bio-Rad Protein Assay (18). Plasma NO levels and cytokines were measured as described below.
For isolation of resident peritoneal macrophages, mice were anesthetized with pentobarbital, and the peritoneal cavity was gently flushed three times with chilled Dulbecco’s modified Eagle’s medium supplemented with 1% fetal calf serum (Highveld). Lavage fluid was pooled together (5–7 ml), centrifuged at 500 × g for 8 min, and resuspended in cold Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 25 mm Hepes, and 10 mm l-glutamine. Cells were seeded into either 24-well plates for measurement of NO production or 6-well plates for Western blot and kinase assays. Following incubation at 37 °C in a 5% CO2 atmosphere for times ranging from 4 h to overnight and removal of any non-adherent cells, the remaining cells (>90% macrophages) were treated with 1 μg/ml LPS or PBS for the indicated times (19) and treated overnight with 200 units/ml polyethylene glycol-superoxide dismutase and 400 units/ml polyethylene glycol-catalase or 50 μm 1400W as indicated in specific experiments. Supernatants were collected for measuring NO levels, and cells were lysed for measuring iNOS protein expression. The number of cells per well was determined by a modified method of Mosmann (20) (Promega), and NO in macrophage supernatants from the Griess assay was normalized to cell numbers.
Subcellular protein fractions were prepared from freshly isolated spleen by homogenization at 4 °C in TES buffer (50 mm Tris, pH 7.4, 1 mm EDTA, and 250 mm sucrose supplemented with 2 μg/ml aprotinin, 1 mm benzamidine, 1 μg/ml pepstatin A, 2 μg/ml leupeptin, 5 μg/ml bestatin, 50 μg/ml TPCK, and 0.1 mm phenylmethylsulfonyl fluoride) or lysis buffer for IKK activity (21). Samples were then centrifuged at 9000 × g for 10 min. Pellets were used for nuclear protein isolation by the NE-PER method (Pierce). Supernatants were further centrifuged at 100,000 × g for 1 h. The resulting supernatants were collected as the cytoplasmic fraction, and membrane proteins in the pellet were obtained by resuspension with 50 μl of lysis buffer as described previously (22).
Nitrite/Nitrate Determination using Griess Assay
NO formation in plasma or macrophage culture supernatants was detected as nitrite/nitrate by the method of Green et al. (23), with slight modification (24).
Western Blot Analysis
All primary antibodies were purchased from Santa Cruz Biotechnology, Inc., with the exception of antibodies against iNOS and β-actin, which were from Sigma. Equal amounts of protein from each sample were separated in 10% Tris-glycine mini gels (Invitrogen) and transferred to nitrocellulose membranes (Amersham Biosciences). The membranes were blocked with 5% nonfat milk in wash buffer (PBS/0.1% Tween 20) for 30 min at 4 °C and probed with primary antibodies overnight at dilutions recommended by the suppliers. Membranes were washed three times with wash buffer (15 min/wash) before incubation with second antibodies (1 h at room temperature). Blots were scanned and visualized with a PhosphorImager (Typhoon; Amersham Biosciences) and quantified by densitometry (Quantity One).
Northern Blot Analysis
Total RNA was isolated from spleen with Tri Reagent (Molecular Research Center, Inc). A total of 10–25 μg of total RNA was fractionated by electrophoresis in 1.2% agarose gels, transferred to nylon membranes, and hybridized to 32P-labeled probes as previously described (25). Images were visualized and quantified in a PhosphorImager. All measurements were normalized to the internal standard cyclophilin or 18S RNA.
Cytokine Measurements by ELISA
Plasma samples for ELISA were collected as described under “Animals and Sample Collection.” Detection of IFN-γ and TNF-α was performed using sandwich ELISA reagents from Chemicon (Temecula, CA).
Microarray Hybridization
Atlas Mouse Inflammatory/Stress cDNA expression microarray analyses were performed according to the manufacturer’s instructions (Clontech). Ucp2+/+ and Ucp2−/− mice for this experiment were treated with 15 μg LPS/g bw or left untreated. All hybridizations were carried out in quadruplicate. Before labeling, RNA samples from two biological replicates were pooled in equal amounts. Briefly, total RNA was converted to [α-32P]dATP-labeled cDNA probe using Moloney murine leukemia virus reverse transcriptase and the Atlas Mouse Inflammatory/Stress CDS primer mix. The 32P-labeled cDNA probe was purified using NucleoSpin columns (Clontech). The membrane was pre-hybridized with Expresshyb (Clontech) for 60 min at 68 °C, followed by hybridization with probe overnight at 68 °C. The arrays were washed four times in 2× SSC/0.1% SDS (30 min each) and two times in 0.1× SSC/0.1% SDS. The arrays were then sealed in plastic wrap and exposed to an Amersham Biosciences PhosphoImage screen or to radiographic film. The images were analyzed densitometrically using AtlasImage software (Clontech). The gene expression intensities were normalized with a total of nine housekeeping genes present on the array. Comparisons between sample groups were made by Student’s t test. p < 0.05 was considered statistically significant.
Electrophoretic Mobility Shift Assays (EMSAs)
Nuclear extracts were prepared as described above and either used immediately or stored at −70 °C. Single-stranded oligonucleotides (Integrated DNA Technologies, Inc.) were annealed by polymerase chain reaction to form consensus oligomers containing the underlined NF-κB-binding site (5′-ATATGAGGGGACTTTCCCAGG-3′ and 5′-ATACCTGGGAAAGTCCCCTCA-3′) and two other oligomers containing the underlined iNOS promoter-specific NF-κB-binding site (NFκB(d), 5′-ACTGGGGACTCTCCCTTTGGG-3′ and 5′-TTCCCAAAGGGAGAGTCCCCA-3′; NFκB(u), 5′-GCTAGGGGGATTTTCCCTCTC-3′ and 5′-GAGAGAGGGAAAATCCCCCTA3′) and their mutant oligonucleotides (italicized) (5′-ACTGCTCACTCTCCCTTTGGG-3′ and 5′-TTCCCAAAGGGAGAGTGAGCA3′; 5′-GCTAGGCTCATTTTCCCTCTC-3′ and 5′-GAGAGAGGGAAAATGAGCCTA3′, respectively) (26). EMSA was performed by incubating nuclear extracts for 20 min at room temperature with 8 fmol of 32P-labeled double-stranded oligonucleotides. Antibodies against NF-κB p50 or p65 were added for the supershift for 30 min at 4 °C before the incubation with oligonucleotides. As a control for nonspecific binding, some samples were processed using antisera against cytochrome c. The DNA-protein complex was separated in 6% native polyacrylamide gels in 0.5% Tris-glycine (Invitrogen) and dried under vacuum.
IκB Kinase Assay
Kinase activity for IKK was measured as previously described (21). Cytoplasmic protein from spleen (1.5 mg) or total protein from macrophage (150 μg) was incubated with 0.75 or 0.4 μg of anti-IKKα/β antibody, respectively, in lysis buffer as previously described (21) for 30 min at 4 °C and then isolated with protein G-Sepharose beads (Sigma) after 3 h of incubation. The immunoprecipitates were washed twice with lysis buffer and once with kinase buffer (20 mm Hepes, pH 7.5, 10 mm MgCl2, 20 mM β-glycerophosphate, and 1 mm Na3VO4). The washed beads were incubated with 30 μl of kinase buffer containing 1.5 μg of IκBα as substrate, 20 μm ATP, 2 mm dithiothreitol, and 7.5 μCi of [γ-32P]ATP for 20 min at 30 °C. The reactions were stopped by addition of 4× SDS-PAGE sample buffer, and proteins were resolved by SDS-14% PAGE. The gels were dried and autoradiographed. Quantification of the radioactive bands was performed with a PhosphorImager.
Measurement of Mitochondrial H2O2 Production
Spleen mitochondria were isolated (Sigma MITO-ISO1) and resuspended in an assay medium based on the method of St-Pierre et al. (27), containing 3 mm Hepes, pH 7.5, 250 mm sucrose, 120 mm KCl, 5 mm KH2PO4, 1 mM ATP, 1 mm EGTA, and 0.3% bovine serum albumin. Mitochondrial protein was determined using Bio-Rad Protein Assay (18). H2O2 levels were measured using Amplex® Red (Hydrogen Peroxide/Peroxidase Assay Kit; Molecular Probes). For each of the three separate experiments conducted, equal amounts of mitochondria (350 μg/100 μl) from two mice of each genotype were incubated at room temperature in assay medium under state 4 conditions in the presence of the Amplex® Red reagent mixture. Mitochondrial production of H2O2 was measured at 10-min intervals over a 30-min period using a SPECTRAmax M2 spectrofluorometer (Molecular Devices). Calibration curves for H2O2 concentrations were generated as previously described (27) in the presence and absence of spleen mitochondria. Independent confirmation of the amount of mitochondria in each reaction sample was verified by quantification of mitochondrial DNA after H2O2 measurement. H2O2 generation was linear through 30 min, and the data are expressed as mol·mg−1 min−1.
Statistical Analysis
Data are expressed as mean ± S.E. A one-way analysis of variance followed by a Tukey’s or Dunnett’s multiple comparisons test was used to compare treated groups with controls. A p value of ≤0.05 was considered significant.
RESULTS
iNOS-derived NO and Cytokine Enhancement in Ucp2−/− Mice upon LPS Treatment
Our original report on Ucp2−/− mice described their exaggerated immune response in vivo and free radical production by macrophages (9), but importantly, it did not distinguish between the relative contributions of ROS and RNS. The first experiments measured the changes in expression patterns of key ROS- and RNS-generating enzymes in spleen from Ucp2+/+ and Ucp2−/− mice, including NADPH oxidase, cyclooxygenase 2, and iNOS. Following LPS treatment, the subcellular distribution of the NADPH oxidase subunits p47 and p67 shifted toward greater membrane association to indicate its activation (28) (Table I), and superoxide levels were increased as measured independently by fluorescence microscopy (data not shown), but there were no significant differences between the two genotypes. Also shown in Table I, the expression of cyclooxygenase 2 was identical in Ucp2+/+ and Ucp2−/− mice and was not affected by LPS. However, Fig. 1A illustrates that upon LPS treatment, total NO (nitrite and nitrate) in plasma from Ucp2−/− mice was increased 2.2 ± 0.8-fold above that produced by similarly treated Ucp2+/+ mice. This difference between the genotypes in the generation of reactive nitrogen species was further reflected by a much higher level of iNOS gene expression in spleen of Ucp2−/− mice as measured by Northern (Fig. 1B) and Western blotting (Fig. 1C). In both genotypes, iNOS expression was not detectable under basal conditions by these methods (data not shown). In addition, macrophages isolated from the two genotypes and stimulated by LPS in vitro gave identical responses to those observed in spleen. As shown in Fig. 2A and B, generation of NO and the induction of iNOS expression were more robust in the cells from Ucp2−/− mice compared with those from Ucp2+/+ mice. Pre-treatment of the cells with the specific iNOS inhibitor 1400W completely prevented NO production (Fig. 2A), confirming that iNOS is the sole source of this NO.
Table I. Subcellular distribution of NADPH oxidase components p47phox and p67phox and cyclooxygenase 2 (COX2) mRNA levels in spleen.
Protein expression data are shown as the fold change relative to Ucp2+/+ without LPS treatment, which is set to 1.00 (italicized) (n = 3). Northern blotting data for RNA are shown as the fold change relative to Ucp2+/+ littermates setting to 1.00 (italicized) (n = 3).
| NADPH oxidase (protein)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p47phox |
p67phox |
|||||||||
| Membrane
|
||||||||||
| LPS (4 μg/g bw, 6 h)
|
Cytosol
|
Membrane
|
Cytosol
|
COX2 (mRNA)
|
||||||
| Genotypes | − | + | − | + | − | + | − | + | − | + |
| Ucp2+/+ | 1.00 ± 0.02 | 1.78 ± 0.02 | 1.00 ± 0.08 | 2.29 ± 0.14 | 1.00 ± 0.05 | 1.95 ± 0.02 | 1.00 ± 0.01 | 1.42 ± 0.02 | 1.00 ± 0.05 | 1.00 ± 0.07 |
| Ucp2−/− | 1.00 ± 0.01 | 1.49 ± 0.02 | 1.08 ± 0.08 | 1.98 ± 0.15 | 1.01 ± 0.01 | 2.09 ± 0.03 | 1.04 ± 0.01 | 1.75± 0.02 | 0.96 ± 0.04 | 1.09 ± 0.11 |
Fig. 1. Nitric oxide production is elevated in Ucp2−/− mice upon stimulation by LPS.
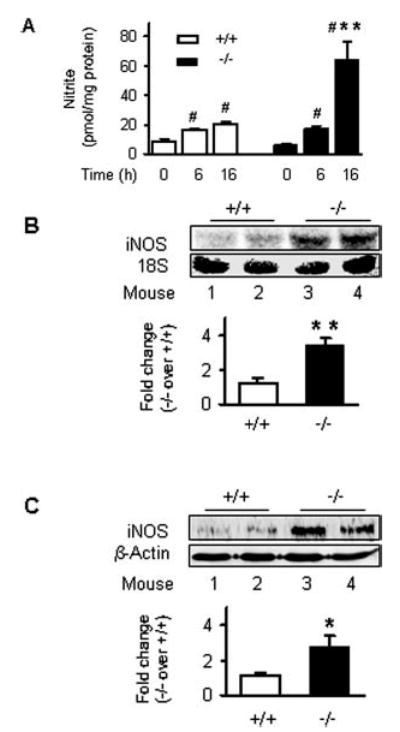
A, NO in plasma. Mice were treated with 4 μg/g bw LPS or PBS. NO levels in plasma were measured and are shown as the mean ± S.E. of three independent experiments (n = 8). B and C, iNOS expression in spleen. Following treatment of mice with 4 μg/g bw LPS, spleen cytoplasmic proteins (16 h) and total RNA (6 h) were prepared. mRNA levels of iNOS were measured by Northern blotting (B), and protein levels of iNOS and β-actin were detected by Western blotting (C). Data are representative of two independent experiments (n = 4–6). #, p < 0.05 for comparison within genotype with respect to PBS vehicle. *, p < 0.05; **, p < 0.01 for comparison between genotypes and relative wild types.
Fig. 2. Nitric oxide production is elevated in resident macrophages from Ucp2−/− mice upon stimulation by LPS.
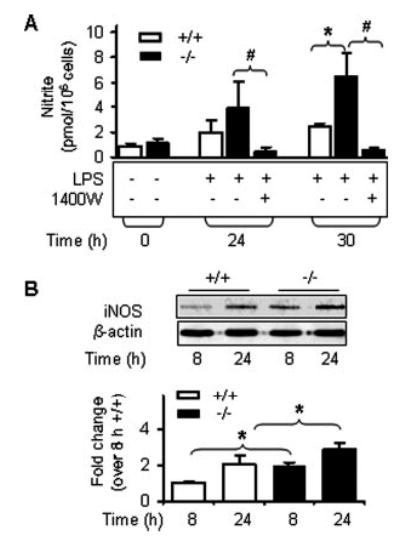
A, NO levels in supernatant of resident peritoneal macrophages. Supernatants of macrophage cultures were collected following treatment with 1 μg/ml LPS in the absence or presence of 50 μm 1400W for NO measurement. Results are shown as the mean ± S.E. of two independent experiments, each representing pooled macrophages from three to four mice. B, iNOS protein levels in peritoneal macrophages. Macrophages were treated with LPS as described in A. Protein levels of iNOS and β-actin were measured by Western blotting. Data are representative of two independent experiments (n = 4). #, p < 0.05 for comparison within genotype with respect to non-inhibitory treatment. *, p < 0.05 for comparison between genotypes and relative wild types.
Because cytokines released from macrophages and other immune cells are important facilitators of the host defense response and augment the induction of iNOS, we next measured the expression of several inflammatory cytokine genes by Northern blotting using total RNA from spleen of wild-type or Ucp2-null mice treated or not treated with LPS. As shown in Fig. 3A, LPS-induced expression of IFN-γ, TNF-α, interleukin-1β, and interleukin-6 was significantly higher in Ucp2−/− mice than in their wild-type littermates following LPS treatment. When we examined plasma levels of two of these key inflammatory cytokines, IFN-γ and TNF-α, both were substantially greater in Ucp2−/− mice than in Ucp2+/+ mice (Fig. 3B and C). Similar results of elevated cytokine gene expression were obtained from microarray analyses of spleen RNA from Ucp2&+/+ and −/− mice (Fig. 4). There was also a greater representation of a subset of tissue-degrading matrix metallo-proteinases and anti-apoptotic gene products in samples from LPS-treated Ucp2−/− mice than from wild-type animals. Gene network analysis (29) illustrates that these molecules regulate the transcription factor NF-κB or are regulated by it.
Fig. 3. Induction of inflammatory cytokines by LPS is increased in Ucp2−/− mice.

A, Northern blotting. Spleen total RNA was isolated from mice treated with 4 μg/g bw LPS or PBS. Levels of indicated cytokine mRNAs, Ucp2, and cyclophilin (Cyclo) were detected by Northern blotting. Data are shown as the fold change of Ucp2−/− mice versus Ucp2+/+ mice (n = 4). *, p < 0.05. B and C, ELISA for IFN-γ and TNF-α, respectively. Mice were treated or not treated with 4 μg/g bw LPS for 6 h, and plasma was collected for ELISA measurement. Each result shown is the mean ± S.E. of plasma sample from four individual mice. #, *, and ** are as defined in the Fig. 1 legend.
Fig. 4. Microarray analysis.
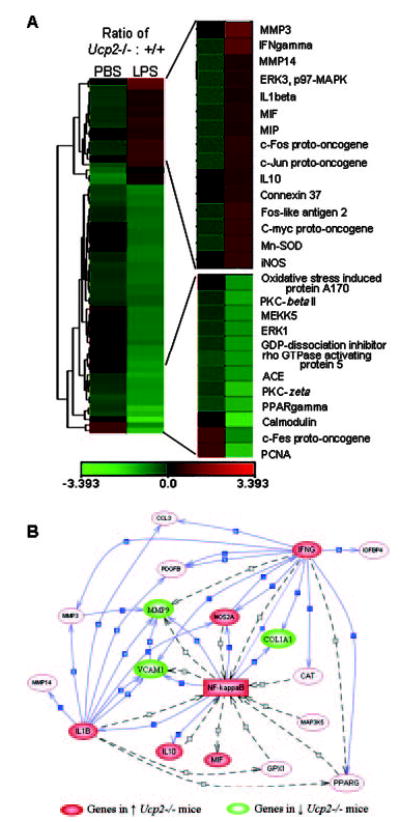
A, a clustering algorithm was used to group the LPS treatment and genes in spleen tissue. Left-hand columns show the ratio of fold change in gene expression from Ucp2−/− versus Ucp2+/+ following PBS treatment (basal condition). Right-hand columns present the ratio of the differences between the genotype (Ucp2−/− versus Ucp2+/+) for the fold change in gene expression between LPS and PBS basal conditions. Green and red rectangles represent repressed and induced values for individual genes, respectively. Genes that are altered significantly between genotypes or by LPS treatment are presented in the figure. The scale at the bottom is expressed as log2 of the relative ratios. B, network analysis of microarray data built up by PathwayAssist™. In this figure, the solid arrows denote regulated expression. The dashed arrows indicate unspecified regulation.
Activation of NF-κB Cascade in Ucp2−/− Mice, Even Under Basal Conditions
In the next series of experiments, we attempted to determine the mechanisms responsible for the exacerbated levels of expression of iNOS and cytokines in Ucp2−/− mice. Because NF-κB is a central regulator of these inflammatory mediators, we examined its relative abundance, subcellular distribution, and functional activity as assessed by its binding to NF-κB DNA response elements in nuclear extracts using electrophoretic mobility shift assays (Fig. 5). Under LPS treatment conditions, binding to oligonucleotides containing the two separate NF-κB response elements from the iNOS gene promoter, NF-κB(d) and NF-κB(u) (26), was elevated in Ucp2−/− mice (Fig. 5A), supporting the idea that the greater increase of iNOS expression in Ucp2−/− mice is NF-κB-dependent. Using a consensus NF-κB response element, we obtained essentially identical results (Fig. 5B). The specificity of these interactions was confirmed by the ability of antisera against the p50 and p65 subunits of NF-κB to retard the mobility (“supershift”) of these binding species (Fig. 5C). Somewhat unexpectedly, we also observed that even under basal conditions, there is a higher relative abundance of activated NF-κB in nuclei from spleen of Ucp2−/− mice compared with Ucp2+/+ mice (Fig. 5D).
Fig. 5. NF-κB activity is increased in Ucp2−/− mice.
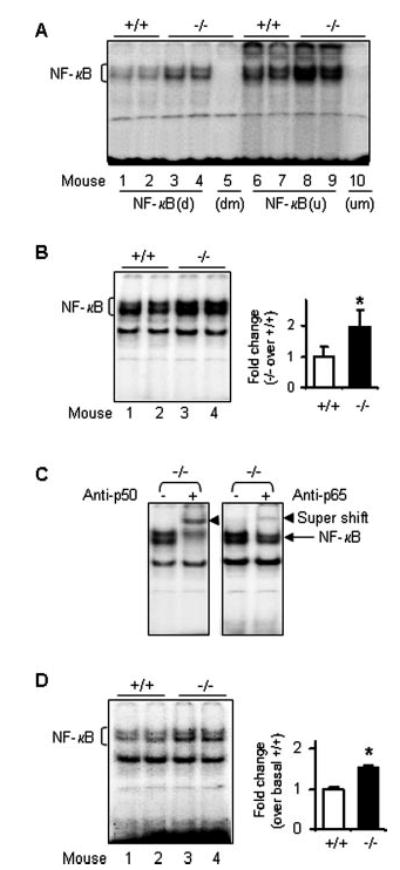
A, LPS stimulated NF-κB activity using oligonucleotides of iNOS-specific NF-κB binding sites designated “d” and “u” according to Ref. 26. Wild-type or Ucp2−/− mice were treated with 4 μg/g bw LPS for 3 h. Spleen nuclear proteins were isolated and used (5 μg) in EMSA with double-stranded DNA oligonucleotides as described under “Experimental Procedures.” For both NF-κB(d) and NF-κB(u) oligonucleotides, the binding from Ucp2−/− mouse extracts was increased 1.8 ± 0.15* and 1.5 ± 0.05*, respectively (*, p < 0.05). B, NF-κB activity stimulated by LPS using consensus oligonucleotides. Spleen nuclear proteins were extracted after LPS treatment and used in EMSA (5 μg) with the double-stranded DNA oligonucleotides containing the consensus sequence for NF-κB binding sites. Antibodies against p50 or p65 of NF-κB were added for the supershift assays (C). D, basal activity of NF-κB. Twelve μg of nuclear proteins were used to perform EMSA with consensus NF-κB binding site oligonucleotides. Results shown represent one of two independent experiments (n = 4).
This result should also be reflected by a shift in the distribution from inactive cytoplasmic NF-κB to activated nuclear NF-κB in Ucp2−/− animals. To examine this issue further, we measured the relative levels of the p50 and p65 subunits of NF-κB in cytoplasmic and nuclear fractions from spleen of both genotypes under basal and stimulated conditions. As shown in Fig. 6A, under basal conditions the levels of nuclear p50 subunit were already higher in Ucp2−/− mice compared with wild-type animals; likewise, they also had relatively less cytoplasmic p50. There was no difference in the distribution of p65 between the genotypes in the absence of acute stimulation. Interestingly, in some instances it has been reported that measurement of increased nuclear accumulation of p50 or its DNA binding is associated with continuous activation of NF-κB in certain pathological conditions (30–32). As shown in Fig. 6B, following LPS treatment the cytoplasmic levels of p50 and p65 subunits were both decreased more significantly in Ucp2−/− mice compared with Ucp2+/+ mice, and their nuclear levels were correspondingly increased.
Fig. 6. Nuclear translocation of NF-κB subunits.
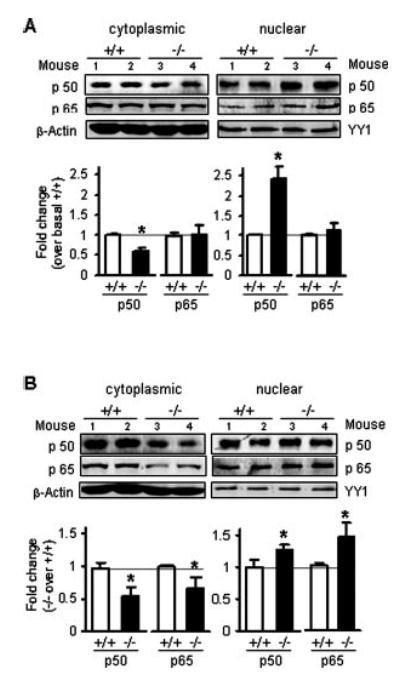
Following treatment of Ucp2+/+ or Ucp2−/− mice with or without 4 μg/g bw LPS for 3 h, spleens were collected for the preparation of cytoplasmic and nuclear protein fractions. Translocation of p50 and p65 under basal conditions (A) or following LPS treatment (B) was detected by Western blotting with specific antibodies against the p50 or p65 subunit and β-actin or nuclear factor YY1. Data shown represent the mean ± S.E. of two independent experiments (n = 4). *, p < 0.05.
The location and activation of the NF-κB subunits are regulated by the inhibitor proteins IκBα and IκBβ, which are in turn regulated by IKK activity. Cytoplasmic levels of IκBα and IκBβ were mildly decreased following LPS treatment in both genotypes, but IκBβ levels declined more significantly in Ucp2−/− mice under both basal and LPS-treated conditions (data not shown). Although the functions of the two IκB subtypes are similar, IkBβ is noted to primarily be regulated by acute cytokine stimulation, whereas IkBβ is mainly involved in the sustained activation of NF-κB by other pathways (33, 34). Together, these results are consistent with a pattern of low-level persistent activation.
Increased IKK Activity in Ucp2−/− Mice in Both Basal and Stimulated Conditions Can Be Suppressed by Inhibitors
The IKK activity in spleen and in isolated macrophages was measured directly by immune complex kinase assay (21). The results shown in Fig. 7A indicate that IKK activity is higher in Ucp2−/− mice both under basal conditions and 2 h after LPS stimulation. Because the antibody used for immuno-precipitation recognizes both IKKα and IKKβ, we cannot distinguish whether one isoform is differentially activated over the other. In any event, together with the previous data, these results show that the increase in IKK activity and the decrease of IkBβ level trigger the translocation and activation of NF-κB in Ucp2−/− mice under basal conditions. Although it is commonly observed that IKK can be activated in response to various stimuli including oxidative stress, cytokines, or ultraviolet radiation, the mechanism by which this occurs remains unclear (35). However, we can rule out any contribution from the classical mitogen-activated protein kinase pathways such as extra-cellular signal-regulated kinase, c-Jun NH2-terminal kinase, and p38 mitogen-activated protein kinase, because there were no differences between Ucp2+/+ and Ucp2−/− mice in either the phosphorylation state or the overall amount of these kinases in spleen under non-stimulated conditions (data not shown).
Fig. 7. IKK activity in spleen mitochondrial and H2O2 production under basal conditions.
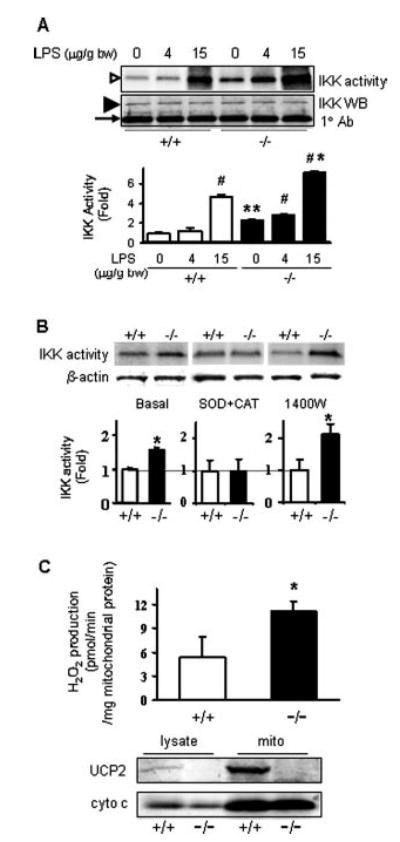
A, IKK activity in spleen. Ucp2+/+ and Ucp2−/− mice were treated with PBS vehicle and LPS (4 or 15 μg/g bw) for 2 h. Spleen cytoplasmic protein was prepared for IKK activity measurement by immune complex kinase assay. The open arrowhead shows γ-32P-labeled IκBα. Total levels of IKKα/IKKβ proteins were measured by Western blotting shown by the filled arrowhead. The bottom band indicated by the arrow shows the H chain of antibody. Data shown represent the mean ± S.E. of two independent experiments (n = 4). #, *, and ** are as defined in the Fig. 1 legend. B, IκB kinase activity in macrophages. Resident macrophages were isolated from three to four wild-type or Ucp2−/− mice and cultured in a 6-well plate overnight with or without the indicated inhibitors. Whole cell protein was extracted for IKK kinase assay. β-Actin was detected in the post-precipitation fraction as a loading control for the kinase assay. Data are representative of two independent experiments and shown as mean ± S.E. (n = 4). *, p < 0.01. C, H2O2 levels in mitochondrial suspensions were examined under basal conditions by Amplex® Red assay. Data shown represent the mean ± S.E. of three independent experiments (n = 5). *, p < 0.01. The Western blot shows the expression of UCP2 (150 μg protein/lane) and cytochrome c (30 μg/lane) in the original spleen lysates and following mitochondrial isolation from Ucp2+/+ and Ucp2−/− mice.
From these findings, it was important to determine whether the increased activity of IKK and subsequent activation of NF-κB observed in Ucp2−/− spleen could be replicated in isolated macrophages cultured in vitro from Ucp2−/− mice. This point is significant for understanding the role of UCP2 in this process because in Ucp2−/− mice we observed increased plasma levels of IFN-γ and higher IFN-γ expression in spleen, but macrophages produce little if any IFN-γ (36). Hence, the elevated IKK activity could be the result of the interaction of multiple cell types and signals within the spleen as opposed to a “cell autonomous” event dependent solely on the lack of UCP2. However, as shown in the left panel of Fig. 7B, we confirmed that under basal conditions, IKK activity is increased in isolated macrophages from Ucp2−/− mice. To distinguish which of these reactive species might be produced to trigger basal activation of IKK, we tested the effect of specific inhibitors for and H2O2 (cell-permeable polyethylene glycol-conjugated superoxide dismutase and catalase) or iNOS (1400W) (we used a combination of superoxide dismutase and catalase in order to completely remove and its immediate breakdown product, H2O2). As shown in the left two panels of Fig. 7B, the and H2O2 inhibitors completely blocked the enhanced IKK activity in isolated primary macrophages of Ucp2−/− mice. The iNOS inhibitor 1400W did not suppress the increased IKK activity and actually tended to increase it somewhat. Because NO can react with to form peroxynitrite, inhibition of NO production might be expected to lead to a net increase in and an elevated cellular response (37). These findings suggest that, under basal conditions, there is a source of superoxide and/or by-products thereof within macrophages of Ucp2-null mice that serves to activate IKK and the NF-κB cascade independent of other circulating factors in vivo.
Increased H2O2 Generation from Isolated Spleen Mitochondria of Ucp2−/− Mice
If superoxide is being generated in macrophages from Ucp2−/− cells to an extent that is able to increase IKK activity, the most likely source is from mitochondria for the following reasons. First, as shown in Table I, there were no differences between genotypes in subcellular distribution of the NADPH oxidase subunits under basal or stimulated conditions. There were also no differences in their relative expression levels as measured by reverse transcription-PCR (data not shown). Second, under physiological resting conditions, mitochondria are presumed to be the major source of ROS (38), and UCP2 is a mitochondria inner membrane protein. To test this idea, mitochondria were isolated from spleen of Ucp2+/+ and Ucp2−/− mice, and H2O2 (as the relatively stable immediate by-product of superoxide) was measured using the fluorometric substrate Amplex® Red. As shown in Fig. 7C, twice as much H2O2 was produced from the mitochondria of Ucp2−/− mice as compared with the mitochondria of wild-type mice.
DISCUSSION
When we first generated UCP2-deficient mice and observed that they were exceptionally resistant to chronic infection with Toxoplasma and had increased bacteriocidal activity (9), we concluded that the lack of UCP2 in macrophage mitochondria led to increased “ROS” in the broad sense of the term, including all reactive “oxygen” species such as superoxide, hydroxyl radical, nitric oxide, or their various by-products. However, at the time, we noted that whereas UCP2 is a mitochondrial protein, we could not rule out a role for NADPH oxidase as a major source of this ROS or even determine which of these ROS was the predominant form produced. In the present study, we first investigated the production of reactive oxygen and nitrogen species from wild-type and Ucp2−/− mice following treatment with LPS. As a membrane glycolipid of Gram-negative bacteria with strong immunostimulating activity, it is a well-established activator of both the “reactive oxygen” and “reactive nitrogen” generating pathways (13, 14). From these experiments, we demonstrated that in response to LPS challenge, iNOS-derived NO production is markedly increased in Ucp2−/− mice in vivo as well as in macrophages isolated from Ucp2-null mice and cultured in vitro. A recent report by Kizaki et al. (39) of reduced NO production in a macrophage cell line engineered to overexpress UCP2 is consistent with our findings (although clonal selection of the cell line could not be ruled out as a basis for their observations). By contrast, there were no significant differences between the two genotypes in the LPS response for the superoxide-generating oxidative burst enzyme, NADPH oxidase, or for cyclooxygenase 2. These data indicate that under LPS-stimulated conditions, the major reactive species that is increased in Ucp2−/− mice compared with wild-type mice is NO, rather than superoxide. Although we did not directly evaluate the contribution of the elevated NO production to the anti-parasitic phenotype of Ucp2−/− mice (9), NO has been implicated in the anti-microbial response to a variety of pathogens including parasites (40–43). Whereas both NO and superoxide are important in resistance to microbes, NO is generally considered more important than super-oxide during the late phase that follows a short-lived, early phase in which superoxide is predominant (44–46).
In this study we also observed that plasma levels of inflammatory cytokines such as IFN-γ and TNF-α were significantly elevated in the Ucp2−/− mouse, as was the expression of these and other cytokines in spleen in response to LPS treatment. Because IFN-γ is produced mainly by T lymphocytes and natural killer cells, rather than macrophages (36), it was reasonable to speculate that the ability of Ucp2−/− mice to generate more NO might be a consequence of a larger overall immune response involving many cell types as opposed to a cell autonomous mechanism of NO generation in macrophages alone. We believe that both processes are probably occurring. Macrophages isolated and cultured in vitro are capable of producing more NO and expressing iNOS to a higher degree than wild-type cells, but that response is probably amplified in vivo in view of the elevated IFN-γ. Therefore, an issue to investigate in the future is whether other cell types in the Ucp2−/− mouse elicit a more robust response to antigenic challenge.
As one of the key transcription factors responsible for increasing the expression of iNOS and various cytokines, we found that upon LPS treatment, the NF-κB pathway was activated in spleen to a greater extent in Ucp2−/− mice at all levels in the activation cascade as far back as IKK activity. Notably this was evident even under basal conditions and in isolated cultured macrophages from the Ucp2−/− animal. NF-κB also regulates the expression of a number of chemo-kines, matrix metalloproteinases, and anti-apoptotic genes either directly or indirectly (47, 48), and our preliminary microarray results found that these genes were also increased in spleen of Ucp2−/− mice. Based on cell fractionation studies, the p50 subunit of NF-κB, but not p65, appeared to be more active in the Ucp2−/− mouse under basal conditions. It also appears that under basal conditions there is a preferential degradation of IκBβ in Ucp2−/− spleen. These two characteristics have both been associated in other studies with the persistent activation of NF-κB (30–34). In the LPS-stimulated situation, even though p50 is more robustly translocated to the nucleus in both Ucp2+/+ and the Ucp2−/− mice, we still observed a significantly greater amount of nuclear p50 in the Ucp2−/− mouse, although the relative difference between genotypes under these conditions is less than that seen under basal conditions. We also observed that, after LPS stimulation, p65 nuclear translocation was increased to a greater extent in Ucp2−/− mice than in wild-type animals. Together, we interpret this behavior of the two subunits, p50 and p65, as indicating a greater response by the Ucp2−/− mouse under both basal and stimulated conditions.
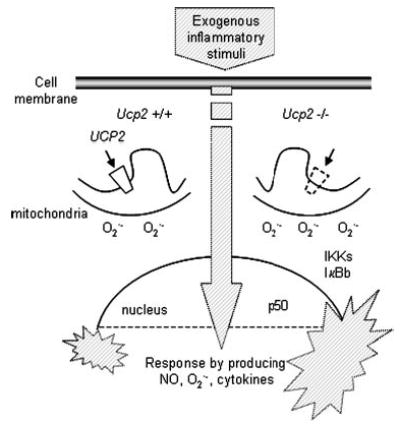
The concept that UCP2 may serve as a modulator of ROS production was first put forth by Casteilla and co-workers (16) and was further supported by our report on the microbicidal phenotype of the Ucp2−/− mouse (9). More recently, superoxide itself was proposed as an activator of UCP2 (49, 50), although it is still unclear exactly what might be “activated” because there are discrepancies in the literature regarding the ability of UCP2 (or UCP3 for that matter) to facilitate proton movement or transport of molecules across the inner membrane (5, 6, 11, 51–53). In any event, our findings here provide an additional pathway to explore how UCP2 controls cell function. Here we show that, at least in macrophages, the absence of UCP2 results in increased mitochondrially derived superoxide and/or hydrogen peroxide, which serves as a stimulus for the activation of IKK, even under basal conditions. As illustrated in Fig. 8, upon antigenic challenge the inflammatory response, particularly the generation of NO and cytokines, is amplified in the Ucp2−/− mouse. UCP2 has a relatively broad tissue distribution (1) and has been negatively associated with metabolic dysfunction in humans and animals (1, 10, 54–57). The increasing evidence that metabolic disorders such as obesity, diabetes, and atherosclerosis have an inflammatory component and oxidative stress in their etiology (52, 58–61) raises the possibility that our finding in macrophages may be relevant to the development of these other diseases.
Fig. 8. Model depicting the mechanism of amplified immune response in Ucp2−/− mice.
Acknowledgments
We thank Drs. Jie Liu and Michael P. Waalkes for assistance with microarray studies; Dr. Longlong Yang for gene network analysis; Drs. Jonathan Stamler, Irwin Fridovich, Paul Gardner, and Julie St-Pierre for valuable advice and suggestions; and Dr. Dennis Thiele, Dr. Steven Shoelson, Dr. James C. Bonner, and members of the Collins laboratory for comments on the manuscript.
Footnotes
This work was supported by National Institutes of Health Award R01-DK54024 (to S. C.) and a Research Award from the American Diabetes Association (to S. C.).
The abbreviations used are: UCP, uncoupling protein; NF, nuclear factor; ROS, reactive oxygen species; RNS, reactive nitrogen species; NO, nitric oxide; LPS, lipopolysaccharide; iNOS, inducible nitric-oxide synthase; IKK, IκB kinase; bw, body weight; PBS, phosphate-buffered saline; IFN, interferon; TNF, tumor necrosis factor; TPCK, as l-1-tosylamido-2-phenylethyl chloromethyl ketone; ELISA, enzyme-linked immunosorbent assay; EMSA, electrophoretic mobility shift assay.
References
- 1.Fleury C, Neverova M, Collins S, Raimbault S, Champigny O, Levi-Meyrueis C, Bouillaud F, Seldin MF, Surwit RS, Ricquier D, Warden CH. Nat Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- 2.Nedergaard J, Matthias A, Golozoubova V, Jacobsson A, Cannon B. J Bioenerg Biomembr. 1999;31:475–491. doi: 10.1023/a:1005400507802. [DOI] [PubMed] [Google Scholar]
- 3.Gimeno RE, Dembski M, Weng X, Deng N, Shyjan AW, Gimeno CJ, Iris F, Ellis SJ, Woolf EA, Tartaglia LA. Diabetes. 1997;46:900–906. doi: 10.2337/diab.46.5.900. [DOI] [PubMed] [Google Scholar]
- 4.Pecqueur C, Couplan E, Bouillaud F, Ricquier D. J Mol Med. 2001;79:48–56. doi: 10.1007/s001090000150. [DOI] [PubMed] [Google Scholar]
- 5.Krauss S, Zhang CY, Lowell BB. Proc Natl Acad Sci U S A. 2002;99:118–122. doi: 10.1073/pnas.012410699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krauss S, Zhang CY, Scorrano L, Dalgaard LT, St-Pierre J, Grey ST, Lowell BB. J Clin Investig. 2003;112:1831–1842. doi: 10.1172/JCI19774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattiasson G, Shamloo M, Gido G, Mathi K, Tomasevic G, Yi S, Warden CH, Castilho RF, Melcher T, Gonzalez-Zulueta M, Nikolich K, Wieloch T. Nat Med. 2003;9:1062–1068. doi: 10.1038/nm903. [DOI] [PubMed] [Google Scholar]
- 8.Dulloo AG, Samec S. Br J Nutr. 2001;86:123–139. doi: 10.1079/bjn2001412. [DOI] [PubMed] [Google Scholar]
- 9.Arsenijevic D, Onuma H, Pecqueur C, Raimbault S, Manning BS, Miroux B, Couplan E, Alves-Guerra MC, Goubern M, Surwit R, Bouillaud F, Richard D, Collins S, Ricquier D. Nat Genet. 2000;26:435–439. doi: 10.1038/82565. [DOI] [PubMed] [Google Scholar]
- 10.Zhang CY, Baffy G, Perret P, Krauss S, Peroni O, Grujic D, Hagen T, Vidal-Puig AJ, Boss O, Kim YB, Zheng XX, Wheeler MB, Shulman GI, Chan CB, Lowell BB. Cell. 2001;105:745–755. doi: 10.1016/s0092-8674(01)00378-6. [DOI] [PubMed] [Google Scholar]
- 11.Couplan E, del Mar Gonzalez-Barroso M, Alves-Guerra MC, Ricquier D, Goubern M, Bouillaud F. J Biol Chem. 2002;277:26268–26275. doi: 10.1074/jbc.M202535200. [DOI] [PubMed] [Google Scholar]
- 12.Liochev SI, Fridovich I. Arch Biochem Biophys. 1995;318:408–410. doi: 10.1006/abbi.1995.1247. [DOI] [PubMed] [Google Scholar]
- 13.Park HS, Jung HY, Park EY, Kim J, Lee WJ, Bae YS. J Immunol. 2004;173:3589–3593. doi: 10.4049/jimmunol.173.6.3589. [DOI] [PubMed] [Google Scholar]
- 14.Stuehr DJ, Marletta MA. Proc Natl Acad Sci U S A. 1985;82:7738–7742. doi: 10.1073/pnas.82.22.7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercurio F, Manning AM. Oncogene. 1999;18:6163–6171. doi: 10.1038/sj.onc.1203174. [DOI] [PubMed] [Google Scholar]
- 16.Negre-Salvayre A, Hirtz C, Carrera G, Cazenave R, Troly M, Salvayre R, Penicaud L, Casteilla L. FASEB J. 1997;11:809–815. [PubMed] [Google Scholar]
- 17.Reichardt HM, Umland T, Bauer A, Kretz O, Schutz G. Mol Cell Biol. 2000;20:9009–9017. doi: 10.1128/mcb.20.23.9009-9017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 19.Werner ER, Bahrami S, Heller R, Werner-Felmayer G. J Biol Chem. 2002;277:10129–10133. doi: 10.1074/jbc.M107326200. [DOI] [PubMed] [Google Scholar]
- 20.Mosmann T. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 21.Bayon Y, Ortiz MA, Lopez-Hernandez FJ, Gao F, Karin M, Pfahl M, Piedrafita FJ. Mol Cell Biol. 2003;23:1061–1074. doi: 10.1128/MCB.23.3.1061-1074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SH, Won SJ, Sohn S, Kwon HJ, Lee JY, Park JH, Gwag BJ. J Cell Biol. 2002;159:821–831. doi: 10.1083/jcb.200112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. Anal Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- 24.Pi J, Horiguchi S, Sun Y, Nikaido M, Shimojo N, Hayashi T, Yamauchi H, Itoh K, Yamamoto M, Sun G, Waalkes MP, Kumagai Y. Free Radic Biol Med. 2003;35:102–113. doi: 10.1016/s0891-5849(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 25.Collins S, Daniel KW, Rohlfs EM, Ramkumar V, Taylor IL, Gettys TW. Mol Endocrinol. 1994;8:518–527. doi: 10.1210/mend.8.4.7914350. [DOI] [PubMed] [Google Scholar]
- 26.Xie QW, Kashiwabara Y, Nathan C. J Biol Chem. 1994;269:4705–4708. [PubMed] [Google Scholar]
- 27.St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 28.Babior BM. Blood. 1999;93:1464–1476. [PubMed] [Google Scholar]
- 29.Bonetta L. Nat Methods. 2004;1:169–176. [Google Scholar]
- 30.De Plaen IG, Tan XD, Chang H, Qu XW, Liu QP, Hsueh W. Biochim Biophys Acta. 1998;1392:185–192. doi: 10.1016/s0005-2760(98)00024-1. [DOI] [PubMed] [Google Scholar]
- 31.Kurland JF, Kodym R, Story MD, Spurgers KB, McDonnell TJ, Meyn RE. J Biol Chem. 2001;276:45380–45386. doi: 10.1074/jbc.M108294200. [DOI] [PubMed] [Google Scholar]
- 32.Vancurova I, Wu R, Miskolci V, Sun S. J Virol. 2002;76:1533–1536. doi: 10.1128/JVI.76.3.1533-1536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bourke E, Kennedy EJ, Moynagh PN. J Biol Chem. 2000;275:39996–40002. doi: 10.1074/jbc.M007693200. [DOI] [PubMed] [Google Scholar]
- 34.Bureau F, Delhalle S, Bonizzi G, Fievez L, Dogne S, Kirschvink N, Vanderplasschen A, Merville MP, Bours V, Lekeux P. J Immunol. 2000;165:5822–5830. doi: 10.4049/jimmunol.165.10.5822. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto Y, Gaynor RB. Trends Biochem Sci. 2004;29:72–79. doi: 10.1016/j.tibs.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Matsuura M, Saito S, Hirai Y, Okamura H. Eur J Biochem. 2003;270:4016–4025. doi: 10.1046/j.1432-1033.2003.03792.x. [DOI] [PubMed] [Google Scholar]
- 37.Radi R, Cassina A, Hodara R, Quijano C, Castro L. Free Radic Biol Med. 2002;33:1451–1464. doi: 10.1016/s0891-5849(02)01111-5. [DOI] [PubMed] [Google Scholar]
- 38.Cadenas E, Davies KJ. Free Radic Biol Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 39.Kizaki T, Suzuki K, Hitomi Y, Taniguchi N, Saitoh D, Watanabe K, Onoe K, Day NK, Good RA, Ohno H. Proc Natl Acad Sci U S A. 2002;99:9392–9397. doi: 10.1073/pnas.142206299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang FC. J Clin Investig. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogdan C. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 42.Wei XQ, Charles IG, Smith A, Ure J, Feng GJ, Huang FP, Xu D, Muller W, Moncada S, Liew FY. Nature. 1995;375:408–411. doi: 10.1038/375408a0. [DOI] [PubMed] [Google Scholar]
- 43.Liew FY, Millott S, Parkinson C, Palmer RM, Moncada S. J Immunol. 1990;144:4794–4797. [PubMed] [Google Scholar]
- 44.Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. J Exp Med. 2000;192:227–236. doi: 10.1084/jem.192.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mastroeni P, Vazquez-Torres A, Fang FC, Xu Y, Khan S, Hormaeche CE, Dougan G. J Exp Med. 2000;192:237–248. doi: 10.1084/jem.192.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray HW, Nathan CF. J Exp Med. 1999;189:741–746. doi: 10.1084/jem.189.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnes PJ, Karin M. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 48.Chase AJ, Bond M, Crook MF, Newby AC. Arterioscler Thromb Vasc Biol. 2002;22:765–771. doi: 10.1161/01.atv.0000015078.09208.92. [DOI] [PubMed] [Google Scholar]
- 49.Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, Harper JA, Roebuck SJ, Morrison A, Pickering S, Clapham JC, Brand MD. Nature. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 50.Echtay KS, Murphy MP, Smith RA, Talbot DA, Brand MD. J Biol Chem. 2002;277:47129–47135. doi: 10.1074/jbc.M208262200. [DOI] [PubMed] [Google Scholar]
- 51.Jaburek M, Varecha M, Gimeno RE, Dembski M, Jezek P, Zhang M, Burn P, Tartaglia LA, Garlid KD. J Biol Chem. 1999;274:26003–26007. doi: 10.1074/jbc.274.37.26003. [DOI] [PubMed] [Google Scholar]
- 52.Jezek P, Zackova M, Ruzicka M, Skobisova E, Jaburek M. Physiol Res. 2004;53(Suppl 1):S199–S211. [PubMed] [Google Scholar]
- 53.Jaburek M, Miyamoto S, Di Mascio P, Garlid KD, Jezek P. J Biol Chem. 2004;279:53097–53102. doi: 10.1074/jbc.M405339200. [DOI] [PubMed] [Google Scholar]
- 54.Surwit RS, Wang S, Petro AE, Sanchis D, Raimbault S, Ricquier D, Collins S. Proc Natl Acad Sci U S A. 1998;95:4061–4065. doi: 10.1073/pnas.95.7.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blanc J, Alves-Guerra MC, Esposito B, Rousset S, Gourdy P, Ricquier D, Tedgui A, Miroux B, Mallat Z. Circulation. 2003;107:388–390. doi: 10.1161/01.cir.0000051722.66074.60. [DOI] [PubMed] [Google Scholar]
- 56.Lembertas AV, Perusse L, Chagnon YC, Fisler JS, Warden CH, Purcell-Huynh DA, Dionne FT, Gagnon J, Nadeau A, Lusis AJ, Bouchard C. J Clin Investig. 1997;100:1240–1247. doi: 10.1172/JCI119637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oberkofler H, Iglseder B, Klein K, Unger J, Haltmayer M, Krempler F, Paulweber B, Patsch W. Arterioscler Thromb Vasc Biol. 2005;25:604–610. doi: 10.1161/01.ATV.0000153141.42033.22. [DOI] [PubMed] [Google Scholar]
- 58.Dandona P, Aljada A, Bandyopadhyay A. Trends Immunol. 2004;25:4–7. doi: 10.1016/j.it.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 59.Willerson JT, Ridker PM. Circulation. 2004;109:II2–II10. doi: 10.1161/01.CIR.0000129535.04194.38. [DOI] [PubMed] [Google Scholar]
- 60.Libby P. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 61.Shoelson SE, Lee J, Yuan M. Int J Obes Relat Metab Disord. 2003;27(Suppl 3):S49–S52. doi: 10.1038/sj.ijo.0802501. [DOI] [PubMed] [Google Scholar]


