Abstract
Leucine rich repeats serve as recognition motifs for surface proteins from bacteria and eukaryotes. The BspA protein from Bacteroides forsythus mediates bacterial binding to fibronectin and contains leucine rich repeats of the Treponema pallidum (TpLRRP) family. Here we show that the protozoan parasite Entamoeba histolytica contains multiple BspA-like proteins, including a family of surface proteins which possess a new form of a leucine rich repeat that differs from the standard Treponema pallidum- like leucine rich repeat (TpLRRP) by possessing two conserved cysteine residues.
Entamoeba histolytica causes amebic dysentery and amebic liver abscess, major sources of morbidity and mortality worldwide. [1] E. histolytica trophozoites invade into submucosal tissues of the colon, which brings them into contact with extracellular matrix proteins, such as collagen, laminin, and fibronectin. Amebic contact with fibronectin induces alterations in actin polymerization, trophozoite motility, and secretion via G-protein coupled receptors and phosphokinase A-dependent signaling. ([2];[3];[4];[5] This signaling reportedly is not mediated by the Gal/Gal NAc surface lectin of ameba, suggesting that other molecules may be involved in the adherence and signaling process. [5]. A 37 kDa “β1-integrin like” membrane protein that binds to fibronectin has been identified, but to date this molecule has not been sequenced. [6]
Proteins with leucine rich repeat (LRR) motifs have been linked to fibronectin binding. LRR motifs consist of structural units, containing a β-strand and an α helix that are linked by loops to form a nonglobular horseshoe-shaped molecule with a curved parallel β sheet lining the inner portion of the horseshoe, while the α helices line the outer circumference. (Reviewed in [10]). There is extensive diversity in the form of the LRR motifs, with at least 7 LRR subfamilies identified. [10,11] The BspA protein of Bacteroides forsythus is a surface molecule that binds both fibronectin and fibrinogen, and has been linked to bacterial colonization of the oral cavity. [7] The BspA protein contains leucine rich repeats of the Treponema pallidum leucine rich repeat (TpLRR) subfamily which have a consensus sequence of (LxxIxIxxxVxxIgxxAFxxCxx) . Recently it was reported that the eukaryotic protozoan, Trichomonas vaginalis, contains a gene encoding a protein with leucine rich repeats (LRR) that resembles the BspA protein of B. forsythus. [9] The authors also reported that a partial sequence containing TpLRR repeats was present in E. histolytica. Wang and colleagues also described the presence of genes encoding leucine rich repeat proteins with TpLRR-like repeats in the related species Entamoeba dispar. [8] Our examination of the E. histolytica genome revealed multiple putative genes encoding proteins with leucine rich repeat (LRR) motifs that resemble BspA-like proteins. Here we describe the cloning, sequencing, and expression of one of the genes encoding an E. histolytica LRR containing protein, (EhLRRP1). We show that EhLRRP1 is a member of a family of proteins that resemble the BspA protein with LRR motifs of the TpLRR (Treponema pallidum leucine rich repeat) subfamily, describe some unique aspects of its sequence, and demonstrate that it is present on the surface of E. histolytica trophozoites.
Searching the E. histolytica genome (www.tigr.org) on 12/2002 we found an initial contig of 350 bp (ENTET35TR) that contained TpLRR domains, but did not encode a full length protein. Using RACE on RNA from E. histolytica HM1:IMSS trophozoites, we were able to obtain a full length clone (EhLRRP1) (GenBank AAW88349.1) This clone contained 1671 nucleotides and encoded a protein with 556 amino acids (figure 1B). Subsequent searching of the E. histolytica genome revealed a new contig (315412) that contained a sequence that was 99% identical at the nucleotide level to the EhLRRP1 sequence. PCR using primers derived from the 5' and 3' ends of the coding region of contig 315412 amplified a transcript in E. histolytica HM1:IMSS with a sequence identical to EhLRRP1. The relationship between the EhLRRP1 sequence and Bsp-A proteins from other species is shown in figure 1A. The EhLRRP1 sequence was most similar to the BspA protein of B. forsythus and other leucine rich repeat proteins from bacteria, consistent with the concept that these genes could have come to E. histolytica via lateral gene transfer from prokaryotes. [8]
Figure 1.
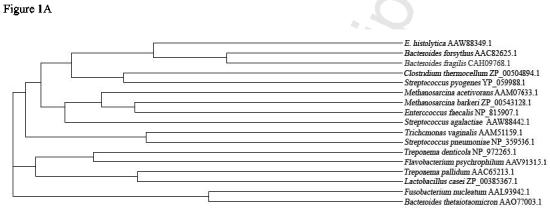
EhLRRP1 and Bsp-A like proteins in E. histolytica. A. Dendrogram showing the relatedness between EhLRRP1 and BspA-like proteins from other organisms. A BLAST homology search (blastp) was run on E. histolytica protein AAW88349.1 against all proteins in the NCBI nonredundant database to identify homologous proteins from organisms other than Entamoeba. Homologous proteins with a calculated expectation value of 3e-10 or above (range 3e-10 to 6e-94) were included in the phylogenetic tree created using ClustalW alignment and Phylip. Only the top-scoring protein sequences were aligned for comparative tree analysis if a single organism (defined by genus and species name) contained multiples proteins which showed expectation values above the cutoff.B. Derived amino acid sequence of EhLRRP1 compared to the 15 most closely related Entamoeba sequences encoding proteins containing BspA-like proteins with TpLRR leucine rich repeats. Darkly shaded residues are conserved among at least 5 sequences, lightly shaded residues are identical in all of the sequences. The classic residues in the TpLRRP repeats (LxxIxIxxxVxxIgxxAFxxCxx) are indicated at the top of the second TpLRRP1 repeat in EhLRRP1. This alignment, as well as the alignment of the entire cohort of 77 E. histolytica BspA-like proteins (http://stanleylab.wustl.edu/ehlrrp/ ) was performed using Clustal software, with some minor manual adjustments.
With the completion of the E. histolytica genome project, we used the InterProScan software (http://www.ebi.ac.uk/InterProScan) to screen for additional E. histolytica sequences that encode putative proteins containing TpLRR leucine rich repeat domains. Remarkably, we found an additional 75 sequences that possessed an initiator methionine and TpLRR repeats. Alignment and a related analysis (dendrogram) of EhLRRP1 (AAW88349.1) with the 75 deposited E. histolytica sequences containing TpLRR leucine rich repeats similar to the BspA protein, demonstrated that EhLRRP1 (AAW88349.1) forms a cluster with a group of 14 other E. histolytica sequences (figure 1B). (Alignment of all 77 E. histolytica sequences is available in supplementary data at http://stanleylab.wustl.edu/ehlrrp/). A search of the currently deposited sequences for Entamoeba dispar, a non-pathogenic ameba that is morphologically identical to E. histolytica, revealed that E. dispar contains a sequence homologous to EhLRRP1 on E. dispar contig 98468. We designated this sequence Ed98648 (figure 1B), and it is 72.1 % identical to EhLRRP1 at the derived amino acid level, but contains an additional stretch of 115 amino acids that are not present in the E. histolytica sequences.
The EhLRRP1 sequence is almost identical to putative E. histolytica gene EAL42510, (differing in only three nucleotides and two derived amino acids), however the derived amino acid sequence of EAL42510 sequence appears to terminate prematurely. This may represent an error in the EAL42510 nucleotide sequence, since EhLRRP1 and the other homologues, EAL49558.1, EAL42734.1, EAL43329.1, EAL44252.1, and Ed98468 display a very conserved N-terminal sequence (figure 1B). A second group of putative BspA-like sequences (particularly EAL42627.1, EAL49268.1, EAL42792.1, EAL48647.1, and EAL42623.1) are very similar to EhLRRP1 through their N-terminal region and initial TpLRRP repeats, but possess only 5 TpLRRP repeats resulting in a significantly smaller protein, with a different C-terminus sequence (figure 1B).
The EhLRRP1 protein appears to contain 22 TpLRR-like repeats of which 12 show near perfect identity to the consensus sequence (LxxIxIxxxVxxIgxxAFxxCxx), while 10 are more degenerate [9,10] [11] (figure 1A). One striking and consistent difference between the E. histolytica EhLRRP1 protein TpLRR repeats and the consensus sequence is the substitution of a cysteine for the consensus alanine at position 17. This substitution is found in all of the TpLRRP repeats in the other E. histolytica Bsp-A like proteins that cluster with EhLRRP1, (figure 1) but there are also groups of E. histolytica proteins with TpLRR repeats that retain alanine or have a different aliphatic residue at position 17 (see supplementary data). The substitution of cysteine at this position is not seen in the BspA protein or the T. vaginalis BspA—like protein. [9] Whether the additional cysteine within the TpLRR repeats of the E. histolytica proteins leads to disulfide bond formation with resultant effects on the conformation of the motifs is unknown. Other differences in the E. histolytica repeats include the occasional substitution of isoleucine for the conserved valine at position 10 in the consensus sequence, and the substitution of leucine for the conserved isoleucine at position 13 in the consensus sequence.
The derived amino acid sequence of EhLRRP1 was used to identify three potentially immunogenic peptides (TLLKSITIPSSISIKL (76-91); IEIPKNLKTINGKKIEKKDIN (334-354); FDGCPNELKKNEVLRKIYYKDD (531-552)) to produce EhLRRP1-specific rabbit antisera (Genemed Synthesis, Inc., South San Francisco, CA). Immunoblotting with the anti-EhLRRP antibody on E. histolytica HM1:IMSS lysates separated by SDS-PAGE under reducing conditions revealed a band at approximately 100 kDa (figure 2). The predicted molecular weight of the EhLRRP1 protein is 60.5 kDa, and the larger size of the peptide recognized in both reducing and non-reducing gels (data not shown) likely represents post-translational modifications to the EhLRRP1 protein. The EhLRRP1 protein has multiple potential phosphorylation sites and one glycosylation site in addition to a prenylation site. Post-translational modifications of EhLRRP1 are consistent with the finding that EhLRRP1 expressed in E. coli as a 6His recombinant fusion protein (which was recognized by the antibodies to EhLRRP1) migrated at a molecular mass of approximately 61 kDa (data not shown).
Figure 2.
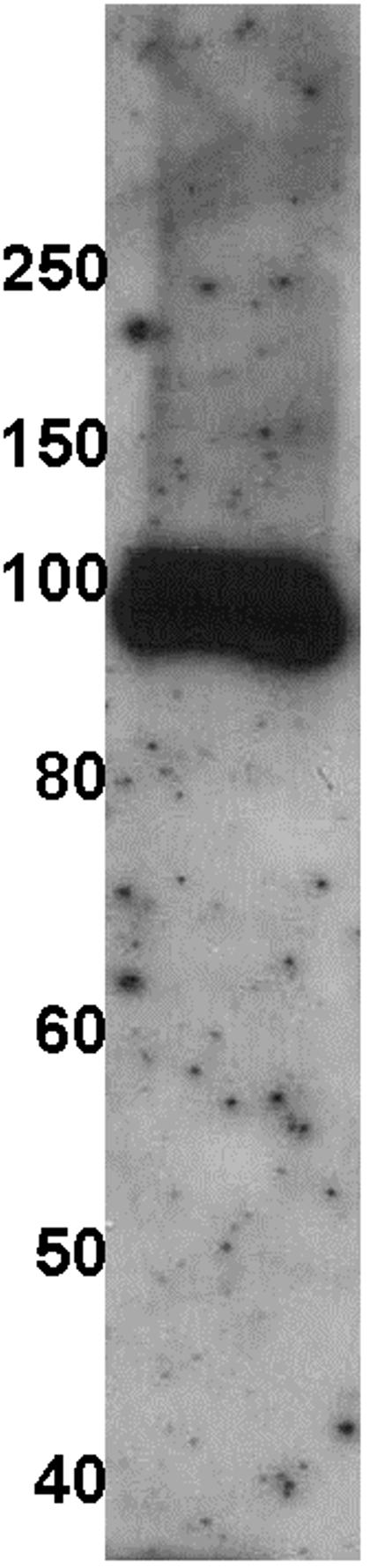
Polyclonal antiserum to three EhLRRP1 peptides detects a single band at approximately 100 kDa. Lysates of E. histolytica HM1:IMSS were prepared as previously described, separated by SDS-PAGE and transferred to nitrocellulose. [12] Antibodies from a 1/500 dilution of rabbit antiserum prepared against peptides TLLKSITIPSSISIKL (76-91); IEIPKNLKTINGKKIEKKDIN (334-354); and FDGCPNELKKNEVLRKIYYKDD (531-552) bound to a single species at approximately 100 kDa.
Two of the EhLRRP1 peptides used for immunization had more than 90% identity with the corresponding sequences in the putative E. dispar protein, and immunoblotting of E. dispar SAW760 lysates separated by SDS-PAGE under reducing conditions with the anti-EhLRRP1 antiserum revealed a single band at approximately 105 kDa (data not shown).
We used the anti-peptide serum to localize the EhLRRP1 protein in E. histolytica HM1:IMSS trophozoites using immunofluorescence and confocal microscopy. Trophozoites, either permeabilized with 0.1% Triton X100, or non-permeabilized, were reacted with polyclonal rabbit anti-EhLRRP1 serum (diluted 1:500) or a control rabbit antiserum at the same dilution, and antibody binding was detected using Cy2-conjugated donkey anti-rabbit IgG (Jackson Immunoresearch, Code 711-225-152). Images were analyzed using a Zeiss LSM510 Meta laser scanning confocal microscope (Carl Zeiss Inc., Thornwood, NY) using the Argon laser at 488 nm. As shown in figure 3, both permeabilized and non-permeabilized E. histolytica trophozoites showed surface membrane staining and there was some cytoplasmic staining in permeabilized trophozoites with anti-EhLRRP1 antibody. Measurements of fluorescent intensity across the diameter of the marked ameba using the Profile option of the LSM510 software showed bimodal peaks, consistent with primarily surface membrane staining. A control rabbit antiserum showed no reactivity with amebic trophozoites (data not shown).
Figure 3.
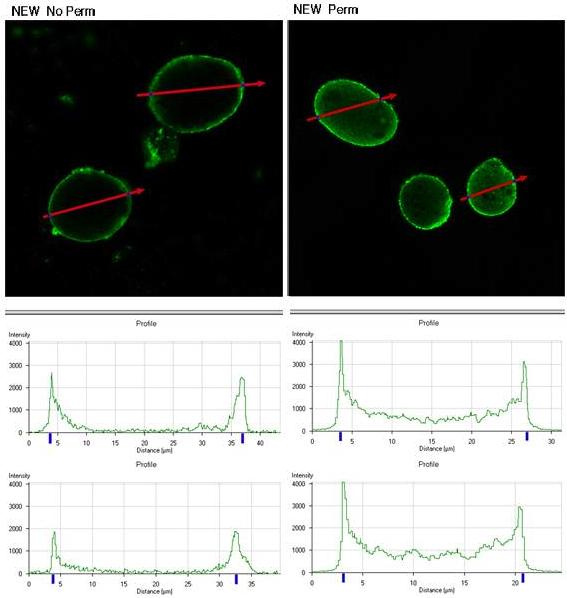
EhLRRP1 is detected on the surface of E. histolytica HM1:IMSS trophozoites by confocal immunoflourescent microscopy. Polyclonal rabbit antiserum to the three EhLRRP1 peptides bound to the surface of non-permeabilized (No perm) or permeabilized (Perm) E. histolytica HM1:IMSS trophozoites. The cross-sectional fluorescent intensity pattern (as measured across the red arrows) for two nonpermeabilized and two permeabilized trophozoites is shown in the graphs underneath each panel.
To confirm the presence of EhLRRP1 on the surface of E. histolytica trophozoites, we used a modified version of the protocol described by Hiken and Steinberg to identify surface proteins by biotinylation [15]. In brief, 5 × 106 live E. histolytica trophozoites were washed, surface labeled using Sulfo-NHS-LC biotin (Pierce, Rockford, IL) for 10 minutes on ice, then quenching buffer [15] containing 20 μM E-64 and 10 μM leupeptin was added. Trophozoites were lysed as previously described [16], and 150 μl of lysate was incubated with 100 μl Ultralink Immobilized NeutrAvidin beads (Pierce) for 2 hr at room temperature. Following 3 washes [15], beads were resuspended in 1X PBS and 4× SDS loading buffer, boiled for 5min, and along with samples of total lysate before reaction with avidin were separated by SDS-PAGE and transferred for immunoblotting. Rabbit polyclonal antiserum against the EhLRRP1 peptides diluted 1:500, and monoclonal antibodies against the serine rich E. histolytica protein (SREHP) or actin (diluted 1:5000) were used for immunoblotting against either the total lysate or the proteins surface-labeled with biotin that had bound to avidin beads. As shown in figure 4, immunoblotting revealed that both SREHP (a known surface antigen) [12] and EhLRRP1 could be detected in the total lysate and the surface labeled fraction, while actin was not detected in the surface labeled fraction.
Figure 4.
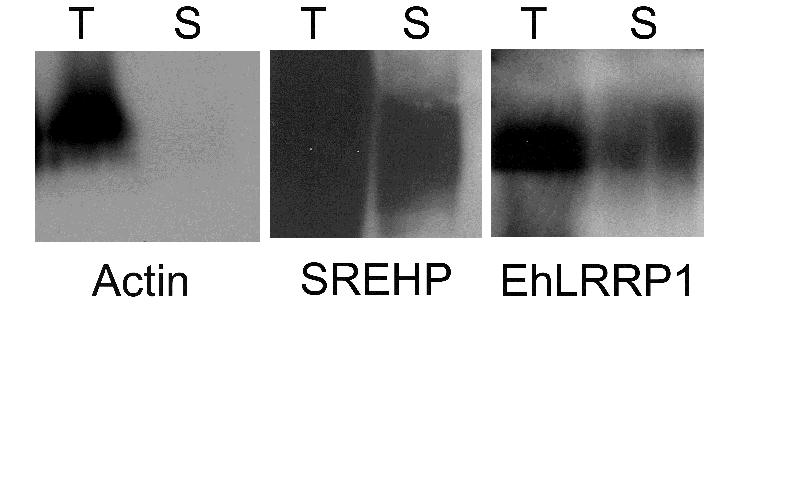
EhLRRP1 is found among surface labeled proteins of E. histolytica. Immunoblotting of total lysate (T) or a biotin-labeled surface protein fraction (S) from E. histolytica trophozoites with either a monoclonal antibody to actin, a monoclonal antibody to SREHP, or polyclonal antiserum to EhLRRP1. Bands at the appropriate molecular weight (41 kDa for actin, approximately 50 kDa for SREHP, and 100 kDa for EhLRRP1) were detected in both fractions for SREHP and EhLRRP1, but only the total lysate for actin.
The EhLRRP1 sequence does not contain a classic signal sequence and was not predicted to have a transmembrane domain based on TMHMM software. [13] Other E. histolytica proteins have been found to exhibit surface location without a classic signal sequence. [14] EhLRRP1, and 9 of the other putative E. histolytica BspA-like complete sequences, do have a predicted prenylation site (CaaX) (where “a” is an aliphatic amino acid) at the C-terminus (CVIV for EhLRRP1 and its homologues, and CVLV for EAL44016.1, EAL42732.1, EAL45834.1). (Figure 1B and supplementary data at http://stanleylab.wustl.edu/ehlrrp/) E. histolytica is known to possess protein prenyltransferases, and the isoprenylation of E. histolytica p21ras and p21rap proteins has been demonstrated in vitro. [15,16] [17] In general, prenylation links intracellular signaling proteins to the plasma membrane, if isoprenylation of EhLRRP1 is confirmed, it could suggest that prenylation may have additional targeting functions in E. histolytica.
In summary, we have identified and characterized one member of a family of BspA-like proteins found in E. histolytica, and found that a similar protein may be present in the non-pathogenic ameba E. dispar. The EhLRRP1 protein and related sequences have unique variants of the TpLRR repeat motifs, with consistent substitution of a cysteine for an alanine in the consensus sequence. Whether this alters the formation of the characteristic horseshoe shape formed by multiple leucine rich repeats [10], and whether disulfide bonds form due to the presence of the additional cysteine within the leucine rich repeat remains unknown. EhLRRP1 is located primarily on the plasma membrane of amebic trophozoites, but further studies will be necessary to determine whether EhLRRP1 or other members of this family mediate E. histolytica interactions with fibronectin or other host proteins.
Supplementary Material
Acknowledgements
This work was supported by NIH grants AI30084 and AI51591 and U54AI57160, the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research. We thank Dr. C. Graham Clark, London School of Hygiene and Tropical Medicine, London for providing E. dispar lysates, and Dr. Wandy Beatty for assistance with the confocal imaging.
Footnotes
Cite this article as: Davis PH, Zhang Z, Chen M, Zhang X, Chakraborty S, Stanley Jr. S.L., Identification of a family of Bsp-A like surface proteins of Entamoeba histolytica with novel leucine rich repeats., Molecular & Biochemical Parasitology, 10.1016/j.molbiopara.2005.08.017
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review before it published in its final citable form. Please note that during the production process errors may be discovered which could affect the content and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Stanley SL., Jr. Amoebiasis. Lancet. 2003;361:1025–34. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- 2.Carbajal ME, Manning-Cela R, Pina A, et al. Fibronectin-induced intracellular calcium rise in Entamoeba histolytica trophozoites: effect on adhesion and the actin cytoskeleton. Exp Parasitol. 1996;82:11–20. doi: 10.1006/expr.1996.0002. [DOI] [PubMed] [Google Scholar]
- 3.Franco E, Manning-Cela R, Meza I. Signal transduction in Entamoeba histolytica induced by interaction with fibronectin: presence and activation of phosphokinase A and its possible relation to invasiveness. Arch Med Res. 2002;33:389–97. doi: 10.1016/s0188-4409(02)00368-5. [DOI] [PubMed] [Google Scholar]
- 4.Manning-Cela R, Marquez C, Franco E, et al. BFA-sensitive and insensitive exocytic pathways in Entamoeba histolytica trophozoites: their relationship to pathogenesis. Cell Microbiol. 2003;5:921–32. doi: 10.1046/j.1462-5822.2003.00332.x. [DOI] [PubMed] [Google Scholar]
- 5.Meza I. Extracellular matrix-induced signaling in Entamoeba histolytica: its role in invasiveness. Parasitol Today. 2000;16:23–8. doi: 10.1016/s0169-4758(99)01586-0. [DOI] [PubMed] [Google Scholar]
- 6.Manning-Cela R, Carbajal EM, Meza I. Secretory pathway activation by interaction of Entamoeba histolytica trophozoites with fibronectin. Arch Med Res. 2000;31:S153–S154. doi: 10.1016/s0188-4409(00)00204-6. [DOI] [PubMed] [Google Scholar]
- 7.Sharma A, Sojar HT, Glurich I, et al. Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect Immun. 1998;66:5703–10. doi: 10.1128/iai.66.12.5703-5710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Samuelson J, Clark CG, et al. Gene discovery in the Entamoeba invadens genome. Mol Biochem Parasitol. 2003;129:23–31. doi: 10.1016/s0166-6851(03)00073-2. [DOI] [PubMed] [Google Scholar]
- 9.Hirt RP, Harriman N, Kajava AV, et al. A novel potential surface protein in Trichomonas vaginalis contains a leucine-rich repeat shared by micro-organisms from all three domains of life. Mol Biochem Parasitol. 2002;125:195–9. doi: 10.1016/s0166-6851(02)00211-6. [DOI] [PubMed] [Google Scholar]
- 10.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Current Opinion in Structural Biology. 2001;11:725–32. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 11.Kajava AV, Kobe B. Assessment of the ability to model proteins with leucine-rich repeats in light of the latest structural information. Protein Sci. 2002;11:1082–90. doi: 10.1110/ps.4010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanley SL, Jr., Tian K, Koester JP, et al. The serine rich Entamoeba histolytica protein (SREHP) is a phosphorylated membrane protein containing O-linked terminal N-acetylglucosamine (O-GlcNAc) residues. J Biol Chem. 1995;270:4121–6. doi: 10.1074/jbc.270.8.4121. [DOI] [PubMed] [Google Scholar]
- 13.Krogh A, Larsson B, Von HG, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 14.Choi MH, Sajed D, Poole L, et al. An unusual surface peroxiredoxin protects invasive Entamoeba histolytica from oxidant attack. Mol Biochem Parasitol. 2005;143:80–9. doi: 10.1016/j.molbiopara.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Maurer-Stroh S, Washietl S, Eisenhaber F. Protein prenyltransferases: anchor size, pseudogenes and parasites. Biol Chem. 2003;384:977–89. doi: 10.1515/BC.2003.110. [DOI] [PubMed] [Google Scholar]
- 16.Maurer-Stroh S, Washietl S, Eisenhaber F. Protein prenyltransferases. Genome Biol. 2003;4:212. doi: 10.1186/gb-2003-4-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen P-S, Sanford JC, Samuelson J. Entamoeba histolytica: Isoprenylation of p21ras and p21ras in vitro. Exp Parasitol. 1996;82:65–8. doi: 10.1006/expr.1996.0008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



